Abstract
The Mouse Genome Database (MGD: http://www.informatics.jax.org) is the primary community data resource for the laboratory mouse. It provides a highly integrated and highly curated system offering a comprehensive view of current knowledge about mouse genes, genetic markers and genomic features as well as the associations of those features with sequence, phenotypes, functional and comparative information, and their relationships to human diseases. MGD continues to enhance access to these data, to extend the scope of data content and visualizations, and to provide infrastructure and user support that ensures effective and efficient use of MGD in the advancement of scientific knowledge. Here, we report on recent enhancements made to the resource and new features.
INTRODUCTION
The Mouse Genome Database (MGD) is the key knowledgebase for integrated and comprehensive access to genetics and genomics data for the laboratory mouse, with a primary goal of facilitating the use of the mouse as a model system for understanding human biology and disease (1,2). MGD develops and supports an integrated representation of mouse genetic, genomic, functional, phenotypic and disease model data essential to biomedical research. MGD maintains the canonical catalog of mouse genes and genome features that is the foundation for connecting genome features with their biological properties. MGD serves as the authoritative source of functional annotations for mouse genome features using molecular function, biological process and cellular location terms in the Gene Ontology and provides authoritative mouse annotations to the Gene Ontology Consortium of which MGD is a founding member. MGD includes the comprehensive catalog of the full range of mouse mutant alleles, and annotates mutant genotypes to phenotypic outcomes using Mammalian Phenotype (MP) Ontology terms, and curates experimentally determined models of human disease. MGD is the primary community resource for official nomenclature for mouse gene names, symbols, alleles and strains. MGD provides data access to its research, translational/clinical and bioinformatics users through flexible, intuitive views and tools for finding, comparing, displaying and downloading relevant data and analyses.
MGD is the core component of the Mouse Genome Informatics (MGI) consortium (http://www.informatics.jax.org). Other database resources coordinated through the MGI consortium include the Gene Expression Database (GXD) (3), the Mouse Tumor Biology Database (MTB) (4), the Gene Ontology project (GO) (5), Mouse Mine (6) and the MouseCyc database of biochemical pathways (7). Data and information for these resources are obtained through a combination of expert curation of the biomedical literature and through semi-automatic processing of data sets downloaded from other data resources. Taken together, these resources provide a combination of data breadth, depth, integration and quality that exists nowhere else for mouse (Table 1).
Table 1. Summary of MGD content September 2016.
| September 2016 | |
|---|---|
| Number of genes and genome features with nucleotide sequence data | 48 285 |
| Number of genes with protein sequence data | 24 682 |
| Number of mouse genes with human orthologs | 17 102 |
| Number of mouse genes with rat orthologs | 18 547 |
| Number of genes with GO annotations | 24 237 |
| Total number of GO annotations | 315 086 |
| Number of mutant alleles in mice genes with targeted mutations | 49 038, 16 832 |
| Number of QTL | 5493 |
| Number of genotypes with phenotype annotation (MP) | 58 370 |
| Total number of MP annotations | 299 961 |
| Number of mouse models (genotypes) associated with human diseases | 5021 |
| Number of references in the MGD bibliography | 228 740 |
Within the last year, MGD focused primarily on improvements to the capture, integration and presentation of human disease data associated with mouse models including the incorporation of the Human Phenotype Ontology (8) terminology, updated the availability and visualization of mouse SNP data by strain, improved the Gene Detail Pages and enhanced GO annotations with spatial and temporal data linked to other biomedical ontologies. These aspects of MGD are focused on below. Our User Support team continues to provide up-to-date documentation for all changes and to provide outreach and to support user requests.
NEW FEATURES AND IMPROVEMENTS
Human mouse disease connection
The Human–Mouse: Disease Connection (HMDC, www.diseasemodel.org) is designed to facilitate the identification of published and potential mouse models of human disease, discovery of candidate genes and investigation of phenotypic similarity between mouse models and human patients. The initial release integrated mouse mutation, phenotype and disease model data from MGI with human gene-to-disease relationships from Online Mendelian Inheritance in Man (OMIM, www.omim.org) (9).
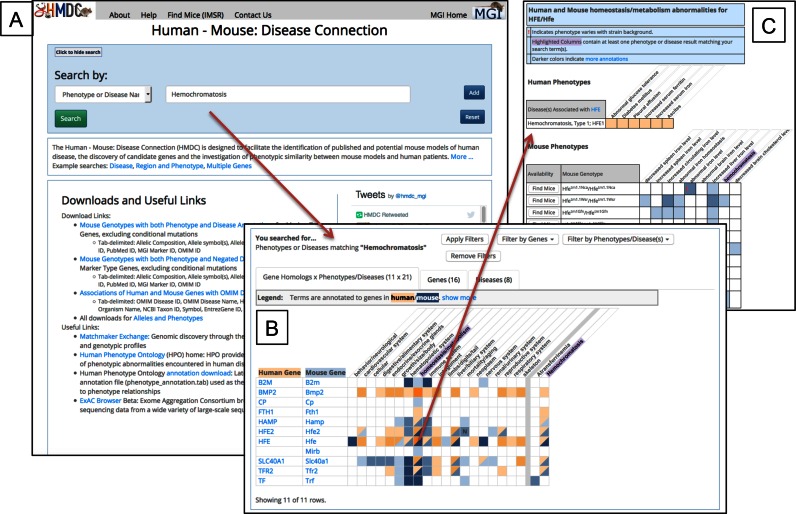
New this year, human phenotype terms and disease-to-phenotype relationships from the Human Phenotype Ontology (HPO, http://human-phenotype-ontology.github.io) Project are integrated into HMDC. In addition, we have significantly redesigned and streamlined the HMDC search form and results pages, and re-implemented the web interface using the AngularJavaScript framework. Users can easily search for combinations of human or mouse genome coordinates, multiple phenotype terms, disease terms, gene symbols and accession identifiers (IDs). An example of a search for the term ‘Hemochromatosis’ is shown in Figure 1. The results of the search include a grid displaying all pairs of mouse and human orthologs that have been annotated to the search term. Filters are available to refine results. Detailed information, including genotype to phenotype terms and availability of mouse models, is available by clicking on the grid cells, or gene and disease information can be viewed in tabular format in additional tab views.
Figure 1.
(A) The Human–Mouse: Disease Connection can be used to search for mouse and human genes, phenotypic similarity between patients and mouse models, and human and mouse model disease data. Multiple search parameters may be used individually or in combination, and include gene names or IDs or lists of genes, phenotype and disease names and IDs, or mouse or human genome location. (B) Sample results for the disease search term ‘Hemochromatosis’. The grid view summarizes gene and ortholog information with high level human and mouse phenotype and disease associations. Additional tabs show detailed gene and disease information in tabular format (not shown). Searches may be further refined by use of filters shown at the top of the grid. (C) Detailed phenotype and disease information may be accessed by clicking on the grid squares.
SNP query enhancement
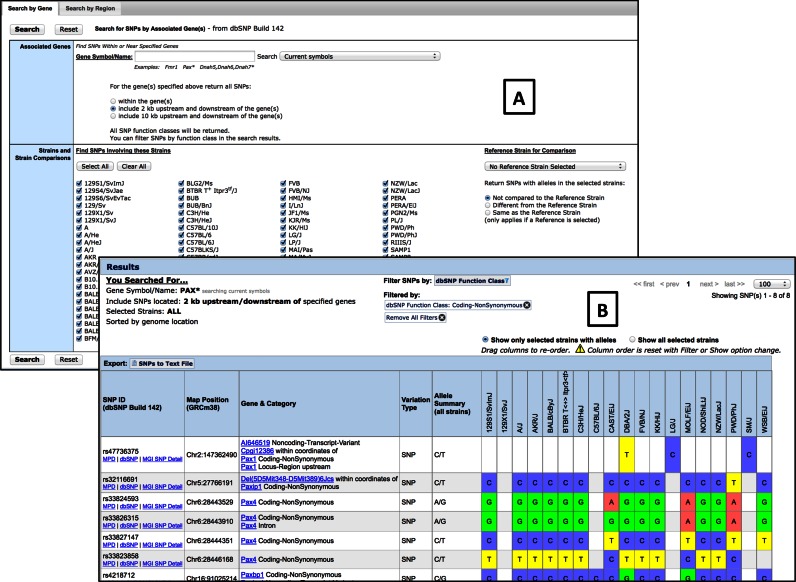
The new MGI Mouse dbSNP Query (http://www.informatics.jax.org/snp) is easier to use and provides vastly improved performance (driven now by Solr indices) and updated data for 88 strains of mice obtained from NCBI's dbSNP resource (Build 142) (10). Search interfaces have been simplified through the implementation of separate interfaces for genes and for genome regions. The user can filter results by dbSNP Function Classes or by Strain, and drag columns to reposition strains of interest. (Figure 2). For queries by gene, results can be filtered by any of (88) strains, and by whether the SNP is located within gene, within 2 kb or within 10 kb upstream and downstream of the gene. A reference strain can be selected, or not, and SNPs can be limited to just those that match (or not) the reference.
Figure 2.
Screenshots of improved SNP data interfaces in MGD. (A) Gene and region search forms are on different tabs to simplify the search interface. Mouse strains for which data are available are shown in alphabetical order and are selected with checkboxes. (B) Search results can be filtered by SNP functional classes. Columns can be manually re-ordered using ‘drag and drop’.
What does this gene do?
In a previous report, we described improvements to the MGI Gene Detail Page, including grid visualizations to summarize and to provide a mechanism to drill down to data about phenotypes, functional and expression data (1). To enhance access to detailed information about the functional characteristics of human orthologs, we have recently implemented links to Wikipedia's pages on human genes. The MyGene.info (11) project organizes these Wikipedia pages and MGI links to these pages only for genes that have a one-to-one mouse/human orthology relationship.
GO data enhancements
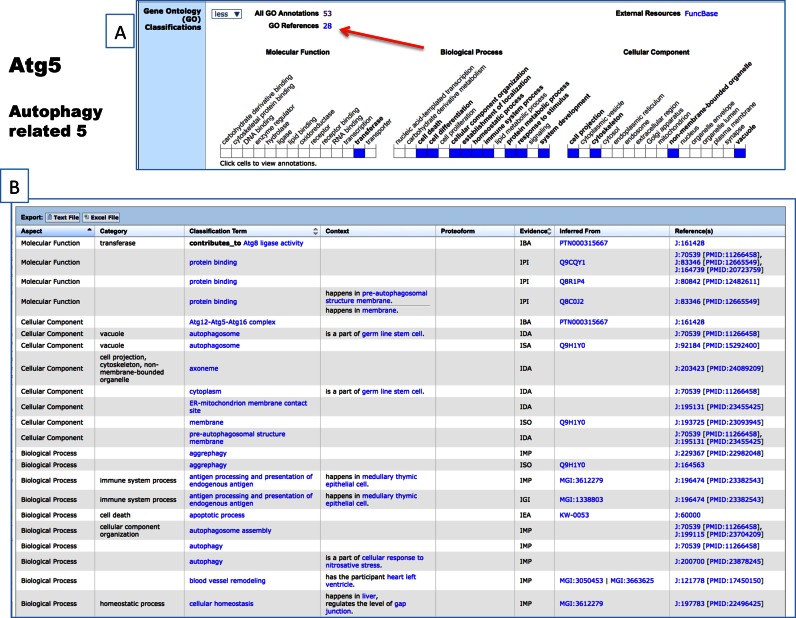
MGD serves as the definitive source for GO annotations for the laboratory mouse. Working as part of the Gene Ontology Consortium (GOC), MGD curators capture functional data through a variety of workflow and curatorial processes (12,13) and are responsible for defining the authoritative set of annotations for the GO community resources. Recent updates have improved the presentation of GO data in MGD. Annotation summary tables now include a new Category column. Categories are from a subset of the Gene Ontologies (a ‘GO slim’) and each category provides an overview of a section of the ontology. In addition, a new GO Context column adds value to the GO classification term by detailing the conditions or refining the tissue used in the experiment (Figure 3).
Figure 3.
Improvements in display of annotations of mouse genes and gene products to the Gene Ontology are here show for the mouse gene Atg5: autophagy related 5. (A) Here the high-level overall summation of annotations for Atg5 are shown across all three aspects of the GO. The blue cells indicated annotations tagging Atg5 as implicated in that component, function or biological process. Clicking on the cell retrieves all the annotations. Clicking on the (red arrow) All GO Annotations (53) brings back the detailed set of annotations (B) with new columns for context and proteoform. Here context from controlled vocabularies for anatomy and cell types as well as the relationship of the gene product to that context are details. A new column for proteoform permits the capture of isoforms participating in the activities and will be populated as these are identified.
MGD now imports data generated through use of the GOC community annotation tool, Noctua, a web-based application for collaborative editing of models of biological processes (14). Noctua is becoming the MGD standard curation tool used to capture and represent causal models and functional annotations for mouse genes. The models are saved in OWL format and converted to GPAD (Gene-Product-Association-Data format) for incorporation into MGD. MGD also provides the list of valid annotatable objects associated with mouse genes via a GPI file (Gene-Product-Information file) that is used to submit gene and gene product information to the GO Consortium. MGD has been a pivotal group in the implementation of Noctua in a production environment.
Future work with the alliance for genome resources
In 2016, MGD joined FlyBase (15), Saccharomyces Genome Database (SGD) (16), WormBase (17), Zebrafish Information Network (ZFIN) (18), Rat Genome Database (RGD) (19) and the Gene Ontology Consortium (GOC) to form the Alliance of Genome Resources (AGR). The Alliance was forged in response to changes in the funding model for biomedical resources at NIH which have been covered in several recent articles Nature (http://www.nature.com/news/funding-for-model-organism-databases-in-trouble-1.20134) and Science(http://science.sciencemag.org/content/351/6268/14.full).
The goal of the AGR is to develop a new model for genome resources that uses a shared modular infrastructure to reduce overall operational costs while preserving the functionality and data quality that the user communities of these resources rely on (https://genestogenomes.org/model-organism-databases-join-forces-announcing-the-alliance-of-genome-resources/). The AGR will implement a single web portal that will allow users to search for information about all of the represented model organisms from a single interface. The goal of the AGR is to represent the union of the existing resources, not only the intersection of data types that are in common. Although the initial implementation of the AGR web portal will focus on the founding resources, the infrastructure will be designed to accommodate data from additional model organisms as well. This approach is essential to support the unique capabilities and qualities of each model system. The current work of the AGR builds on several ongoing collaborative efforts among the current members to develop and use common tools that predate the formal launch of the consortium. However there are many challenges ahead as methods and infrastructure for data acquisition, curation, search and visualization at each resource have been developed in response to the needs of different research communities over many years.
OTHER INFORMATION
Mouse gene, genome feature, allele and strain nomenclature
Under the guidelines set by the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/nomen), MGD is the authoritative source for the international scientific community for nomenclature for mouse genes, genome features, alleles, mutations and strains. Official nomenclature and persistent IDs for mouse genome features, alleles, and strains are distributed worldwide through the MGD web site and through regular data exchanges with other bioinformatics resources. MGD actively promotes adherence to nomenclature standards in publications and online sites and works with journal editors and other data resources to ensure use of nomenclature standards. In addition, MGD, RGD and the HUGO Human Gene Nomenclature Committee (20) collaborate to co-assign genome feature symbols that are consistent for orthologs across species. To contact the MGD nomenclature coordinator for assistance with nomenclature, use email: nomen@jax.org.
Bulk and programmatic data access
MGD provides access to its data by a number of tools and including web services (see links on http://www.informatics.jax.org/software.shtml). In addition, bulk data sets are available as downloadable reports (http://www.informatics.jax.org/downloads/reports/index.html) and via the MGD Batch Query tool (http://www.informatics.jax.org/batch), which allows users to customize data sets.
Electronic data submission
MGD accepts contributed data sets from individuals and organizations for any type of data maintained by the database. The most frequent types of contributed data are mutant and phenotypic allele information originating with the large mouse mutagenesis centers and strain data from repositories that contribute to the IMSR (http://www.findmice.org) (21). Each electronic submission receives a permanent database accession ID. All data sets are associated with their source, either a publication or an electronic submission reference. Details about data submission procedures can be found at http://www.informatics.jax.org/submit.shtml.
MGD also provides a ‘Your Input Welcome’ link in the upper right hand corner of gene and allele detail pages. Users are encouraged to submit corrections and additions to data through this page. MGD staff will follow up if there are questions about the submission.
Community outreach and user support
The MGD resource has full time staff members who are dedicated to user support and training. Members of the User Support team can be contacted via email, web requests, phone or fax.
World wide web: http://www.informatics.jax.org/mgihome/support/mgi_inbox.shtml.
Facebook: https://www.facebook.com/mgi.informatics.
Twitter: https://twitter.com/mgi_mouse.
Email access: mgi-help@jax.org.
Telephone access: +1 207 288 6445.
Fax access: +1 207 288 6830.
MGD User Support staff are available for on-site help and training on the use of MGD and other MGI data resources. MGD provides off-site workshop/tutorial programs (roadshows) that include lectures, demos and hands-on tutorials and can be customized to the research interests of the audience. To inquire about sponsoring a MGD roadshow, email mgi-help@jax.org.
On-line training materials for MGD and other MGI data resources are available as FAQs and on-demand help documents.
Other outreach
MGI-LIST (http://www.informatics.jax.org/mgihome/lists/lists.shtml) is a moderated and active email bulletin board for the scientific community supported by the MGD User Support group. The MGI-LIST has over 1800 subscribers. A second list service, MGI-TECHNICAL-LIST, is maintained for technical information for software developers and bioinformaticians accessing MGI data, using APIs and making links to MGI.
Included in our outreach and training activities are summer and academic year research internships in which MGD staff members mentor high school and college students from diverse backgrounds to foster interest in biomedical informatics and data science (https://www.jax.org/education-and-learning/high-school-students-and-undergraduates/learn-earn-and-explore).
IMPLEMENTATION AND PUBLIC ACCESS
The master internal MGD database resides in a PostgreSQL normalized relational database and is the workplace for integration of MGD data. This database is optimized for data loading, curation and integration processes. As data are prepared for the weekly public release, they are migrated to a public database instance in PostgreSQL that is denormalized and supplemented by a set of Solr/Lucene indexes. This public instance of MGD has excellent performance qualities for supporting searches and web displays. Keeping distinct versions of MGD for internal data loading, curation and integration and for public access on the web via a denormalized search-optimized version also helps us to manage the impact of changes required to either the internal or public MGD versions.
MGD provides free public access to data from http://www.informatics.jax.org. The web interface provides a simple ‘Quick Search’, available from all web pages in the system and is the most used first entry point for users. Various query forms are provided that allow more precise parameter searching. For example, using ‘caveolin’ as the keyword in the ‘Quick Search’ box returns 125 genome features (as of September 2016). In contrast, using the Genes and Markers Query form and entering ‘prostaglandin’ in the gene name box, choosing feature type of protein-coding gene and location of Chr 8 returns nine results. Query forms for specific parameter searching are available for Genes and Markers; Phenotypes, Alleles and Diseases; SNPs; and References.
Browsers are provided for exploring various vocabularies used in MGD (e.g. GO, Mammalian Phenotype (MP) ontology and OMIM disease terms) and terms from these vocabularies are linked to relevant MGD annotated data. Genome browsing is accomplished with our installation of JBrowse (http://jbrowse.org), a JavaScript-based interactive genome browser with extensive features for navigation and track selection (22).
MGD offers additional batch methods for data querying and downloads for users wishing to retrieve data in bulk. The Batch Query tool (http://www.informatics.jax.org/batch) (23,24) is used for retrieving bulk data about lists of genome features that can be typed in or uploaded as lists of gene symbols or gene IDs. Gene IDs from MGI, NCBI's GENE, Ensembl, VEGA, UniProt and other resources can be used. Users can select information they wish to retrieve, such as genome location, GO annotations, list of mutant alleles, MP annotations, RefSNP IDs and OMIM terms. Results are returned as a web display or in tab delimited text or Excel format.
MGD access is also powered through MouseMine (http://www.mousemine.org) (6), an instance of InterMine that offers flexible querying, templates, iterative refinement of results and linking to other model organism InterMine instances. MouseMine contains many data sets from MGD, including genes and genome features, alleles, strains and annotations to GO, MP and OMIM.
MGD also provides a large set of regularly updated database reports (http://www.informatics.jax.org/downloads/reports/index.html), and direct SQL access to a read-only copy of the database (contact MGI user support for an account). MGI User Support is also available to assist users in generating custom reports on request.
CITING MGD
For a general citation of the MGI resource, researchers should cite this article. In addition, the following citation format is suggested when referring to datasets specific to the MGD component of MGI: mouse genome database (MGD), MGI, The Jackson Laboratory, Bar Harbor, Maine (URL: http://www.informatics.jax.org). [Type in date (month, year) when you retrieved the data cited.]
MOUSE GENOME DATABASE GROUP
A. Anagnostopoulos, R.M. Baldarelli, J.S. Beal, S.M. Bello, O. Blodgett, N.E. Butler, L.E. Corbani, H. Dene, H.J. Drabkin, K.L. Forthofer, S.L. Giannatto, P. Hale, D.P. Hill, L. Hutchins, M. Knowlton, A. Lavertu, M. Law, J.R. Lewis, V. Lopez, D. Maghini, D. Perry, M. McAndrews, D. Miers, H. Montenko, L. Ni, H. Onda, J.M. Recla, D.J. Reed, B. Richards-Smith, D. Sitnikov, C.L. Smith, M. Tomczuk, L. Wilming and Y. Zhu.
Acknowledgments
The authors thank David Shaw for his contributions and leadership in providing User Support services to the community of scientists who use MGD and all MGI data resources. The use of Human Phenotype Ontology and Human Phenotype Annotation files from Dr Peter Robinson (The Jackson Laboratory) is gratefully acknowledged.
FUNDING
Mouse Genome Database is funded by NIH/NHGRI [HG000330, HG007053]. Funding for open access charge: NIH/NHGRI [HG000330].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bult C.J., Eppig J.T., Blake J.A., Kadin J.A., Richardson J.E., Mouse Genome Database, G. Mouse genome database 2016. Nucleic Acids Res. 2016;44:D840–D847. doi: 10.1093/nar/gkv1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake J.A., Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Mouse Genome Database, G. The mouse genome database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith C.M., Finger J.H., Hayamizu T.F., McCright I.J., Xu J., Eppig J.T., Kadin J.A., Richardson J.E., Ringwald M. GXD: a community resource of mouse gene expression data. Mamm. Genome. 2015;26:314–324. doi: 10.1007/s00335-015-9563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bult C.J., Krupke D.M., Begley D.A., Richardson J.E., Neuhauser S.B., Sundberg J.P., Eppig J.T. Mouse tumor biology (MTB): a database of mouse models for human cancer. Nucleic Acids Res. 2015;43:D818–D824. doi: 10.1093/nar/gku987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motenko H., Neuhauser S.B., O'Keefe M., Richardson J.E. MouseMine: a new data warehouse for MGI. Mamm. Genome. 2015;26:325–330. doi: 10.1007/s00335-015-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evsikov A.V., Dolan M.E., Genrich M.P., Patek E., Bult C.J. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J., et al. The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NCBI_Resource_Coordinators. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C., Macleod I., Su A.I. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drabkin H.J., Blake J.A., Mouse Genome Informatics, D. Manual gene ontology annotation workflow at the mouse genome informatics database. Database. 2012;2012:bas045. doi: 10.1093/database/bas045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drabkin H.J., Christie K.R., Dolan M.E., Hill D.P., Ni L., Sitnikov D., Blake J.A. Application of comparative biology in GO functional annotation: the mouse model. Mamm. Genome. 2015;26:574–583. doi: 10.1007/s00335-015-9580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gene Ontology, C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attrill H., Falls K., Goodman J.L., Millburn G.H., Antonazzo G., Rey A.J., Marygold S.J., FlyBase C. FlyBase: establishing a gene group resource for drosophila melanogaster. Nucleic Acids Res. 2016;44:D786–D792. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheppard T.K., Hitz B.C., Engel S.R., Song G., Balakrishnan R., Binkley G., Costanzo M.C., Dalusag K.S., Demeter J., Hellerstedt S.T., et al. The saccharomyces genome database variant viewer. Nucleic Acids Res. 2016;44:D698–D702. doi: 10.1093/nar/gkv1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C., et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruzicka L., Bradford Y.M., Frazer K., Howe D.G., Paddock H., Ramachandran S., Singer A., Toro S., Van Slyke C.E., Eagle A.E., et al. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015;53:498–509. doi: 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.J., et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eppig J.T., Motenko H., Richardson J.E., Richards-Smith B., Smith C.L. The international mouse strain resource (IMSR): cataloging worldwide mouse and ES cell line resources. Mamm. Genome. 2015;26:448–455. doi: 10.1007/s00335-015-9600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bult C.J., Kadin J.A., Richardson J.E., Blake J.A., Eppig J.T., Mouse Genome Database, G. The mouse genome database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bult C.J., Eppig J.T., Kadin J.A., Richardson J.E., Blake J.A., Mouse Genome Database, G. The mouse genome database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]