Abstract
All cellular life contains an extensive array of membrane transport proteins. The vast majority of these transporters have not been experimentally characterized. We have developed a bioinformatic pipeline to identify and annotate complete sets of transporters in any sequenced genome. This pipeline is now fully automated enabling it to better keep pace with the accelerating rate of genome sequencing. This manuscript describes TransportDB 2.0 (http://www.membranetransport.org/transportDB2/), a completely updated version of TransportDB, which provides access to the large volumes of data generated by our automated transporter annotation pipeline. The TransportDB 2.0 web portal has been rebuilt to utilize contemporary JavaScript libraries, providing a highly interactive interface to the annotation information, and incorporates analysis tools that enable users to query the database on a number of levels. For example, TransportDB 2.0 includes tools that allow users to select annotated genomes of interest from the thousands of species held in the database and compare their complete transporter complements.
INTRODUCTION
All living cells have proteins in their cell membrane(s), generally referred to as transport proteins, that play crucial roles in fundamental processes such as the uptake of nutrients, the efflux of toxic and other compounds and in ion homeostasis. Transport proteins can be simple channels or pores created in the membrane, that facilitate diffusions of compounds down their concentration gradient, or active transporters that utilize either ATP and PEP hydrolysis, or chemiosmotic energy in the form of an electrochemical proton, ion or solute gradient, to drive the movement of solutes against their concentration gradient (1,2).
Genomic DNA sequencing efforts are generating data on predicted genes many orders of magnitude faster than laboratory experimentalists can investigate. This has resulted in an increased requirement for high throughput and high quality in silico bioinformatic annotation that is not accommodated by basic genome annotation pipelines. For example, thousands of membrane transport proteins have been experimentally characterized over the course of the last few decades (3), but we now have on the order of millions of predicted transporters. Furthermore, the number and type of transport systems varies widely between genomes (4). However, experimentally characterized transporters can be utilized as a reference set from which the functions of the putative transporters can be computationally inferred based on primary sequence identity, predicted secondary structural homology and topology.
TransportDB is a MySQL database with the objective of providing annotations for predicted transporters across sequenced genomes. In 2004, TransportDB was initially released with membrane transport analysis conducted on 121 genomes (5), and subsequently updated in 2007 (3) to 248 genomes. The analyses were conducted by a semi-automated bioinformatic pipeline with additional manual curation. Subsequently a further 117 genomes were analyzed and the results included in TransportDB, facilitating large-scale analysis of membrane transporter distribution (3,4). There are tens of thousands of complete and/or draft genome sequences in the major public databases and these numbers continue to increase at an accelerating rate (6,7). The semi-automatic bioinformatic characterisation, combined with a degree of manual curation, utilized by the original TransportDB, does not scale well and is inadequate at keeping pace with the rapid rate of sequence data generation.
To the best of our knowledge there are no other databases that currently describe transporters amongst large numbers of sequenced genomes. Other transporter tools or databases that exist include TransportTP, an annotation pipeline optimized for eukaryotic organisms, that uses homology modeling approaches followed by machine learning methods, which requires a model organism to be selected to refine the predictions (8). There are some databases that focus on only one type of transporter (e.g. the Ligand-Gated Ion Channel database; (9)) or include the transporters only of a single model organism (e.g. Human Transporter Database, (10)). The Transporter Classification Database, TCDB (11), is a very well developed resource for classification of transporters, whose main focus is to classify model proteins into families (analogous to the enzyme commission system) but it does not list annotations for whole organisms.
In this paper, we present the latest iteration of the database, TransportDB 2.0, which now utilizes an augmented pipeline with completely automated transporter annotation, and hence is able to better keep pace with the rate of data acquisition. We have populated TransportDB 2.0 with the membrane transporter annotation for over 2700 closed genomes, including the representative sequences available from NCBI's RefSeq database, and this is being added to on an ongoing basis. Furthermore, we have modernized the web portal for TransportDB 2.0, to better visualize data generated from the increased numbers of genomes now available, to expedite the future inclusion of more interactive and engaging ways of viewing the data presented, and to facilitate a range of comparative analyses.
Data sources
The transporter annotation pipeline uses data sources including NCBI's RefSeq (ftp://ftp.ncbi.nih.gov/genomes/refseq/bacteria) (12) and COG (13) databases, the Transporter Classification Database (TCDB; www.tcdb.org) (11) as well as selected HMMs for transporter protein families from the TIGRfam and Pfam databases (14,15). An additional important datasource is the manually curated set of over 100 000 membrane transport proteins from the original TransportDB database. RefSeq is used as the source for new genome sequences to provide a diverse set of annotations for organisms with genome sequences available, while avoiding the potential redundancy of GenBank and other International Nucleotide Sequence Database Collaboration databases. All data sources are updated regularly to ensure that transporter annotations are current.
A suite of Perl and shell scripts extracts protein sequences, general information and taxonomic data from the genome files obtained from RefSeq. The general and taxonomic data then form the basis of an SQL insertion command contained in a file which is uploaded to the TransportDB MySQL database and forms the link to the associated transporters subsequently determined. The protein sequences are used to create a FASTA file for each organism, which is subsequently used to run all relevant searches.
Sequence characterization and annotation
An automated annotation pipeline, TransAAP is employed to generate membrane transporter predictions. In brief, a Perl script splits each FASTA file into smaller files and allocates these for distributed execution on a dedicated high-powered processor (an 18-node Centos Linux cluster utilizing the Perceus cluster software and Torque Portable Batch System) (16,17). The pipeline runs blastp and rpsblast version 2.2.25 (18), hmmscan HMMER3 (19) and TMHMM 2.0c (20) searches for each protein sequence, using the databases listed above where relevant.
Subsequently a combination of Perl, bash and PHP scripts identify and load all potential transport protein candidates into a MySQL database, based on them having potentially significant identity to transporter candidates in the TransportDB, TCDB, Pfam, TIGRfam and COG datasets, as well as putative transmembrane regions as determined using TMHMM. These searches are designed to be inclusive to minimize false negatives in this first stage of the selection process. Subsequently, a combination of PHP and Perl scripts are used to drive a rigorous filtering of the candidates, using empirically derived rules to eliminate false positives.
The above processes result in the generation of three SQL files per genome, encoding: (i) the genome data (taxonomy, size, transporter number), (ii) the identified transporters and (iii) the history of the searches (the nearest database matches to the identified transporters). Each of these SQL files are then uploaded to corresponding tables in the MySQL database on the TransportDB 2.0 server (http://www.membranetransport.org/transportDB2/) and are accessible through the TransportDB 2.0 portal.
Web interface
The previous version of the website serving TransportDB was primarily server-side driven, with PHP scripts providing service of the MySQL database to access to the raw transporter annotation data. In order to move toward providing more engaging and dynamic ways of viewing content, and incorporate new analytical tools which facilitate greater user interaction, we have developed a new website utilizing the more extensible, and client-side based resources provided by the jQuery (21) and D3 (22) JavaScript libraries.
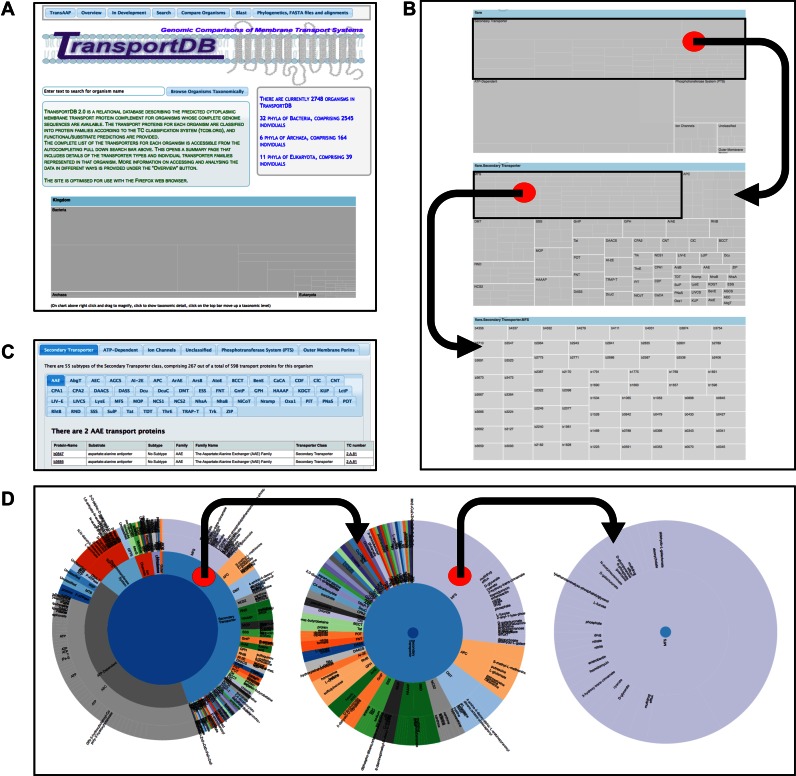
The front page of the new site includes several panels of summary information and mechanisms to access data. The top of the page has links to data or analysis tools (detailed below), as well as an overview describing the current contents of the database, including the number of genomes that have been analyzed from the broad taxonomic divisions (bacteria, archaea, eukaryota). An interactive treemap diagram showing the taxonomic breakdown of organisms represented in the database is shown at the bottom of the front page, a mouse click on elements within each division will zoom the view to a lower taxonomic grouping. Following the ‘Browse Organisms Taxonomically’ button will access complete lists of all organisms in each taxonomic group.
The front page also presents the user with an autocompleting pull-down encompassing all members of the database. Selection of an organism presents the user with the transporter page (Figure 1), which illustrates the transporter annotation for the organism. This is presented visually in the form of a zoomable sunburst graphic that shows the distribution of the transporters by their predicted substrate specificity. A zoomable treemap diagram shows the distribution of transporters in terms of individual proteins, clicking on this permits the user to ‘drill-down’ to the individual representatives of each annotated transporter class.
Figure 1.
TransportDB 2.0 home page and core dynamic elements of the organism pages facilitating interactive data access. (A) Screenshot of the TransportDB 2.0 web portal home page. The home page provides an overview of the database and the current database content in both text format and as an interactive zoomable treemap image at the bottom of the page, which operates in the same manner as the organism treemaps (see B). The organism pages can be accessed by typing into the autocompleting search bar, or by following the ‘Browse Organisms Taxonomically’ button. Selecting an organism will open the organism page, which provides a textual overview of the transporter content of that organism, as well as three alternative avenues to view the transporter annotation data for that organism (panels B–D). (B) A zoomable treemap is present in the top right corner of the organism page. At the top level (top panel) the treemap shows transporter classes, clicking in the box for any class (e.g. Secondary Transporter) shows the transporter families or superfamilies within that class (middle panel), and subsequent clicking on a transporter family (e.g. MFS) shows the constituent members of that family (bottom panel). Clicking on any transporter locus tag will open up the annotation evidence for that transporter. (C) The middle section of the organism pages contain nested tabbed lists of all transport proteins within each family and their accompanying details. (D) The lower section of the page shows a zoomable sunburst diagram depicting the distribution of transporter classes in the inner ring, and transporter families in the middle ring, broken down by substrate specificity at the outer ring. As with the treemap zooming in is achieved by clicking on the desired region. Zooming out is achieved by clicking in the centre of the circle.
The visual information is supplemented by lists of putative transporter proteins embedded in nested tabs, and a summary box under the autocomplete search box (Figure 1). These provide information on the total number of transporters, the size of the genome and the total number of proteins encoded by each organism. Furthermore, each protein family is hyperlinked to the corresponding TCDB family (1). The complete list of transporters annotated for the organism can be downloaded in a CSV format, accessible via the ‘Print CSV of Transporters’ button.
Rather than view all putative transport proteins belonging to a particular organism, users can conduct a detailed search of the database by selecting the ‘Search’ button at the top of the front page. Here users are presented with the capacity to search by transporter type, transporter family, transporter protein or substrate. The search will return a list of proteins meeting the search criteria. Clicking on the protein names in the list will result in a pop-up page showing details of the transporter, including its amino acid sequence and its TransAAP transporter annotation evidence.
To compare the transporter content of two or more organisms in the database, users can select the ‘Compare Organisms’ button on the front page. This will present a list of all organisms in the database and allow users to select their organisms using a checkbox to run the comparison. The comparison will return a table with one column per organism selected to list the number of organisms in each transporter family.
The ‘Blast’ button links to a blast query page set up using the SequenceServer 1.0.8 (Priyam et al., 2015. BioRxiv: http://dx.doi.org/10.1101/033142). Users can submit a DNA or amino acid sequence query sequence to query against all protein sequences in TransportDB, using Blastx or Blastp, respectively. Following the ‘Phylogenetics, FASTA files and alignments’ button will facilitate access to files of information for each transport protein family or superfamily, including fasta formatted files containing all protein sequences, multiple sequence alignment files of all sequences within a family, as well as, phylogenetic trees showing the relationships of all sequences within each family, using the software tool Archaeopteryx (23).
Contents
TransportDB 2.0 now serves the membrane transporter annotation of 2777 complete genomes, comprising 2557 eubacteria, 164 archaea and 56 eukaryota, and is currently available at http://www.membranetransport.org/transportDB2. The database currently contains 860 252 individual transporters, classified into 168 distinct families of transporters. In terms of each class of transporter, the database currently includes 457 183 ATP-Dependent transport proteins, 32 952 phosphotransferase system, 318 233 secondary transporters, 29 874 ion channels, 3273 outer membrane porins and 18 731 unclassified transporter proteins.
In addition to the data currently listed in TransportDB 2.0, we provide a free transporter annotation service to the academic community, via the ‘TransAAP’ button on the TransportDB 2.0 front page. Users can create their own user login and upload the amino acid sequence file(s) for their organism(s), for annotation by TransAAP. The content generated will be accessible to the user via their unique login and will not be made accessible to the public until permission is given by requesting groups, or the data is published.
CONCLUSIONS
The analysis process implemented in TransportDB 2.0 is now fully automated and scalable, allowing for more rapid processing of annotated genomes, and facilitating future developments such as the outsourcing of CPU intensive parts of the processing to external clustered or cloud based resources. The updated website allows an increased level of user interaction with the database, as well as providing a basis for a readily extensible platform for addition of features.
Future development plans include expanding TransportDB 2.0 to include a greater number of the available complete bacterial, archaeal and eukaryotic genomes. We are also developing a variant of the pipeline to work effectively on short reads from metagenomic datasets, which will allow the subsequent development of MetaTransportDB. In terms of the web portal, a short term goal is the addition of more comparative visualisation tools to allow comparison of transporter distribution, substrate specificity, energization and phylogeny between individual organisms and larger taxonomic groups.
Acknowledgments
Thanks to Mr Brendan O'Dea, for technical assistance. Thanks also to Dr Qinghu Ren for his work on the previous iterations of TransportDB. Thanks to Mr Karl Elbourne for some gifs in a jiffy!
FUNDING
Macquarie University Research Development Grant [9201401563 to L.D.H.E. and K.A.H.]. Australian Research Council Fellowship Grant [DE150100009 to S.G.T.]
Conflict of interest statement. None declared.
REFERENCES
- 1.Busch W., Saier M.H. The IUBMB-endorsed transporter classification system. Mol. Biotechnol. 2004;27:253–262. doi: 10.1385/mb:27:3:253. [DOI] [PubMed] [Google Scholar]
- 2.Saier M.H. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren Q., Chen K., Paulsen I.T. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren Q., Paulsen I.T. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren Q., Kang K.H., Paulsen I.T. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 2004;32:D284–D288. doi: 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagani I., Liolios K., Jansson J., Chen I.-M.A., Smirnova T., Nosrat B., Markowitz V.M., Kyrpides N.C. The genomes OnLine database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loman N.J., Constantinidou C., Chan J.Z.M., Halachev M., Sergeant M., Penn C.W., Robinson E.R., Pallen M.J. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat. Rev. Microbiol. 2012;10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Benedito V.A., Udvardi M.K., Zhao P.X. TransportTP: A two-phase classification approach for membrane transporter prediction and characterization. BMC Bioinformatics. 2009;10:418. doi: 10.1186/1471-2105-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donizelli M., Djite M.-A., Le Novère N. LGICdb: a manually curated sequence database after the genomes. Nucleic Acids Res. 2006;34:D267–D269. doi: 10.1093/nar/gkj104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye A.Y., Liu Q.-R., Li C.-Y., Zhao M., Qu H. Human transporter database: comprehensive knowledge and discovery tools in the human transporter genes. PLoS ONE. 2014;9:e88883. doi: 10.1371/journal.pone.0088883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saier M.H., Reddy V.S., Tsu B.V., Ahmed M.S., Li C., Moreno-Hagelsieb G. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft D.H., Selengut J.D., Richter R.A., Harkins D., Basu M.K., Beck E. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res. 2013;41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichev K., Stork S., Keller R., Fernández E. MPI support on the grid. Comput. Informatics. 2012;27:213–222. [Google Scholar]
- 17.Kulkarni A., Lumsdaine A. Stateless clustering using Oscar and Perceus. High Performance Computing Systems and Applications, 2008. HPCS 2008. 22nd International Symposium on. 2008:26–32. [Google Scholar]
- 18.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 21.Chaffer J., Swedberg K. JQuery reference guide: a comprehensive exploration of the popular ... - Jonathan Chaffer, Karl Swedberg - Google Books. 2010.
- 22.Bostock M., Ogievetsky V., Heer J. D3: Data-Driven Documents. IEEE Trans. Visual. Comput. Graph. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 23.Han M.V., Zmasek C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]