Abstract
The IPD-MHC Database project (http://www.ebi.ac.uk/ipd/mhc/) collects and expertly curates sequences of the major histocompatibility complex from non-human species and provides the infrastructure and tools to enable accurate analysis. Since the first release of the database in 2003, IPD-MHC has grown and currently hosts a number of specific sections, with more than 7000 alleles from 70 species, including non-human primates, canines, felines, equids, ovids, suids, bovins, salmonids and murids. These sequences are expertly curated and made publicly available through an open access website. The IPD-MHC Database is a key resource in its field, and this has led to an average of 1500 unique visitors and more than 5000 viewed pages per month. As the database has grown in size and complexity, it has created a number of challenges in maintaining and organizing information, particularly the need to standardize nomenclature and taxonomic classification, while incorporating new allele submissions. Here, we describe the latest database release, the IPD-MHC 2.0 and discuss planned developments. This release incorporates sequence updates and new tools that enhance database queries and improve the submission procedure by utilizing common tools that are able to handle the varied requirements of each MHC-group.
INTRODUCTION
The major histocompatibility complex (MHC) represents the most variable region between vertebrate genomes, encoding numerous genes, including the highly polymorphic class I and class II. The genes encode the MHC class I and class II molecules that present intracellular and extracellular peptides respectively, which are presented to T cell and NK cell receptors in order to activate the immune response. Therefore, these molecules are at the heart of the immune response, orchestrating and influencing an enormous range of both adaptive and innate responses (1). In particular, class I molecules are present on the surface of every nucleated cell in an organism while class II has a distribution restricted to the professional antigen presenting cells. Consequently, the products of these genes are at the heart of the immune system and allow discrimination between self and non-self.
Early in comparative MHC research it became clear that to prevent a confusing array of sequences, using different nomenclature systems with high levels of redundancy and variable quality, a standardized nomenclature and curated databases were needed. Consequently, non-human MHC nomenclature for genes and alleles has been overseen informally by research groups involved in allele sequencing and by formal nomenclature committees set up by the International Society for Animal Genetics (ISAG). In order to standardize non-human MHC research and allele nomenclature (2), this work is now overseen by the Comparative MHC Nomenclature Committee (3), supported by ISAG and the Veterinary Immunology Committee of the International Union of Immunological Societies. The IPD-MHC Database (4) is a centralized resource that collects, organizes, curates and manages current and future MHC gene and allele sequences from non-human organisms (4) addressing the needs of the comparative MHC community for such a system. The initial version in 2003 was modelled on the IPD-IMGT/HLA Database, and involved the work of groups specializing in non-human primates (NHP) (5), canids (DLA) (6) and felids (7). Since then the database underwent various updates and expanded to include bovins (BoLA) (8), equids (ELA) (9), salmonids (10), murids (RT1) (11), ovids (12) and suids (13).
With the advent of new high-throughput sequencing technologies, which have massively reduced the per-base sequencing cost, the number of sequences submitted to public databases has enormously increased. These sequencing methods have also allowed the targeting of MHC genes and alleles from a wide range of species, but generating large amounts of data from highly polymorphic genes can often lead to a confusing range of local allelic nomenclature. The manual curation of such data by experts has huge importance for improving data quality by resolving ambiguities difficult to detect from automatic systems, preventing redundancy within a single system and therefore maximizing the value of individual research efforts. In the IPD-MHC Database, experts assigned by the MHC nomenclature committee for the relevant species further curate the submitted sequences to ensure quality and provide an allele name within the official nomenclature system. This allows for improvements in data quality as well as the addition of more specialized information, such as taxonomic information, and particular sequence features (exonic and intronic regions, coding sequences and translation variants).
As the database expanded with new taxonomic groups and species, while also receiving more sequence submissions, it became essential to reorganize and create a uniform structure that preserved the ability of the nomenclature committees to curate and oversee each section. The growing interest from the research community now requires a flexible and expandable database, able to accommodate future taxonomic groups and incorporate more analysis tools. Therefore, we are reporting this database release as it is a significant advance that provides IPD-MHC with a much improved submission system and an expanded and integrated range of analysis tools.
IPD-MHC ORGANIZATION
The underlying database behind the IPD-MHC Database project is organized in taxonomic groups, each one listing MHC alleles from a number of related species. The analyses and comparison of different species requires a common data organization that allows the simultaneous collection of common features and group-specific annotation data. However, the coordination of a high volume of data from different groups each with bespoke requirements is challenging. In previous versions of the IPD-MHC Database, submitted data were collected centrally and then locally curated by each MHC group into flat files. These are plain text files that comprise of a series of strictly controlled data entries, presented in a tabular manner and defined by EBI (http://www.ebi.ac.uk/ena/submit/data-formats). Flat files were then regularly uploaded to the centralized system by the different groups located all over the world. Until recently, submitted data were processed, inserted into a relational database before an analysis pipeline calculated reports, amino acid translation products and alignments on a daily basis and reported this back to the curators. The curators, however, did not have access to the central repository and all data manipulation was managed through changes to the flat files. Given that the overall success was dependent on the quality of the data, and that the flat file curation is the most sensitive step in this procedure, simple errors like misspelled or missing information were common problems, requiring flat file correction, and re-importing that takes time and effort. Furthermore, groups with higher volumes of data (i.e. non-human primates) require more frequent updates, and data checks. To address these issues and make the process more efficient and scalable, the IPD-MHC Database and the curation process has been completely revamped to accommodate a centralized structure, unifying all the data and tools within a single unique system.
IPD-MHC DATABASE 2.0 CURATION AND CONTENT
For each new submission, flat files are generated and sent from the public side of the database, hosted at the European Molecular Biology Laboratory's Bioinformatics Institute (EMBL-EBI), to a core curation server, where they are automatically validated and stored in a relational database. MHC group curators can access a private section of this database in order to amend or validate the submitted data. Data validation and processing is done in real-time, the submitted data only being publically available after the next scheduled release date. The IPD-MHC Database has been designed to accommodate the large amounts of data on sequence variants and be easily expandable. Curators can edit and update each MHC group's section, and new taxonomic groups can be simply assigned. Furthermore, the centralized organization allows curators to easily manage and create unique organism names, facilitating the insertion of new species and taxonomic groups. For each organism, a common name as well as a scientific name and a 4-letters unique name are provided. In addition, a taxon ID, automatically retrieved by the server, allows synchronizing taxonomic data from the NCBI Taxonomy Database (14), keeping the organism information as well as the lineage up-to-date. The submission procedure has been updated to alleviate and improve the curation procedure by supplying curators high-quality submissions with automatically validated data. The new submission tool allows simple editing in all the mandatory fields using a convenient wizard-like interface. As several fields are MHC group specific, some additional mandatory information are required while a consistent number of fields are optional. At the end of the submission procedure, a unique ID is assigned to the new sequence. Novel sequences are aligned against those in the database and an initial name is automatically assigned based on nucleotide and protein sequence identity. However, final names are manually reviewed, and only officially assigned by curators, maintaining the overall high-level data quality through curation. For each MHC taxonomic group, a member of the committee has access to the database backend, where a set of tools allows the curation of newly submitted sequences, the assigning of alleles and report issues or corrections to the sequence submitter. Once validated, the sequence is available on the public side of the database after the next scheduled release. At each scheduled release, the data from the core curation server is synced with the open access public facing system at EMBL-EBI. As the new database becomes established, a unified versioning system will record changes in stored data as well as new features and database fixes. A list of changes and improvements to date is already available at http://www.ebi.ac.uk/ipd/mhc/version.
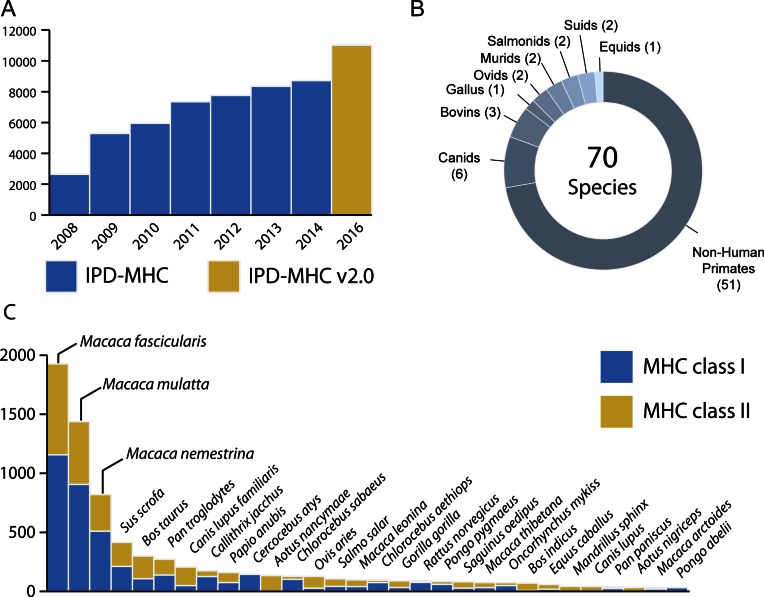
This latest release includes data from the previously available nine taxonomic groups, comprising a total of 70 species (Figure 1). All data from the previous version of the IPD-MHC Database, including sequences awaiting validation, have been incorporated. Moreover, this release includes for the first time alleles from chicken, further expanding the number of taxonomic groups hosted by the IPD-MHC Database. As a result, nearly 10000 expertly curated sequences, derived from 11061 unique International Nucleotide Sequence Database Collaboration (INSDC) (15) entries organized into 7794 alleles (Figure 1A–C), are publicly available for the research community. Moreover, the dynamic and scalable structure of the database has created the ability to simply expand the number of organisms and loci hosted as new sequences are submitted.
Figure 1.
An overview of the current state of IPD-MHC Database. (A) Number of submitted sequences over the years (blue, original IPD-MHC Database; yellow, IPD-MHC Database v2.0); (B) Species distribution in the IDP-MHC Database v2.0; (C) Distribution of alleles per species in IPD-MHC Database (blue, class I; yellow, class II); species covering the 95% of all the alleles are shown.
The analysis tools available to the research community have been expanded and integrated with EBI existing tools to work from the new structure. An overview of all the data and tools, together with statistics and bibliographic references, is available for each MHC group. The nomenclature report allows searching for alleles within groups or loci, and provides details of alleles, EMBL-ENA/GenBank/DDBJ accession numbers and references. An entirely new and improved version of the alignment tool provides a convenient way to visualize sequence similarities for both nucleotide and protein sequences within a particular locus. The alignments follow the same format as that used in the IPD-IMGT/HLA Database, and it's available both for printing and downloading. Furthermore, in this release, the alignment tool is expanded for inter- and intra- species locus alignment, allowing comparing and identifying evolutionary tracts in MHC sequences. The improvement of the alignment tool relies on its ability to compare not only taxonomically and evolutionally related loci, but also every locus in the database against every other, explained in more detail below. While this initial release contains all the basic tools already present in the IPD-MHC Database, as the database grows, many of the tools present in the IPD-IMGT/HLA system will be added, and new analysis tools will be developed, specially focussing on the analysis of conserved motifs within protein sequences.
MATERIALS AND METHODS
Data preparation
The IPD-MHC Database curation processes makes use of flat files for the submission, curation and storage of data, while a relational database is used for consultation and analysis of MHC sequences. In order to populate the novel database structure, a pipeline for the analysis, formatting and parsing of flat files has been developed. All data from previous versions of the IPD-MHC Database were analysed and manually amended when needed. A final number of 11t061 flat files were parsed, comprising 7794 alleles distributed over 532 loci in 70 organisms.
Database structure
The underlying database was designed using relational tables, and implemented using MySQL. The website was built using a combination of PHP, javascript and CSS. In particular, the server side was built using silex (http://silex.sensiolabs.org/), the popular micro-framework, together with twig (http://twig.sensiolabs.org/), a template engine for PHP. In-house pipelines were developed for data manipulation and analysis. In particular, contig alignments as well as allele alignments for nucleotide and protein sequences were calculated with MUSCLE (16).
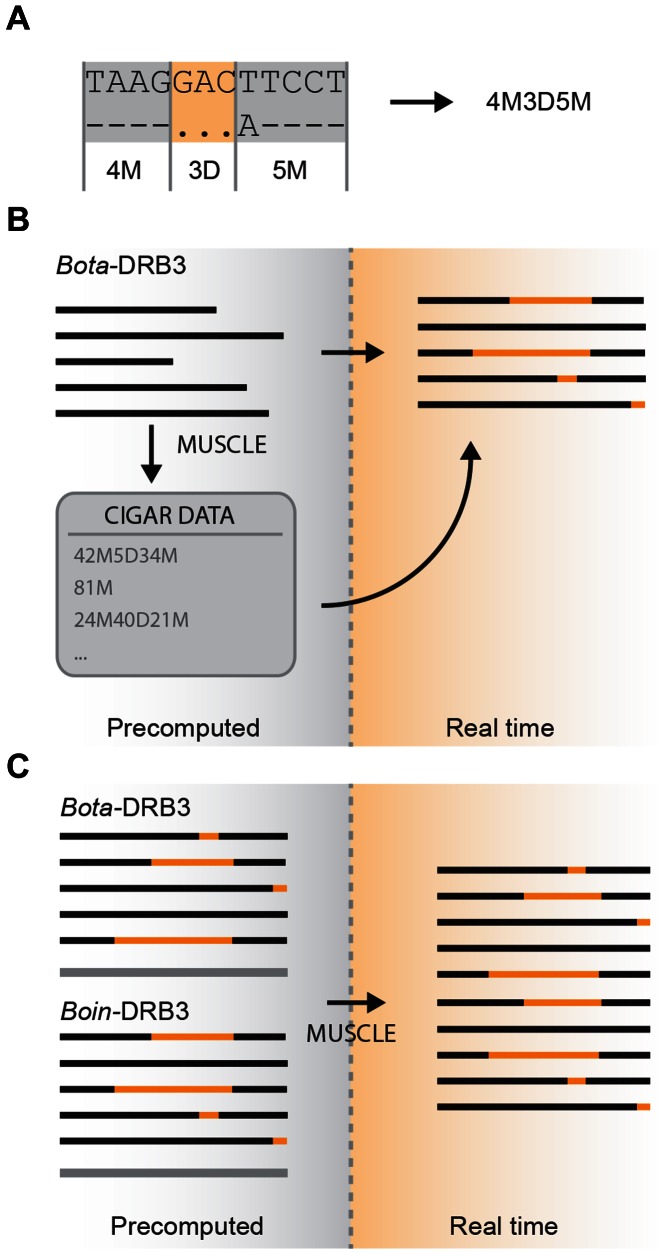
Multi-species alignment
The newly introduced multi-locus alignment allows the comparison of loci from different species in real time, giving users a powerful tool for phylogenetic and structural sequence analysis. As in the previous IPD-MHC Database versions, alignments are pre-computed and stored for each locus of every organism in the database. In this version of the database, alignments are computed with MUSCLE (16), and stored in the database as CIGAR (Compact Idiosyncratic Gapped Alignment Report) strings. The CIGAR string is the result of the sequence alignments, defining the sequence of matches/mismatches and deletions (or gaps) compared to the reference sequence (Figure 2A). CIGAR strings, together with the allele sequences, are used to generate a visualization of the loci alignment. Furthermore, the IPD-MHC Database allows generating multi-locus alignments on the fly, by choosing two or more loci from the ones available. This is achieved by a hybrid approach that combines pre-computed alignments of single loci with dynamic inter- and intra- species sequence alignments. Consensus sequences from the pre-computed alignment of each selected locus are aligned with MUSCLE and a CIGAR string is generated for each aligned locus. The newly generated CIGAR is combined with the information stored in the database of the pre-computed alignments, thus allowing to properly rendering the multiple locus alignment (Figure 2B and C).
Figure 2.
Single- and multi-locus alignment. (A) For each computed alignment, a CIGAR (Compact Idiosyncratic Gapped Alignment Report) string defining the sequence of matches/mismatches (M) and deletions or gaps (D) compared to the reference sequence is stored in the database. (B) For each locus in the database, the nucleotide and protein allele alignment is pre-computed and the CIGAR string is stored in the database to correctly represent the sequence alignment. (C) In multi-locus alignment, the consensus sequence of each locus is aligned in real time and the previously calculated single-locus aligned are assembled and rendered as one.
DISCUSSION
The IPD-MHC Database 2.0 introduces the new data organization and submission pipeline, providing a scalable and manageable system, while maintaining the high level of curation that has always characterized the IPD-MHC Database. The new centralized organization of the database allows a convenient and highly accurate comparison and analysis of data. Furthermore, by accommodating related systems in a common structure, smaller projects will benefit from the same tools and pipelines of the larger MHC groups. As the number of submitted sequences increases, the benefit of having manually curated sequences from experts in the relevant species becomes more apparent. This release represents an improved version of the IPD-MHC Database, providing a powerful tool for the study of polymorphic genes central to vertebrate animal immune systems. The centralized system allows an easier inter- and intra-species alleles comparison, making the curation process more consistent through the different MHC-groups and at the same time presenting novel levels of analysis. In particular, the newly introduced multi-locus alignment allows the comparison of loci from different species in real time, and the download of the aligned sequences for further studies and analysis. The next planned developments will significantly expand the range of analysis tools, including some of the features already available in the IPD-IMGT/HLA database, and developing bespoke tools for the analysis of conserved sequence and structural motifs. In addition, other polymorphic loci could be included in the database as the new infrastructure is now easily scalable.
AVAILABILITY
The IPD-MHC Database is publicly available at http://ebi.ac.uk/ipd/mhc/. If you are interested in contributing to the IPD-MHC project, please contact James Robinson, james.robinson@anthonynolan.org for further information.
Acknowledgments
The authors would like to acknowledge the work of all the individual MHC nomenclature committees. The authors would also like to acknowledge the support provided by the External Services Group at the European Bioinformatics Institute, which allows the IPD project to be hosted within the EBI infrastructure. The authors would also like to acknowledge the work of the comparative MHC community in sequencing, identifying and submitting the many sequences, which form the core data that the IPD-MHC Database represents. In particular, the authors would like to acknowledge the work of Dr Lorna Kennedy for curating DLA data, Nel Otting for NHP data curation, Prof. Shirley Ellis for BoLA data curation, Prof. Lutz Walter for RT1 data curation, Dr Donald Miller and Prof Douglas Antczak for ELA data curation and Dr Hans Dijkstra and the late René Stet for their contributions to salmonoid fish.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [BB/M011488/1 to G.M. and J.H.]; Anthony Nolan [to J.R. and S.M.]; European Molecular Biology Laboratories, International Society for Animal Genetics (ISAG) and the Veterinary Immunology Committee (VIC) [to IPD-MHC Databases]; Scottish Government [to K.B.]. Funding for open access charge: Anthony Nolan, a charitable organisation [to J.R. and S.M.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Klein J., Satta Y., O'hUigin C., Takahata N. The molecular descent of the major histocompatibility complex. Annu. Rev. Immunol. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- 2.Klein J., Bontrop R.E., Dawkins R.L., Erlich H.A., Gyllensten U.B., Heise E.R., Jones P.P., Parham P., Wakeland E.K., Watkins D.I. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- 3.Ellis S.A., Bontrop R.E., Antczak D.F., Ballingall K., Davies C.J., Kaufman J., Kennedy L.J., Robinson J., Smith D.M., Stear M.J., et al. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- 4.Robinson J., Halliwell J.A., McWilliam H., Lopez R., Marsh S.G.E. IPD–the Immuno Polymorphism Database. Nucleic Acids Res. 2013;41:D1234–D1240. doi: 10.1093/nar/gks1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot N.G., Otting N., Robinson J., Blancher A., Lafont B.A.P., Marsh S.G.E., O'Connor D.H., Shiina T., Walter L., Watkins D.I., et al. Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kennedy L.J., Angles J.M., Barnes A., Carter S.D., Francino O., Gerlach J.A., Happ G.M., Ollier W.E.R., Thomson W., Wagner J.L. Nomenclature for factors of the dog major histocompatibility system (DLA), 2000: second report of the ISAG DLA Nomenclature Committee. Anim. Genet. 2001;32:193–199. doi: 10.1046/j.1365-2052.2001.00762.x. [DOI] [PubMed] [Google Scholar]
- 7.Drake G.J.C., Kennedy L.J., Auty H.K., Ryvar R., Ollier W.E.R., Kitchener A.C., Freeman A.R., Radford A.D. The use of reference strand-mediated conformational analysis for the study of cheetah (Acinonyx jubatus) feline leucocyte antigen class II DRB polymorphisms. Mol. Ecol. 2004;13:221–229. doi: 10.1046/j.1365-294x.2003.02027.x. [DOI] [PubMed] [Google Scholar]
- 8.Hammond J.A., Marsh S.G.E., Robinson J., Davies C.J., Stear M.J., Ellis S.A. Cattle MHC nomenclature: is it possible to assign sequences to discrete class I genes. Immunogenetics. 2012;64:475–480. doi: 10.1007/s00251-012-0611-7. [DOI] [PubMed] [Google Scholar]
- 9.Tseng C.T., Miller D., Cassano J., Bailey E., Antczak D.F. Identification of equine major histocompatibility complex haplotypes using polymorphic microsatellites. Anim. Genet. 2010;41(Suppl. 2):150–153. doi: 10.1111/j.1365-2052.2010.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimholt U. MHC and evolution in Teleosts. Biology (Basel) 2016;5:6. doi: 10.3390/biology5010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dressel R., Walter L., Günther E. Genomic and funtional aspects of the rat MHC, the RT1 complex. Immunol. Rev. 2001;184:82–95. doi: 10.1034/j.1600-065x.2001.1840108.x. [DOI] [PubMed] [Google Scholar]
- 12.Ballingall K.T., Herrmann-Hoesing L., Robinson J., Marsh S.G.E., Stear M.J. A single nomenclature and associated database for alleles at the major histocompatibility complex class II DRB1 locus of sheep. Tissue Antigens. 2011;77:546–553. doi: 10.1111/j.1399-0039.2011.01637.x. [DOI] [PubMed] [Google Scholar]
- 13.Ho C.-S., Lunney J.K., Ando A., Rogel-Gaillard C., Lee J.-H., Schook L.B., Smith D.M. Nomenclature for factors of the SLA system, update 2008. Tissue Antigens. 2009;73:307–315. doi: 10.1111/j.1399-0039.2009.01213.x. [DOI] [PubMed] [Google Scholar]
- 14.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochrane G., Karsch-Mizrachi I., Takagi T., International Nucleotide Sequence Database Collaboration The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2016;44:D48–D50. doi: 10.1093/nar/gkv1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]