Abstract
Biological data are generated at unprecedentedly exponential rates, posing considerable challenges in big data deposition, integration and translation. The BIG Data Center, established at Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, provides a suite of database resources, including (i) Genome Sequence Archive, a data repository specialized for archiving raw sequence reads, (ii) Gene Expression Nebulas, a data portal of gene expression profiles based entirely on RNA-Seq data, (iii) Genome Variation Map, a comprehensive collection of genome variations for featured species, (iv) Genome Warehouse, a centralized resource housing genome-scale data with particular focus on economically important animals and plants, (v) Methylation Bank, an integrated database of whole-genome single-base resolution methylomes and (vi) Science Wikis, a central access point for biological wikis developed for community annotations. The BIG Data Center is dedicated to constructing and maintaining biological databases through big data integration and value-added curation, conducting basic research to translate big data into big knowledge and providing freely open access to a variety of data resources in support of worldwide research activities in both academia and industry. All of these resources are publicly available and can be found at http://bigd.big.ac.cn.
INTRODUCTION
The rapid advancements of high-throughout sequencing technologies provide us with formidable capacity in genome sequencing, accordingly producing biological data at an unprecedentedly exponential rate and resultantly accumulating huge amounts of biological data at multiple omics levels (1). To address the most important and complex biological questions, it is often required to provide researchers with open access to various data resources (2). Nowadays, China has become a powerhouse in generating vast quantities of biological data, but is in the embarrassing situation of lacking a centralized data center that is committed to opening data in this big data world and to making data well-organized and publicly accessible to worldwide scientific communities (3).
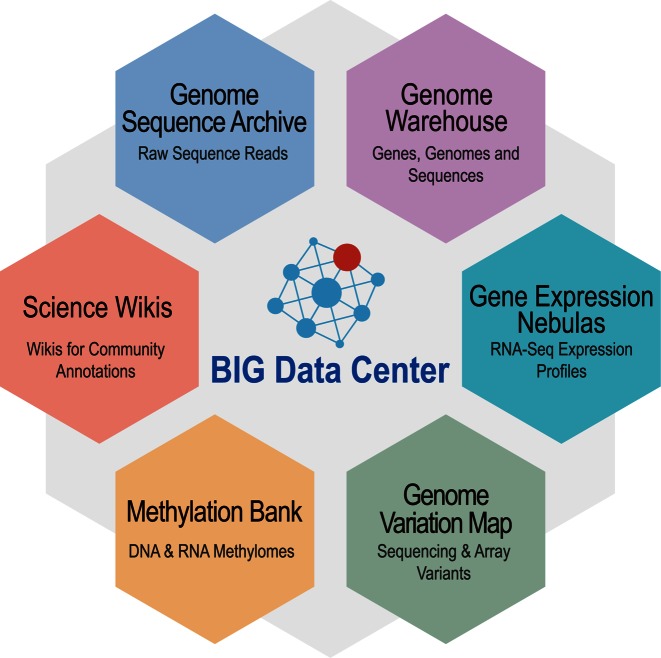
The BIG Data Center, established at Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, takes full advantages of valuable resources and experiences in BIG as well as partner institutions to provide sustainable and reliable services in aid of research activities throughout the world. Specially, BIG features important achievements in not only actively participating in the International Human Genome Project China Part (4) but also presiding several prestigious national research projects (e.g. the Chinese Superhybrid Rice Genome Project (5), the Chicken (6), Silkworm (7), Date Palm (8), Common Carp (9), Cassava (10) and Rubber Tree (11) Genome Projects), pioneers the Chinese Population Precision Medicine Initiative (http://news.xinhuanet.com/english/2016-01/09/c_134993997.htm) and possesses rich experiences in developing and maintaining biological databases. In addition, it is well equipped with facilities including both DNA sequencers and high performance computing resources. Therefore, the BIG Data Center is dedicated to constructing and maintaining biological databases by big data integration and value-added curation, performing basic research by development of advanced methods to aid translation of big data into big discovery and providing freely open access to a suite of featured data resources in support of worldwide activities in both academia and industry (http://bigd.big.ac.cn; Figure 1).
Figure 1.
The BIG Data Center's core data resources. A full list of data resources, which contains links to each resource, is available at http://bigd.big.ac.cn/databases.
GENOME SEQUENCE ARCHIVE
The Genome Sequence Archive (GSA; http://gsa.big.ac.cn) is a data repository specialized for archiving raw sequence reads. It supports data generated from a variety of sequencing platforms ranging from Sanger sequencing machines to single-cell sequencing machines and provides data storing and sharing services free of charge for worldwide scientific communities. In addition to raw sequencing data, GSA also accommodates secondary analyzed files in acceptable formats (like BAM, VCF). Its user-friendly web interfaces simplify data entry and submitted data are roughly organized as two parts, viz., Metadata and File, where the former can be further assorted into BioProject, BioSample, Experiment and Run, and the latter contains raw sequence reads. Since its inception in August 2015, GSA, as of October 2016, houses a total of 155 projects, 7140 samples, 7646 experiments and 8433 runs for more than 50 species, and stores compressed sequence files that are more than 100TB in size.
GENE EXPRESSION NEBULAS
High-throughput sequencing technologies provide a revolutionary way for transcriptome profiling, enable facile generation of large-scale RNA sequencing (RNA-Seq) data and accordingly facilitate high-resolution quantification of gene expression levels across a variety of tissues and treatments (12–14). Thus, gene expression profiling from RNA-Seq data is of fundamental significance for deciphering functional elements under diverse conditions and characterizing the dynamics of transcriptomic regulation. The Gene Expression Nebulas (GEN; http://bigd.big.ac.cn/gen) is a data portal of gene expression profiles based entirely on RNA-Seq data (that are retrieved from NCBI SRA (15)), which currently hosts two featured resources, namely, Mammalian Transcriptomic Database (16) and Rice Expression Database (RED) (17).
Mammalian transcriptomic database (MTD)
Mammalian transcriptomic database (MTD) (http://bigd.big.ac.cn/mtd) (16) is a mammalian transcriptomic database that is based on large quantities of RNA-Seq data across various tissues/cell lines. In the current version, it incorporates a wealth of transcriptomes from human, mouse, rat and pig, which are all obtained from NCBI SRA (15). MTD features easy-to-use web interfaces for exploration of transcriptomic profiling for genes or for a specific genomic region, characterization of detailed expression profiles at the levels of exon, transcript, and gene and visualization of transcriptomic data in an interactive manner powered by a genome browser. In addition, MTD allows users to search for genes or isoforms with customized transcriptional features, such as housekeeping genes, expression profiles of tissues/cell lines and isoforms undergoing an ‘exon skipped’ alternative splicing event. Moreover, it supports comparative transcriptomic analysis not only within a species but also across species, bearing the potential to reveal the dynamics of gene expression regulation. Together, MTD is a valuable resource for mammalian transcriptomic and evolutionary studies.
Rice expression database (RED)
RED (http://expression.ic4r.org), a committed project of Information Commons for Rice (IC4R) (17), is an integrated database hosting rice gene expression profiles derived entirely from high-quality RNA-Seq data. Unlike extant related databases that are mostly based on microarray data and/or contain limited RNA-Seq data, RED contains a comprehensive collection of 284 high-quality RNA-Seq experiments obtained from NCBI SRA (15) and thus houses a large number of gene expression profiles that span a broad range of rice growth stages and cover a wide variety of biotic and abiotic treatments. Powered by AJAX (Asynchronous JavaScript and XML, a collection of web development technologies for creating highly interactive web applications) and HighChart (a JavaScript-based library for setting up interactive charts in web pages), RED also features interactive search and display of expression profiles of concerned genes across different tissues and treatments. In addition, RED provides online tools for construction and visualization of gene co-expression networks, which can be achieved simply by specifying genes of interest. Ongoing efforts include integration of more high-quality RNA-Seq data and characterization of transcriptomic profiles by association with important agronomic traits in rice. Moreover, we plan to use RED as a framework to incorporate data from other plants, such as maize (Zea mays), rubber tree (Hevea brasiliensis), potato (Solanum tuberosum) and cotton (Gossypium raimondii), and to extend it into a generalized gene expression database for multiple economically important plants.
GENOME VARIATION MAP
The Genome Variation Map (GVM; http://bigd.big.ac.cn/gvm) is a data repository and retrieval system of genome variations, including single nucleotide polymorphisms (SNPs) and small insertions and deletions (indels). Currently, GVM focuses on genome variations for human as well as domesticated animals (e.g. dog) and cultivated plants (e.g. rice), which are of great importance for in-depth exploration of favorable traits (e.g. drought resistance in plant) and investigation of species domestication and evolution. The current version of GVM integrates a collection of variation data from several featured species including human, dog, rice and sorghum.
Dog genome SNP database (DoGSD)
Dog genome SNP database (DoGSD) (http://bigd.big.ac.cn/dogsd) (18) is a Canidae-specific SNP database for domesticated dogs and gray wolves, comprising ∼19 million high-quality whole genome SNPs of 77 individual samples (that are obtained from published studies (19–21) and NCBI dbSNP (22)). DoGSD integrates a comprehensive collection of SNP related information, including SNP annotation, associated genes, synonymous or non-synonymous SNPs, sample location, breed information, together with the population genetic statistics (FST) for online analysis. DoGSD provides friendly interfaces to browse detailed information for each SNP, to retrieve a list of SNPs for any given sample referencing to specific chromosome and to obtain SNP statistics for multiple different samples of interest. As a committed sub-project of Dog 10K Genomes Project (dog10K; http://dog10k.big.ac.cn), DoGSD is committed to incorporating more comprehensive variation data for the canine research community, serving as a critical resource for better understanding the evolutionary history of dogs, investigating genetic changes associated with domestication and relating genetic changes to phenotype.
Rice variation database (RVD)
As rice is not only a key model organism for plant studies but also the most widely consumed staple food for a large number of global human population, thousands of rice accessions have been re-sequenced to date. Rice variation database (RVD) (http://variation.ic4r.org) (17) is built based on a large collection of 5152 re-sequenced rice accessions, which are mainly from published literatures (23–27) and the 3K Rice Genomes Project (28), and accordingly includes ∼18 million SNPs against Os-Nipponbare-Reference-IRGSP-1.0 pseudomolecule identified by using unified standard SNP-calling pipeline. RVD provides detailed annotations, including SNP consequence, gene function and results from association studies, and hyperlinks to other external database resources. Besides, RVD is equipped with online analysis tools, viz., RiceClustalW for multiple sequence alignment against specific rice accession(s), Population Genetic Analysis for computing population genetic parameters for any specific region, and Gene Haplotype Analysis for calculating gene haplotype diversity and structure.
Sorghum genome SNP database (SorGSD)
Sorghum is not only one of the most important crops but also a potential bio-energy feedstock. SorGSD (http://bigd.big.ac.cn/sorgsd) (29) is a sorghum genome SNP database that is of great significance in genetic characterization of important quantitative traits in sorghum. SorGSD covers a diverse collection of 48 sorghum lines (30,31) that fall into four groups, viz., improved varieties, landraces, wild and weedy sorghums, and a wild relative Sorghum propinquum. Totally, SorGSD includes ∼62.9 million SNPs identified from the whole genome re-sequencing data of these individuals by mapping to the S. bicolor reference genome (v3). SorGSD provides a detailed summary of SNP information and their relevant annotations for all individual accessions, such as allele information, gene information, SNP density and external links to other resources. In addition, it allows comparison of SNP data among two or more sorghum lines, equips with easy-to-use visualization interfaces by integrating the GBrowse package and collects other sorghum-related resources and literature references, providing a valuable repository for sorghum genetic and molecular breeding studies.
Virtual chinese genome database (VCGDB)
Virtual Chinese Genome Database (VCGDB) (http://bigd.big.ac.cn/vcg) (32) is a dynamic genome database of Chinese populations based on whole-genome sequencing data of 194 individuals that are publicly available in the 1000 Genomes Project (33). VCGDB presents two types of genetic variations: virtual and dynamic; virtual variations are those shared by all collected individuals, reflecting what is common to the Chinese populations, whereas dynamic variations are those that vary among individuals, revealing genetic differences specific to the individuals. As a result, VCGDB houses a large variety of dynamic genomic variations including 35 million single nucleotide variations (SNVs), 0.5 million indels and 29 million rare variations. In addition, a highly interactive user-friendly interface is provided in VCGDB to display the virtual and dynamic variations and a web search engine is also installed in VCGDB to support online real-time high-performance queries. Based on the ongoing project of the Chinese Population Precision Medicine Initiative we lead, VCGDB will incorporate more Chinese population genomes and provide a more precise Chinese reference genome.
GENOME WAREHOUSE
The Genome Warehouse (GWH; http://bigd.big.ac.cn/gwh) is a centralized resource housing genome-scale data, with the purpose to archive high-quality genome sequences and gene annotation information. Currently, GWH offers users with open access to a featured collection of 26 genomes from economically important plants and animals, which are either publicly available in NCBI (34) or sequenced in-house; among them, the genome of Hevea brasiliensis (rubber tree) (11) that has been released recently and sequenced by our institution is a representative example. For each species, GWH contains detailed genome-related information including species metadata, genome assembly, sequence data and the corresponding annotations. Additionally, a functionality of ‘Tree View’ is provided to depict the evolutionary relationship of all species collected in GWH. For convenience, sequence data of individual genomes as well as their gene annotations are downloadable via File Transfer Protocol (FTP). Future directions of GWH include continuous integration of newly sequenced genomes and development of enhanced interfaces for data presentation and visualization.
METHYLATION BANK
The Methylation Bank (MethBank; http://bigd.big.ac.cn/methbank) (35) is a repository that integrates whole-genome single-base resolution methylomes and provides an interactive browser for visualization of high-resolution DNA methylation data. It incorporates high-quality whole-genome bisulfite sequencing methylome maps for five economically important crops (Oryza sativa, Glycine max, Manihot esculenta, Phaseolus vulgaris and Solanum lycopersicum) as well as two model animals (Danio rerio and Mus musculus) (36,37). Specifically, to quality-control all collected methylomes (that are publicly available in NCBI SRA (15) till May 2016), MethBank discards low-quality methylomes by considering genome coverage and bisulfite conversion rate, and as a result, obtains 42 high-quality methylomes for O. sativa, 21 for G. max, 1 for M. esculenta, 1 for P. vulgaris, 7 for S. lycopersicum, 9 for D. rerio and 9 for M. musculus. MethBank features genome-wide profiling of methylation levels across chromosomes, identification of differentially methylated promoters (DMP) between a range of conditions, and visualization of methylation profiles for genes, regions and CpG Islands under multiple different samples. In addition, MethBank offers intuitive interfaces for data browse and retrieval; it is able to provide a genome-wide methylation view and to retrieve gene methylation profiles and regional methylation levels across all collected samples. It is also equipped with interactive interfaces to facilitate search of methylation levels for any given gene that is related to DMP or highly-methylated CpG islands. In addition to DNA methylation, evidence has accumulated that RNA methylation is closely related with various biological processes and human diseases. Therefore, our ongoing efforts not only incorporate more types of DNA methylation from diverse species, but also integrate a wide range of RNA methylation data.
SCIENCE WIKIS
Community curation—harnessing community intelligence for biological knowledge curation, in contrast to expert curation that is heavily based on dedicated experts and vulnerably threatened by funding cuts (38)—promises to be a solution to deal with the deluge of biological data (39). A case in point is Wikipedia, an online encyclopedia that allows any user to create/edit any content and features community integration, huge coverage, up-to-date content as well as low cost for maintenance. Spirited by its extraordinary success, Science Wikis (http://bigd.big.ac.cn/sciencewikis) are a series of biological databases wikified for community curation (40), among which LncRNAWiki and RiceWiki are two featured resources that exploit the full potential of worldwide scientific communities for big data collection, integration and management.
LncRNAWiki
LncRNAWiki (http://bigd.big.ac.cn/lncrnawiki) (41) is a wiki-based, open-content and publicly editable platform that employs collective efforts in community curation of human long non-coding RNAs (lncRNAs). In addition, it quantifies community-curated efforts and provides explicit authorship based on quantitative contributions (41), which potentially attracts more people to share their knowledge and accordingly enables LncRNAWiki to serve as an up-to-date and comprehensive knowledgebase for human lncRNAs. As of September 2016, LncRNAWiki houses a total of 105 824 non-redundant lncRNAs that are integrated from GENCODE (42), NONCODE (43) and LNCipedia (44). Among them, 719 lncRNAs have been manually community-curated based on published literatures and 290 of them have been experimentally validated to be associated with cancer and other diseases. Moreover, considering the functional significance of lncRNA-encoded small proteins as reported in (45,46), we developed computational approaches for identification of small proteins in all collected human lncRNAs, identified 9387 lncRNAs potentially encoding small proteins and revealed that 2246 out of them have higher confidence by taking account of protein instability, secondary structure and transmembrane helix. As a result, all these identified lncRNAs as well as their associated small proteins are incorporated into LncRNAWiki. Since this July, LncRNAWiki has become a member of RNAcentral (47), further facilitating data exchange and sharing between LncRNAWiki and other related databases.
RiceWiki
RiceWiki (http://wiki.ic4r.org) (48) is a wiki-based, publicly editable platform for community curation of rice genes. Compared with other relevant databases, RiceWiki features collective intelligence on knowledge integration and annotation and explicit authorship in terms of quantified community-curated contributions (49). Since its inception in 2014, RiceWiki has been continuously updated, expanded and enriched, leading to more than 400 genes community-curated and covering over 3000 rice-related scientific articles. In addition, several MediaWiki extensions that are a bunch of codes for fulfilling customized functionalities are developed and deployed in RiceWiki, which aid to incorporate different types of rice-related data, such as RNA-Seq-based gene expression profiles from RED and rice-related literatures from Rice Literature Miner (http://literature.ic4r.org). Furthermore, a lightweight BLAST module is also implemented as a MediaWiki extension that enables community curators to conduct sequence alignment and facilitate gene annotation. Based on community curation, RiceWiki has the potential to cover a larger scope of rice-related knowledge and function as a comprehensive and up-to-date encyclopedia that are constantly improved and broadly shared by the rice research community.
TRAINING
The need for personnel training across diverse biological disciplines is high, especially in the face of critical challenges posed by big data generated in life and health sciences. We engage in the Genomics and Bioinformatics Training (GBT; http://bigd.big.ac.cn/training/gbt) at various levels ranging from introductory to in-depth and provide GBT courses for researchers and biomedical professionals at postgraduate level and above. Since the first GBT in 2008, we have delivered more than 20 GBT events (2–3× per year) over the past several years and more than 830 individuals have been trained, including graduate students and junior faculty members from the field of biology, medicine, agriculture and forestry. In the big data era where a range of data operations (including deposition, curation, processing, analysis and visualization) become routine and increasingly daunting, we solicit feedbacks from all trainers as well as colleagues and peers to keep pace with practical needs and improve our training programs. In addition, we are open to suggestions and worldwide collaborations to make the training programs more useful and better targeted.
CONCLUDING REMARKS
The BIG Data Center provides freely open access to a variety of database resources in support of research activities in both academia and industry throughout the world. With the ultimate goal to advance life and health sciences, therefore, it is dedicated to constructing and maintaining biological databases by value-added curation and performing basic research to address critical challenges in big data deposition, integration and translation. The BIG Data Center, albeit relatively young, will grow to be indispensable for worldwide biological studies as more data are integrated and its associated services are mature.
Acknowledgments
We thank Jun Yu, Songnian Hu, Xiangdong Fang, Jiang Liu, Hongxing Lei, Fei Chen, Guodong Wang, Hong Luo, Hang He, Lingling Chen, Yu Xue, Xiangfeng Wang and Yunchao Ling for constructive comments on development of database resources. We also thank a number of users for sending suggestions and reporting bugs.
APPENDIX
BIG Data Center Members (Participants are arranged by project role and then by contribution except for Group Leader, as indicated; DCA = Data Curation & Analysis; DSD = Database System Development; GL = Group Leader)
Corresponding authors: Zhang Zhang1,2,3,*, Wenming Zhao1,*, Jingfa Xiao1,2,3,*
GSA Group: DSD: Yanqing Wang1,#, Fuhai Song2,#, Junwei Zhu1,#, Bixia Tang1,3, Yadong Yang2,3; DCA: Tingting Chen1,#, Sisi Zhang1, Lili Dong1; GL: Wenming Zhao1,*
GEN Group: DCA: Xin Sheng1,2,3,#, Lin Xia1,2,3,#, Chen Gao1,2,3, Jian Sang1,2,3, Lijuan Zhang1,2,3, Wan Fang1,2,3, Mingming Lu1,2,3, Zhewen Zhang1,2, Jingfa Xiao1,2,3,*; DSD: Dong Zou1,2,#, Xin Sheng1,2,3,#, Xingjian Xu1,2,3; GL: Lili Hao1,2,#, Meili Chen1,2,#
GVM Group: DCA: Dongmei Tian1,#, Cuiping Li1,#, Lili Dong1,#, Na Yuan1, Jingyao Zeng1, Jinyue Wang1,2,3, Shuo Shi1,2,3, Yadong Zhang1,2,3; DSD: Bixia Tang1,3,#, Xingjian Xu1,2,3,#, Dong Zou1,2; GL: Zhenglin Du1,#, Shuhui Song1,#
GWH Group: DCA: Jian Sang1,2,3,#, Xiaomeng Ge2,#, Shixiang Sun1,2,3,#, Man Li1,2,3, Mengwei Li1,2,3, Lin Liu1,2,3, Ting Wei2,3, Zilong He2,3, Chunlei Yu1,2,3, Hongyan Yin1,2,3, Guangyu Wang1,2,3, Lin Xia1,2,3, Yan Sun2,3, Shanshan Zou2,3, Yuan Liang2,3, Shuangyang Wu2,3; DSD: Xingjian Xu1,2,3,#, Dong Zou1,2,#, Fan Wang1,2; GL: Lili Hao1,2,#
MethBank Group: DCA: Shixiang Sun1,2,3,#, Fang Liang1,#, Mengwei Li1,2,3; DSD: Dong Zou1,2,3,#; GL: Rujiao Li1,#
ScienceWikis Group: DCA: Jian Sang1,2,3,#, Guangyu Wang1,2,3,#, Chunlei Yu1,2,3,#, Lin Liu1,2,3, Man Li1,2,3, Guangyi Niu1,2,3; DSD: Jian Sang1,2,3,#, Dong Zou1,2,#, Zhang Zhang1,2,3,*; GL: Lina Ma1,2,#
Hardware & System Administration Group: Huanxin Chen1 (GL), Yubin Sun1, Lei Yu1, Shuang Zhai1, Mingyuan Sun1
Writing Group: Zhang Zhang1,2,3,*, Wenming Zhao1,*, Jingfa Xiao1,2,3,*, Shuhui Song1,#, Lili Hao1,2,#, Rujiao Li1,#, Lina Ma1,2,#, Xin Sheng1,2,3,#, Jian Sang1,2,3,#, Yanqing Wang1,#, Bixia Tang1,3,#, Zhewen Zhang1,2,#
1BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, Beijing 100101, China
2CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
3University of Chinese Academy of Sciences, Beijing 100049, China
*To whom correspondence should be addressed to Zhang Zhang (zhangzhang@big.ac.cn). Correspondence may also be addressed to Wenming Zhao (zhaowm@big.ac.cn) and Jingfa Xiao (xiaojingfa@big.ac.cn).
#The authors wish it to be known that, in their opinion, these authors should be regarded as Joint First Authors.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040500 to W.Z, Z.Z.; XDA08020102 to Z.Z.]; National Key Research Program of China [2016YFC0901603 to W.Z.; 2016YFB0201702, 2016YFC0901903 to J.X.]; National Programs for High Technology Research and Development [863 Program; 2015AA020108 to Z.Z.]; National Natural Science Foundation of China [31100915 to L.H.; 31200978 to L.M.; 31271386, 31471248 to J.X.]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008]; Key Program of the Chinese Academy of Sciences [KJZD-EW-L14 to J.X.]; Key Technology Talent Program of the Chinese Academy of Sciences (to W.Z.); The 100 Talent Program of the Chinese Academy of Sciences (to Z.Z.). Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marx V. Biology: the big challenges of big data. Nature. 2013;498:255–260. doi: 10.1038/498255a. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z., Bajic V.B., Yu J., Cheung K.-H., Townsend J.P. Data integration in bioinformatics: current efforts and challenges. In: Mahdavi MA, editor. Bioinformatics—Trends and Methodologies. Vol. 1. Rijeka: InTech; 2011. pp. 41–56. [Google Scholar]
- 3.Zhang Z., Zhu W., Luo J. Bringing biocuration to China. Genomics Proteomics Bioinformatics. 2014;12:153–155. doi: 10.1016/j.gpb.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 6.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 7.Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., Zhao P., Zha X., Cheng T., Chai C., et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- 8.Al-Mssallem I.S., Hu S., Zhang X., Lin Q., Liu W., Tan J., Yu X., Liu J., Pan L., Zhang T., et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013;4:2274–2282. doi: 10.1038/ncomms3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu P., Zhang X., Wang X., Li J., Liu G., Kuang Y., Xu J., Zheng X., Ren L., Wang G., et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Genet. 2014;46:1212–1219. doi: 10.1038/ng.3098. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Feng B., Xiao J., Xia Z., Zhou X., Li P., Zhang W., Wang Y., Moller B.L., Zhang P., et al. Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 2014;5:5110–5118. doi: 10.1038/ncomms6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang C., Yang M., Fang Y., Luo Y., Gao S., Xiao X., An Z., Zhou B., Zhang B., Tan X., et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants. 2016;2:16073–16082. doi: 10.1038/nplants.2016.73. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M., Xiao J., Zhang Z., Liu J., Wu J., Yu J. Identification of human HK genes and gene expression regulation study in cancer from transcriptomics data analysis. PLoS One. 2013;8:e54082. doi: 10.1371/journal.pone.0054082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams J. Transcriptome: connecting the genome to gene function. Nat. Educ. 2008;1:195. [Google Scholar]
- 15.Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database, C. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng X., Wu J., Sun Q., Li X., Xian F., Sun M., Fang W., Chen M., Yu J., Xiao J. MTD: a mammalian transcriptomic database to explore gene expression and regulation. Brief Bioinform. 2016:1–9. doi: 10.1093/bib/bbv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The IC4R Project Consortium. Information commons for rice (IC4R) Nucleic Acids Res. 2016;44:D1172–D1180. doi: 10.1093/nar/gkv1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai B., Zhao W.M., Tang B.X., Wang Y.Q., Wang L., Zhang Z., Yang H.C., Liu Y.H., Zhu J.W., Irwin D.M., et al. DoGSD: the dog and wolf genome SNP database. Nucleic Acids Res. 2015;43:D777–D783. doi: 10.1093/nar/gku1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gou X., Wang Z., Li N., Qiu F., Xu Z., Yan D., Yang S., Jia J., Kong X., Wei Z., et al. Whole-genome sequencing of six dog breeds from continuous altitudes reveals adaptation to high-altitude hypoxia. Genome Res. 2014;24:1308–1315. doi: 10.1101/gr.171876.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freedman A.H., Gronau I., Schweizer R.M., Ortega-Del Vecchyo D., Han E., Silva P.M., Galaverni M., Fan Z., Marx P., Lorente-Galdos B, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G.D., Zhai W., Yang H.C., Fan R.X., Cao X., Zhong L., Wang L., Liu F., Wu H., Cheng L.G., et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat. Commun. 2013;4:1860–1868. doi: 10.1038/ncomms2814. [DOI] [PubMed] [Google Scholar]
- 22.Saccone S.F., Quan J., Mehta G., Bolze R., Thomas P., Deelman E., Tischfield J.A., Rice J.P. New tools and methods for direct programmatic access to the dbSNP relational database. Nucleic Acids Res. 2011;39:D901–D907. doi: 10.1093/nar/gkq1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., Li C., Zhu C., Lu T., Zhang Z., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 24.Huang X., Kurata N., Wei X., Wang Z.X., Wang A., Zhao Q., Zhao Y., Liu K., Lu H., Li W., et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., Li W., Guo Y., Deng L., Zhu C., et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 26.Xu X., Liu X., Ge S., Jensen J.D., Hu F., Li X., Dong Y., Gutenkunst R.N., Fang L., Huang L., et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 27.Zhao H., Yao W., Ouyang Y., Yang W., Wang G., Lian X., Xing Y., Chen L., Xie W. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res. 2015;43:D1018–D1022. doi: 10.1093/nar/gku894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The 3000 Rice Genomes Project. The 3,000 rice genomes project. Gigascience. 2014;3:7–12. doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo H., Zhao W., Wang Y., Xia Y., Wu X., Zhang L., Tang B., Zhu J., Fang L., Du Z., et al. SorGSD: a sorghum genome SNP database. Biotechnol. Biofuels. 2016;9:6–14. doi: 10.1186/s13068-015-0415-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mace E.S., Tai S., Gilding E.K., Li Y., Prentis P.J., Bian L., Campbell B.C., Hu W., Innes D.J., Han X., et al. Whole-genome sequencing reveals untapped genetic potential in Africa's indigenous cereal crop sorghum. Nat. Commun. 2013;4:2320–2328. doi: 10.1038/ncomms3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng L.Y., Guo X.S., He B., Sun L.J., Peng Y., Dong S.S., Liu T.F., Jiang S., Ramachandran S., Liu C.M., et al. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor) Genome Biol. 2011;12:R114–R127. doi: 10.1186/gb-2011-12-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling Y., Jin Z., Su M., Zhong J., Zhao Y., Yu J., Wu J., Xiao J. VCGDB: a dynamic genome database of the Chinese population. BMC Genomics. 2014;15:265–277. doi: 10.1186/1471-2164-15-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayers E.W., Barrett T., Benson D.A., Bolton E., Bryant S.H., Canese K., Chetvernin V., Church D.M., Dicuccio M., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou D., Sun S., Li R., Liu J., Zhang J., Zhang Z. MethBank: a database integrating next-generation sequencing single-base-resolution DNA methylation programming data. Nucleic Acids Res. 2015;43:D54–D58. doi: 10.1093/nar/gku920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang L., Zhang J., Wang J., Wang L., Zhang L., Li G., Yang X., Ma X., Sun X., Cai J., et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Zhang J., Duan J., Gao X., Zhu W., Lu X., Yang L., Zhang J., Li G., Ci W., et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157:979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baker M. Databases fight funding cuts. Nature. 2012;489:19. doi: 10.1038/489019a. [DOI] [PubMed] [Google Scholar]
- 39.Howe D., Costanzo M., Fey P., Gojobori T., Hannick L., Hide W., Hill D.P., Kania R., Schaeffer M., St Pierre S, et al. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J.C., Aramayo R., Bolser D., Conway T., Elsik C.G., Gribskov M., Kelder T., Kihara D., Knight T.F., Jr, Pico A.R., et al. The emerging world of wikis. Science. 2008;320:1289–1290. doi: 10.1126/science.320.5881.1289b. [DOI] [PubMed] [Google Scholar]
- 41.Ma L.N., Li A., Zou D., Xu X.J., Xia L., Yu J., Bajic V.B., Zhang Z. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015;43:D187–D192. doi: 10.1093/nar/gku1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:D174–D180. doi: 10.1093/nar/gku1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T., et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The RNAcentral Consortium. RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015;43:D123–D129. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z., Sang J., Ma L., Wu G., Wu H., Huang D., Zou D., Liu S., Li A., Hao L., et al. RiceWiki: a wiki-based database for community curation of rice genes. Nucleic Acids Res. 2014;42:D1222–D1228. doi: 10.1093/nar/gkt926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai L., Tian M., Wu J., Xiao J., Wang X., Townsend J.P., Zhang Z. AuthorReward: increasing community curation in biological knowledge wikis through automated authorship quantification. Bioinformatics. 2013;29:1837–1839. doi: 10.1093/bioinformatics/btt284. [DOI] [PMC free article] [PubMed] [Google Scholar]