Abstract
The neXtProt human protein knowledgebase (https://www.nextprot.org) continues to add new content and tools, with a focus on proteomics and genetic variation data. neXtProt now has proteomics data for over 85% of the human proteins, as well as new tools tailored to the proteomics community.
Moreover, the neXtProt release 2016-08-25 includes over 8000 phenotypic observations for over 4000 variations in a number of genes involved in hereditary cancers and channelopathies. These changes are presented in the current neXtProt update. All of the neXtProt data are available via our user interface and FTP site. We also provide an API access and a SPARQL endpoint for more technical applications.
INTRODUCTION
neXtProt (https://www.nextprot.org; (1–3)) is a knowledge platform that represents the current state of knowledge on human proteins. It complements UniProtKB (4) by extending the content and tools, with the aim of supporting applications specifically relevant to human proteins. To integrate data accurately and develop tools that are relevant to the users, we collaborate closely with all our data and tools providers. neXtProt values high quality data, which is achieved by manual annotation or review of all data and a stringent quality control process. Whenever possible, we provide our data, tools and code freely to all users.
Since our last neXtProt update (1), we have continued to expand the database, focusing mainly on proteomics and genetic variations. One major new development is a large set of manual annotations that we have created to capture the phenotypic effect of genetic variations as described in the literature, as well as a corresponding new view to present the impact of changes at the amino acid sequence on various characteristics of the protein. We have also developed tools to support our proteomics user community. This article describes the major features of our most recent release.
neXtProt contents overview
Our main data sources are listed in Table 1: UniProtKB (4), Bgee (5), HPA (6), PeptideAtlas (7), SRMAtlas (8), GOA (9), dbSNP (10), Ensembl (11), COSMIC (12), DKF GFP-cDNA localization (13,14), Weizmann Institute of Science's Kahn Dynamic Proteomics Database (15) and IntAct (16). In the past year we have worked closely with PeptideAtlas (17) to integrate their processed data (2016-01 build), including new phosphorylation data (2015-09 build). Moreover, for the first time ADP-ribosylation sites have been integrated (18), and new acetylation sites with their corresponding peptides have also been loaded (19). With this data, neXtProt now contains 142,453 post-translational modification sites and 1,150,170 peptides. Our own curation efforts have also led to the annotation of more than 4000 variants associated with more than 8000 observations at the molecular, cellular and organism levels. These data can be viewed on our user interface in a new phenotype view (see Section II) in each annotated entry, or on our new ‘Portal’ pages (see Section III).
Table 1. Data content of the neXtProt 2016-08-25 release.
| Entries | Statistics | Change since previous release | Source |
|---|---|---|---|
| Protein entries / isoforms | 22 061/42 024 | +6/+32 | UniProtKB |
| Binary interactions | 140 270 | +18 351 | IntAct |
| Post-translational modifications (PTMs) | 142 453 | +172 | PeptideAtlas, UniProtKB, neXtProt |
| Variants (including disease mutations) | 4 943 914 | +2 461 938 | UniProtKB, COSMIC, dbSNP |
| Entries with an experimental 3D structure | 5740 | +119 | PDB via UniProtKB |
| Entries with proteomics data | 17 279 | +340 | PeptideAtlas |
| Entries with a disease | 3916 | +336 | UniProtKB |
| Phenotypic annotations | 8014 | +8014 | neXtProt |
| Cited publications | 99 922 | +22 850 | All resources |
Phenotypic impact of genetic variations
In the course of two separate projects to characterize genetic variants involved in hereditary cancers and channelopathies, we have annotated the phenotypic impact of genetic variants for proteins with known causative roles in these diseases: BRCA1, BRCA2, EPCAM, MLH1, MLH3, MSH2, MSH6, PMS2, SCN1A, SCN2A, SCN3A, SCN4A, SCN5A, SCN8A, SCN9A, SCN10A and SCN11A.
The neXtProt variation phenotypes are curated in a highly structured model with complete traceability to the original experimental results. Our annotation statements are triplets composed of (i) a subject, corresponding to the protein variation being annotated; (ii) a predicate (or relation) describing how the object is affected (Table 2); and (iii) an object describing the phenotype tested. This phenotype can correspond to the protein's molecular function or its localization (captured with Gene Ontology (GO) terms (20)), effects at the level of the organism (captured with the mammalian phenotype ontology (21)), interactions with proteins (represented by neXtProt entries) or small molecules (captured using the ChEBI dictionary of molecular entities (22)). Changes in protein or mRNA stability are captured with in-house vocabularies available on our FTP site: ftp://ftp.nextprot.org/pub/current_release/controlled_vocabularies/cv_protein_property.obo. Finally, for ion channels, the impact on electrophysiological properties is captured with the ICEPO ontology (23). Experimental evidence for each statement is provided, including a reference, an evidence code from the Evidence and Conclusion Ontology (24), and, importantly, a qualitative assessment of the phenotype intensity: mild, moderate or severe. Variations producing no significant difference compared to the wild-type are annotated as having no impact. A detailed description of our annotation model and data will be published elsewhere.
Table 2. Relations used in the neXtProt phenotype annotations.
| Relation | Definition |
|---|---|
| No impact | No significant effect observed compared to wild-type control |
| Impact | A significant effect observed compared to wild-type control |
| Increase | A significant increase observed in a measured parameter compared to wild-type control |
| Decrease | A significant decrease observed in a measured parameter compared to wild-type control |
| Gain of function | Mutant protein acquires a property absent from the wild-type (new substrate, new cellular localization, etc.) |
New phenotype view
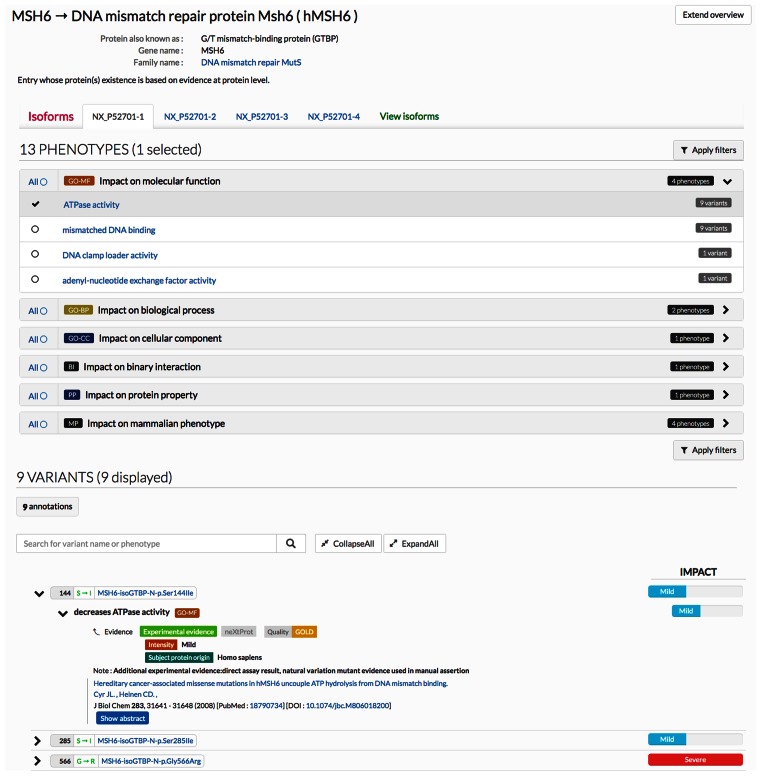
We have deployed a new view to display the phenotype data. The view contains two main sections: Phenotypes and Variants. The Phenotypes section (Figure 1A) lists the different phenotypes observed for the protein, grouped by object type: GO molecular function, biological process, cellular component, binary interaction, protein property and mammalian phenotype, as well as the number of phenotypes in each group. Clicking on the object type opens the list of phenotypes annotated in that category. For example the effect of MSH6 variants on four different GO molecular functions have been tested: ATPase activity, mismatched DNA binding, DNA clamp loader activity and adenyl–nucleotide exchange factor activity. The number of variants assayed for each specific phenotype is given: as shown in Figure 1A.
Figure 1.
New Phenotype View for the MSH6 entry. (A) Phenotypes section. (B) Variants sections.
Nine MSH6 variants have been tested for ATPase activity, including Ser144Ile, Ser285Ile and Gly566Arg. Note that the names are shown with the specific isoform selected: for MSH6, the isoform displayed by default is MSH6-isoGTBP-N.
Any number of phenotypes can be selected. The variants associated with these phenotypes can be viewed by clicking on either ‘Apply filter’ button, on the top and bottom right corner of the phenotype box.
The Variants Section (Figure 1B) lists all variants ordered by their position along the sequence. Different depth of information can be viewed. At the top-most level, the variant names are shown with the intensity of its most deleterious phenotype on the right. The phenotype(s) assayed for a variant can be viewed by clicking on the arrow on the left, which can also be opened in more details to view the experimental evidence supporting the annotation: an evidence code (‘experimental evidence’ or ‘sequence similarity evidence’ for experiments carried out in non-human model systems), whether the evidence is Gold or Silver (general criteria are described in (1)), the intensity of the phenotype, the species from which the test protein was derived, and the reference. Alternatively, all details can be opened for all the variants at once by clicking the button ‘Expand all’.
Variant portals
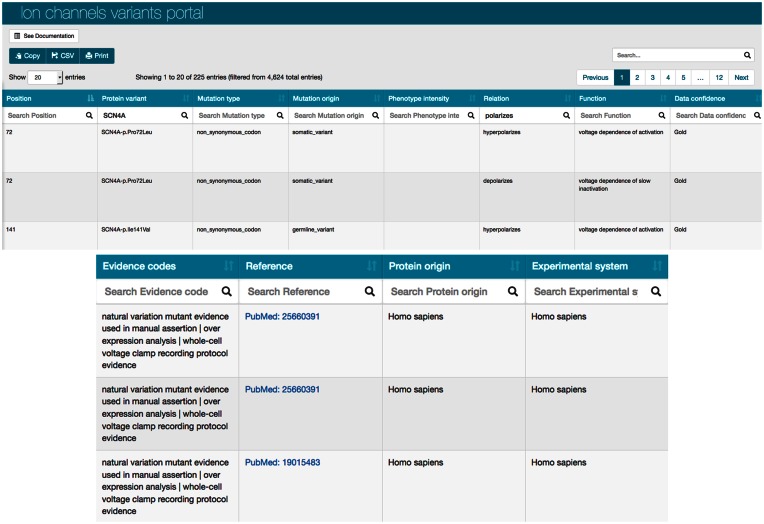
For convenience, our phenotypic data are also available on our new data portals: the neXtProt Cancer variants portal and the Ion channels variants portal, both accessible from the top menu ‘Portals’. These portals contain the variants, their associated molecular, cellular and organism level phenotypes, and the associated experimental evidence for each observation (Figure 2). These data are presented in table format, in which each column is searchable and sortable. The data can also be downloaded in CSV format, copied or printed.
Figure 2.
Excerpt of the Ion channel variants portal.
Improvements to proteomics data representation and peptide unicity checker
In entries having proteomics data, we have implemented a new peptide view that lists all the peptides that match the entry, their position on the sequence, an indication as to whether or not they are unique to that entry (‘proteotypic’), and whether they have been detected in biological samples (‘natural’) or chemically synthesized as reagents for selected reaction monitoring experiments (‘synthetic’).
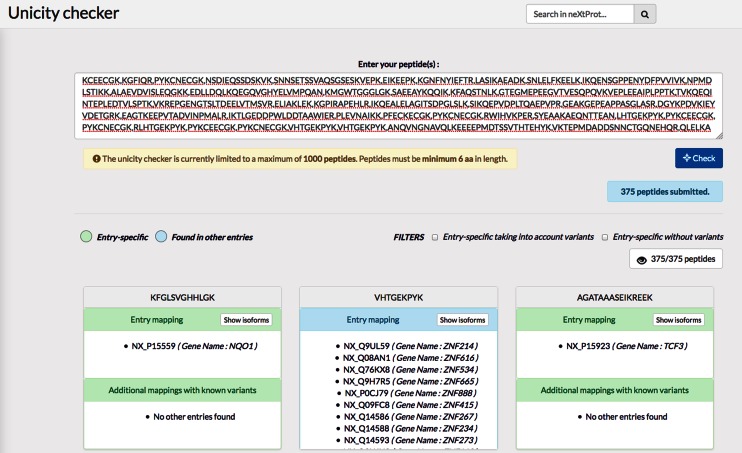
One important need of the proteomics community is to be able to determine whether a peptide is unique to a protein or not. The ‘unicity checker’, which uses the pepx program (available at https://github.com/calipho-sib/pepx), was designed to help scientists determine which peptides are unambiguous and can thus be used to confidently identify protein entries. The ‘additional mappings with known variants’ mode takes into account all the SNPs and disease mutations in neXtProt to increase the search space. Making use of this tool for mass spectrometry data interpretation is now part of the recommendations of the HUPO Human Proteome Project (25). An example of the unicity checker user interface is shown in Figure 3.
Figure 3.
Peptide unicity checker user interface. A list of peptides provided by the user (separated by a space, a comma, a semi-colon or a carriage return) is verified for their unicity in neXtProt sequences by hitting the ‘Check’ button. Users have the option to take into account variants for determining unicity.
Revamped neXtProt website
Our latest release also features a renewed user interface. The neXtProt home page has been re-organized so that information and data are easier to find and use. Navigation in our website has also been re-organized such that the user has access to all the neXtProt content via menus in the page headers and footers. The new header menu offers quick access to tools, data and new documentation describing the neXtProt data model, how to access data (instructions for searching and downloading) and how to use our tools and API. Information regarding neXtProt, the human proteome, the current data release including the best evidence for protein existence of entries broken down by chromosome and how to cite neXtProt is available from the About menu.
Data and software availability
All neXtProt annotations are available as XML and PEFF files (3) on our FTP site (ftp://ftp.nextprot.org/). Note that our XML format has changed to accommodate the new phenotypic data. Changes are documented in a comment at the beginning of the new XSD file (version 2), also on the FTP site. The former XML files are no longer provided due to technical constraints. Annotations can also be accessed through our API at https://api.nextprot.org and our SPARQL endpoint (https://www.nextprot.org/proteins/search?mode=advanced). The Cellosaurus – a knowledge resource on cell lines is available at ftp://ftp.expasy.org/databases/cellosaurus/ (note that the files are no longer on the neXtProt FTP site). The neXtProt content is available under the Creative Commons Attribution-NoDerivs License. Our software is freely available from the GitHub repository (https://github.com/calipho-sib) or biojs (http://www.biojs.io/), as described in our documentation (https://www.nextprot.org/help/technical-corner/).
CONCLUSION
The neXtProt human protein knowledgebase integrates data to provide comprehensive, up-to-date, high quality information organized in such a way so as to provide scientists around the world with a resource that facilitates their research. neXtProt is continually evolving and, in terms of content, the focus will continue to be the incorporation of new variant and proteomics data in the immediate future. Concerning the quality of the data, global checks complementing the spot checks described in our previous paper (1) have been introduced. While content is important, it is just as critical that users be able to view, analyze and export the data in a useful manner. We have thus developed a number of tools, starting with the private lists and queries, and the latest being the peptide unicity checker. We recently started introducing more user interaction features in our web interface so as to improve usability. In the new Phenotype view, the user can select the content that should be displayed both in the Phenotype and Variant sections, thereby focusing exclusively on the data of interest. Another new feature allowing more flexibility for the user is the possibility to download all or part of the data, in XML, JSON or FASTA format, for the entry currently being viewed by simply clicking on the ‘download’ icon at the top right of each page. While all these developments are undertaken in the hope of improving user access and use of our knowledgebase, we count on feedback from our users to provide a high quality resource.
Acknowledgments
The authors thank the UniProt groups at SIB, EBI and PIR for their dedication in providing up-to-date high-quality annotations for the human proteins in UniProtKB/Swiss-Prot thus providing neXtProt with a solid foundation. The authors thank the PeptideAtlas team for fruitful exchanges regarding proteomics data handling. The authors also thank Google Summer of Code for supporting J.J.L. for the summer 2016.
FUNDING
The neXtProt server is hosted by Vital-IT, the SIB Swiss Institute of Bioinformatics’ Competence Centre in Bioinformatics and Computational Biology; SIB Swiss Institute of Bioinformatics, University of Geneva; Recherche Suisse Contre le Cancer (RSCC) [KFS-3297-08-2013 to A.B.]; Swiss National Science Fund [CR33I3_156233 to A.B.]. Funding for open access charge: SIB Swiss Institute of Bioinformatics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gaudet P., Michel P.A., Zahn-Zabal M., Cusin I., Duek P.D., Evalet O., Gateau A., Gleizes A., Pereira M., Teixeira D., et al. The neXtProt knowledgebase on human proteins: current status. Nucleic Acids Res. 2015;43:D764–D770. doi: 10.1093/nar/gku1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaudet P., Argoud-Puy G., Cusin I., Duek P., Evalet O., Gateau A., Gleizes A., Pereira M., Zahn-Zabal M., Zwahlen C., et al. neXtProt: organizing protein knowledge in the context of human proteome projects. J. Proteome Res. 2013;12:293–298. doi: 10.1021/pr300830v. [DOI] [PubMed] [Google Scholar]
- 3.Lane L., Argoud-Puy G., Britan A., Cusin I., Duek P.D., Evalet O., Gateau A., Gaudet P., Gleizes A., Masselot A., et al. neXtProt: a knowledge platform for human proteins. Nucleic Acids Res. 2012;40:D76–D83. doi: 10.1093/nar/gkr1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breuza L., Poux S., Estreicher A., Famiglietti M.L., Magrane M., Tognolli M., Bridge A., Baratin D., Redaschi N. The UniProtKB guide to the human proteome. Database (Oxford) 2016;2016:bav120. doi: 10.1093/database/bav120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosikiewicz M., Comte A., Niknejad A., Robinson-Rechavi M., Bastian F.B. Uncovering hidden duplicated content in public transcriptomics data. Database (Oxford) 2013;2013:bat010. doi: 10.1093/database/bat010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 7.Farrah T., Deutsch E.W., Omenn G.S., Sun Z., Watts J.D., Yamamoto T., Shteynberg D., Harris M.M., Moritz R.L. State of the human proteome in 2013 as viewed through PeptideAtlas: comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven human proteome project. J. Proteome Res. 2014;13:60–75. doi: 10.1021/pr4010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kusebauch U., Campbell D.S., Deutsch E.W., Chu C.S., Spicer D.A., Brusniak M.Y., Slagel J., Sun Z., Stevens J., Grimes B., et al. Human SRMAtlas: A resource of targeted assays to quantify the complete human proteome. Cell. 2016;166:766–778. doi: 10.1016/j.cell.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J., O'Donovan C. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:D1057–D1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatusova T. Update on genomic databases and resources at the national center for biotechnology information. Methods Mol. Biol. 2016;1415:3–30. doi: 10.1007/978-1-4939-3572-7_1. [DOI] [PubMed] [Google Scholar]
- 11.Aken B.L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez Banet J., Billis K., Garcia Giron C., Hourlier T., et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebel U., Starkuviene V., Erfle H., Simpson J.C., Poustka A., Wiemann S., Pepperkok R. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 2003;554:394–398. doi: 10.1016/s0014-5793(03)01197-9. [DOI] [PubMed] [Google Scholar]
- 14.Simpson J.C., Wellenreuther R., Poustka A., Pepperkok R., Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigal A., Danon T., Cohen A., Milo R., Geva-Zatorsky N., Lustig G., Liron Y., Alon U., Perzov N. Generation of a fluorescently labeled endogenous protein library in living human cells. Nat. Protoc. 2007;2:1515–1527. doi: 10.1038/nprot.2007.197. [DOI] [PubMed] [Google Scholar]
- 16.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deutsch E.W., Sun Z., Campbell D., Kusebauch U., Chu C.S., Mendoza L., Shteynberg D., Omenn G.S., Moritz R.L. State of the human proteome in 2014/2015 as viewed through peptideatlas: enhancing accuracy and coverage through the AtlasProphet. J. Proteome Res. 2015;14:3461–3473. doi: 10.1021/acs.jproteome.5b00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Wang J., Ding M., Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 19.Sun G., Jiang M., Zhou T., Guo Y., Cui Y., Guo X., Sha J. Insights into the lysine acetylproteome of human sperm. J. Proteomics. 2014;109:199–211. doi: 10.1016/j.jprot.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 20.The Gene Ontology Consortium Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith C.L., Eppig J.T. The Mammalian Phenotype Ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm. Genome. 2012;23:653–668. doi: 10.1007/s00335-012-9421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinard V., Britan A., Rougier J.S., Bairoch A., Abriel H., Gaudet P. ICEPO: the ion channel electrophysiology ontology. Database (Oxford) 2016;2016:baw017. doi: 10.1093/database/baw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chibucos M.C., Mungall C.J., Balakrishnan R., Christie K.R., Huntley R.P., White O., Blake J.A., Lewis S.E., Giglio M. Standardized description of scientific evidence using the evidence ontology (ECO) Database (Oxford) 2014;2014:bau075. doi: 10.1093/database/bau075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deutsch E.W., Overall C.M., Van Eyk J.E., Baker M.S., Paik Y.K., Weintraub S.T., Lane L., Martens L., Vandenbrouck Y., Kusebauch U., et al. Human proteome project mass spectrometry data interpretation guidelines 2.1. J. Proteome Res. 2016;15:3961–3970. doi: 10.1021/acs.jproteome.6b00392. [DOI] [PMC free article] [PubMed] [Google Scholar]