Abstract
We present an update of the Eukaryotic Promoter Database EPD (http://epd.vital-it.ch), more specifically on the EPDnew division, which contains comprehensive organisms-specific transcription start site (TSS) collections automatically derived from next generation sequencing (NGS) data. Thanks to the abundant release of new high-throughput transcript mapping data (CAGE, TSS-seq, GRO-cap) the database could be extended to plant and fungal species. We further report on the expansion of the mass genome annotation (MGA) repository containing promoter-relevant chromatin profiling data and on improvements for the EPD entry viewers. Finally, we present a new data access tool, ChIP-Extract, which enables computational biologists to extract diverse types of promoter-associated data in numerical table formats that are readily imported into statistical analysis platforms such as R.
INTRODUCTION
EPD is an old promoter resource first published as a table in a journal article (1) and shortly afterwards distributed in machine-readable form (first on magnetic tapes then via the internet). Promoters are conceptually and operationally defined as transcription start sites or initiation regions. EPD was initially a manually compiled and curated database, strictly relying on critical assessment of experimental data published in journal articles. From the beginning, it was a sequence annotation resource not a sequence collection. The representative TSS of a promoter was defined by an accession number and a sequence position in an EMBL Nucleotide Sequence Library entry. A detailed description of the scope, contents, format and maintenance procedures of the old, manually compiled part of EPD can be found in (2).
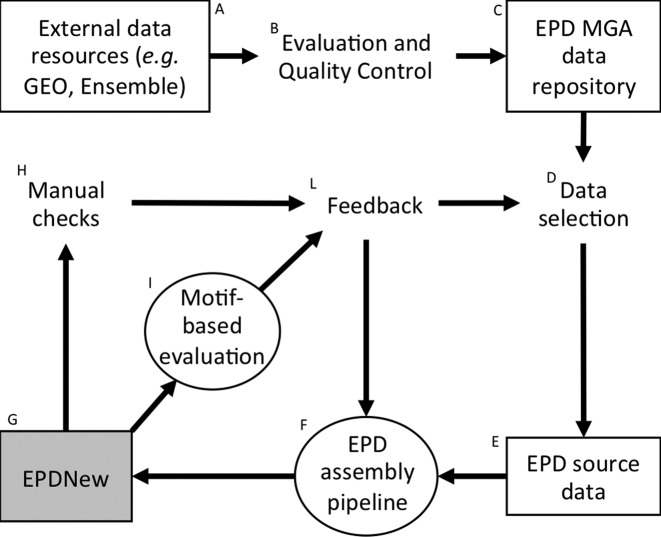
The advent of ultra-high-throughput protocols for genome-wide TSS mapping forced us to completely revise our data acquisition and curation procedures. The result of this major redesign is EPDnew, a computationally generated database derived from electronically distributed primary data. EPD thus now consists of two parts: (i) the old, manually curated part containing promoters from more than 100 different species all contained in a single file and (ii) EPDnew, which consists of multiple files, each containing a comprehensive TSS collection for an important eukaryotic model organisms. These modules are independent entities conforming to minimal data representation standards. For instance, each model organism has its own entry viewer displaying different types of promoter-associated genomic features and hyperlinking to different external data resources. The design principles of EPDnew were already explained in (3). Here, we present only a short summary in form of a flowchart shown in Figure 1. The development, generation and quality control of an EPDnew module is shortly explained in the accompanying Figure legend.
Figure 1.
Schematic representation of the EPDnew development and production pipeline. (A) Download of authoritative gene catalogs and primary TSS mapping data from public databases, data repositories and consortium websites. (B) Quality control (QC) of incoming data (e.g. read mapping efficiency, contaminations, etc.). (C) Data passing QC are reformatted and incorporated into the MGA repository. (D) Selection of a subset of TSS mapping experiments for generating a new organism-specific TSS collection. (E) Input data for a new module of EPDnew. (F) Organism-specific automatic database assembly pipeline tailored to the input data, see (3) for a detailed description of the human EPDnew assembly pipeline. (G) Preliminary or final TSS collection (H) Manual sanity checks of individual randomly selected promoter entries using the corresponding entry viewer, see Figure 2D for an example of an entry view. (I) Automatic quality evaluation of the TSS collections as a whole by motif enrichment tests, see Figure 1A for an example and ref (22) for an explanation of the method. (L) Feedback is collected from quality evaluation steps H and I. This may lead to the exclusion, replacement or addition of source data sets or modifications (e.g. program parameter fine-tuning) of the computational database generation pipeline. Note that the development of a final, publicly released EPDNnew module typically involves several evaluation-modification cycles.
EPDnew is tightly integrated with two accessory bioinformatics resources, the Signal Search Analysis (SSA) (4) and ChIP-Seq servers (5). The former offers tools for DNA motif-oriented analysis, the latter for exploring and downloading promoter-associated functional genomics data. More about the use of these resources in conjunction with EPDnew can be found in (6). The reason why we keep these tools separate, is because they are useful in many other contexts, for instance for ChIP-seq data analysis.
RECENT DEVELOPMENTS
Extension of EPDnew to plants and fungi
The content of EPDnew has substantially increased over the last two years. In our previous paper (6), we presented promoter collections for five model organisms, all animals (Homo sapiens, Mus musculus, Drosophila melanogaster, Danio rerio and Caenorhabditis elegans), totaling together more than 75 thousand entries. In the meantime, the number of promoters for H. sapiens and D. melanogaster has increased; both databases are approaching complete coverage with >92% of protein coding genes covered by at least one validated promoter (Table 1). In addition, we were able to extend EPDnew to five new organisms: a new insect (Apis mellifera), two plant species (a dicotyledonous, Arabidopsis thaliana and a monocotyledonous, Zea mays) and two fungi (Saccharomyces cerevisiae and Schizosaccharomyces pombe). These five new databases greatly extend EPDnew to non-animal species allowing scientists to perform comparative studies of promoter features and organization (7). The expansion of EPDnew to novel organisms has prompted us to add links to species-specific databases such as TAIR for A. thaliana (8) or PomBase for S. pombe (9). Moreover, in order to facilitate the conversion of promoter lists to different genome assemblies, we recently added a genome coordinate conversion (liftOver) tool (10) to our promoter selection and download pages for all EPDnew databases corresponding to an organism that is supported by the UCSC Genome Browser.
Table 1. Current contents of EPDnew.
| Organism, version | Promoters, genesa | TSS librariesb | Chromatin data MNase–Dnasec | ChIP-seq samples histones–PIC–TFsd |
|---|---|---|---|---|
| H. sapiens (4) | 25 503, 17 785 (95%) | 1088 | 23–998 | 2231–491–3794 |
| M. musculus (2) | 21 239, 17 565 (90%) | 339 | 4–0 | 174–60–384 |
| D. melanogaster (2) | 15 073, 12 603 (92%) | 57 | 6–23 | 29–12–189 |
| D. rerio (1) | 10 728, 10 235 (43%) | 12 | 4–4 | 12–3–1 |
| C. elegans (1) | 7120, 6363 (32%) | 8 | 6–6 | 2–1–3 |
| A. mellifera (1) | 6493, 5712 (53%) | 16 | 0–0 | 0–0 |
| A. thaliana (1) | 10 229, 10 177 (37%) | 1 | 0–0 | 0–0–32 |
| Z. mays (1) | 17 081, 15 828 (59%) | 8 | 0–0 | 8–0–0 |
| S. cerevisiae (2) | 5117, 5110 (88%) | 19 | 1–27 | 0–8–17 |
| S. pombe (1) | 3440, 3438 (67%) | 1 | 8–8 | 6–0–51 |
aIn parenthesis is indicated the percentage of genes coverage.
bCAGE, GRO-cap and TSS-seq samples used to build the relative database.
cMNase-seq and DNase-seq samples that are present in the MGA repository.
dChIP-seq samples for histone marks and variants (such as H3K4me3, H2A.Z, H3), components of the PIC (such as Pol-II, TFIID, TFIIB, TBP, etc.) and Transcription Factors that are present in the MGA repository.
Increased precision of the human promoter collection
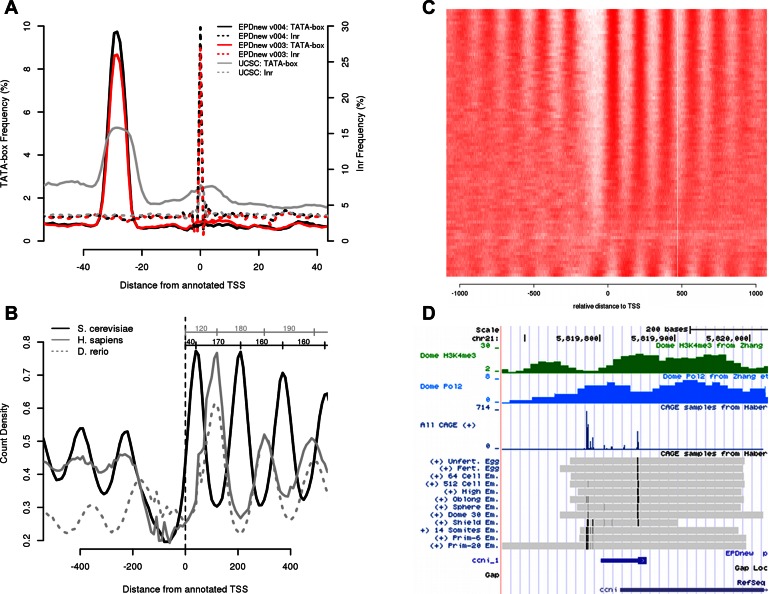
The H. sapiens database is at its fourth release and it has been generated using more than a thousand samples totaling >20 billion reads (data from ENCODE (11) and FANTOM5 (12) consortia). It has the largest collection of data among EPDnew databases and can be taken as a model on how other EPDnew databases will evolve in the near future. Although the number of samples used in this release is more than six times the previous, the database is reaching saturation (coverage of 95%) and as a consequence the increase in promoter numbers and gene coverage is not as significant (25 503 promoters for v004, 23 360 for v003). In this case, the addition of many more samples did not lead to the finding of many new promoters but to an overall increase in TSS mapping precision. This can be seen in the positional distributions of core promoter elements, which are expected to be found at fixed distance from the TSS (Figure 2A). The distribution of both the TATA-box and the Inr motifs within the different promoter collections show an increased frequency at the expected positions for the newer version compared to the other, indicating an increased quality for the latest version. We can predict that the M. musculus and D. melanogaster databases will soon follow the same trend. As their coverage surpasses the 90% limit, the addition of new samples will not lead to more promoters validated but to better estimates of TSS positions for existing promoters. Note that the curves shown in Figure 2A were generated with the OProf tool from the SSA server, which is directly accessible from the EPD web site. Detailed instructions how to reproduce the results are given in Supplemental Data.
Figure 2.
EPDnew analysis and tools. (Instructions how to generate these figures via the EPD web server are given in Supplementary Data.) (A) TATA-box (continuous lines) and Initiatior (Inr, dotted lines) occurrence profiles in three H. sapiens promoters databases. This picture has been obtained with the use of OProf from the SSA program package for two EPDnew versions (3 and 4) and from a list gene starts from the UCSC Gene list, which was used as input for the generation of the EPDnew collections. (B) Distribution of nucleosomes around S. cerevisiae, H. sapiens and D. rerio promoters. The Figure is based on MNase-seq data from (13–15) and has been made with the ChIP-Cor tool from the ChIP-Seq server (5). The MNase-seq data are stored in the MGA repository and are directly accessible via a pull-down menu from the ChIP-Cor input form. This comparative analysis shows the differences in the position of the N+1 (+40 from the TSS for S. cerevisiae and +120 for H. sapiens and D. rerio), distance between two consecutive nucleosomes (+160 in S. cerevisiae and +180 in H. sapiens and D. rerio) and length of the nucleosome-free region for the three organisms. (C) An example of ChIP-Extract output to study nucleosome maps around S. cerevisiae promoters. Each row in the matrix represents a promoter whereas each column the counts of MNase-seq reads found at a specific distance from the TSS. In this example the ordering option in ChIP-Extract has been turned on, which orders rows according to their similarity with the average signal (shown in Figure 2B). This simple procedure shows that some yeast promoters do not have the expected chromatin organization. Data is from (15). (D) An example of D. rerio EPD Hub visualized at the UCSC Genome Browser for the promoter of the ccni gene. The single experiment CAGE tracks shows the promoter shifting from maternally induced TSS (top lines) to zygotic specific TSS (bottom lines) (21). The blue icon near the bottom shows the TSS assignment of the corresponding promoter entry in EPDnew. Note that the two narrow TSS clusters are represented by only one promoter entry since they are too close to each other. The minimum distance requirement for two separate alternative promoters in EPDnew is 100 bp. In such cases, the EPDviewer provides essential information to users interested in the very details of the transcription initiation patterns.
New data in the MGA repository
As usual, the source data from which the current versions of EPDnew were derived is available in standardized format in the MGA repository (3), the back-end data archive used by EPD and the other tools developed by our group. This repository is not restricted to TSS-related data only (such as CAGE, GRO-cap, etc.) but can potentially contain any data set that can be represented as single coordinates in the genome. Examples are genome annotations (TSS, CDS, Intron-exon boundaries, transcripts ends, etc.), ChIP-seq samples (transcription factors, histones marks, etc.), MNase-seq samples, SNPs and conservation scores. Currently, it contains >11 000 samples. The recent addition of samples related to chromatin structure and promoter activity, such as ChIP-seq experiments on histone marks, Pol-II and components of the pre-initiation complex (PIC), gives substantial value to EPD as well (Table 1), as all these samples are accessible by the EPD accessory data analysis tools and can be used to study promoter function in greater details. One example of the use of MGA samples combined with EPDnew is shown in Figure 2B. It involves public MNase-seq data from human, zebra fish and yeast (13–15) and addresses the question whether the canonical nucleosome organization of promoters differs between eukaryotic species as has been reported previously (16,17). The most striking difference revealed by this analysis is the position of the first nucleosome downstream of the TSS: in vertebrate it is centered at about pos. +120 and does not cover the TSS whereas in yeast it occurs at +40 and thus includes the TSS. These results were generated with the ChIP-Cor tool from the ChIP-Seq server. ChIP-Cor computes the distribution of a chromatin feature (here MNase-seq reads) relative to a set of genomic positions (here TSSs). The analysis can be reproduced with a few mouse clicks starting from the EPDnew home page (see Supplementary Data).
ChIP-extract: a new tool to download promoter data in numerical table format
Recently we added a new tool called ChIP-Extract to the ChIP-Seq resource. ChIP-Extract enables computational biologists to extract promoter relevant data from the MGA repository in table format for downstream processing with other tools (e.g. R software). The output is a matrix with each row representing a promoter and each column a distance range relative to the TSS. Each cell then contains the number of sequence reads (or any other kind of genomic feature) that are found at a particular distance from the TSS in a particular promoter. In addition to a tab-delimited text file, the ChIP-Extract server returns a graphical representation of the data as a heatmap. Figure 2C shows the distribution of MNase-seq reads around S. cerevisiae promoters. Note that in this picture, the rows have been reordered according to their similarity to the average MNase-seq profile. However, the main purpose of the ChIP-Extract tool is to export the data for analysis with locally installed software tools. An example of such downstream analysis of promoter/MNase-seq data can be found in Figure 4 of (18). There, a probabilistic partitioning algorithm was used for the identification of human promoter subclasses based on nucleosome distribution.
Improvement and reorganization of the EPD viewer
In 2013 we first introduced the EPD viewer for H. sapiens (3) based on a careful selection of the tracks to be visualized in the UCSC Genome Browser. We developed it with the intent to provide a customizable visualization platform to explore promoter-relevant genomic features (experimental, computationally derived, and manually annotated) of individual promoters. As the number of EPDnew databases grew, we reorganized and extended the viewer to all other organisms that were supported by the UCSC Genome Browser. To achieve this, we developed a track hub (19) as a web-accessible directory tree containing the genomic data visualized in the Genome Browser. The hub has a minimal composition of 3 EPD-specific tracks: the combined TSS mapping samples used in the EPD assembly pipeline at single base pair resolution for the plus and minus strands separately and the EPD promoter track. Other computationally derived and annotation tracks are often present such as a gene track, the conservation scores and repetitive element tracks and, when available, a CpG island track. Additionally, other data might be visualized if the corresponding samples are present in the MGA repository such as promoter specific ChIP-seq samples (Pol-II and H3K4me3); enhancer specific markers (H3K4me1); selected CAGE samples from representative cell lines or tissues organized as a track set. Following these lines, the human viewer has been updated with new global CAGE tracks and single CAGE samples for several cell lines (11), the viewers for mouse and D. melanogaster with CAGE samples for different tissues (12,20), and the D. rerio viewer with CAGE samples for early embryonic developmental stages (21). Figure 2D shows an EPD viewer snapshot for D. rerio with chromatin and CAGE tracks displayed for the ccni_1 promoter. This example was adopted from a recent paper (21) reporting that TSS positions of some zebra fish genes shift during early development. As seen in the picture, zygotic transcripts of the ccni_1 initiate about 60 bp upstream of the maternal TSS.
ACCESS
EPD and EPDnew are freely accessible without need for preregistration. Web-based access is provided via the EPD web site at http://epd.vital-it.ch/. Data files can be downloaded via FTP from ftp://ccg.vital-it.ch/.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss Government (to E.P.D.); Swiss National Science Foundation [31003A_125193 to G.A.]. Funding for open access charge: Swiss Government.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bucher P., Trifonov E.N. Compilation and analysis of eukaryotic POL II promoter sequences. Nucleic Acids Res. 1986;14:10009–10026. doi: 10.1093/nar/14.24.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavin Perier R., Junier T., Bucher P. The eukaryotic promoter database EPD. Nucleic Acids Res. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreos R., Ambrosini G., Cavin Perier R., Bucher P. EPD and EPDnew, high-quality promoter resources in the next-generation sequencing era. Nucleic Acids Res. 2013;41:D157–D164. doi: 10.1093/nar/gks1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambrosini G., Praz V., Jagannathan V., Bucher P. Signal search analysis server. Nucleic Acids Res. 2003;31:3618–3620. doi: 10.1093/nar/gkg611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambrosini G., Dreos R., Bucher P. Principles of ChIP-seq data analysis illustrated with examples. Proceedings Iwbbio 2014: International Work-Conference on Bioinformatics and Biomedical Engineering. 2014;1 and 2:682–694. [Google Scholar]
- 6.Dreos R., Ambrosini G., Perier R.C., Bucher P. The eukaryotic promoter database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015;43:D92–D96. doi: 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreos R., Ambrosini G., Bucher P. Influence of rotational nucleosome positioning on Transcription Start Site Selection in Animal Promoters. PLoS Comput. Biol. 2016;12:e1005144. doi: 10.1371/journal.pcbi.1005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood V., Harris M.A., McDowall M.D., Rutherford K., Vaughan B.W., Staines D.M., Aslett M., Lock A., Bahler J., Kersey P.J., et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karolchik D., Hinrichs A.S., Kent W.J. Current Protocols in Bioinformatics / Editoral Board, Andreas D. Baxevanis … [et al.] 2009. The UCSC genome browser. Chapter 1, Unit1.4. [Google Scholar]
- 11.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FANTOM Consortium and the RIKEN PMI and CLST. Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nepal C., Hadzhiev Y., Previti C., Haberle V., Li N., Takahashi H., Suzuki A.M.M., Sheng Y., Abdelhamid R.F., Anand S., et al. Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res. 2013;23:1938–1950. doi: 10.1101/gr.153692.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaffney D.J., McVicker G., Pai A.A., Fondufe-Mittendorf Y.N., Lewellen N., Michelini K., Widom J., Gilad Y., Pritchard J.K. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J., et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozsolak F., Song J.S., Liu X.S., Fisher D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 17.Weiner A., Hughes A., Yassour M., Rando O.J., Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair N.U., Kumar S., Moret B.M.E., Bucher P. Probabilistic partitioning methods to find significant patterns in ChIP-Seq data. Bioinformatics. 2014;30:2406–2413. doi: 10.1093/bioinformatics/btu318. [DOI] [PubMed] [Google Scholar]
- 19.Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D., et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahsan B., Saito T.L., Hashimoto S., Muramatsu K., Tsuda M., Sasaki A., Matsushima K., Aigaki T., Morishita S. MachiBase: a Drosophila melanogaster 5′-end mRNA transcription database. Nucleic Acids Res. 2009;37:D49–D53. doi: 10.1093/nar/gkn694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haberle V., Li N., Hadzhiev Y., Plessy C., Previti C., Nepal C., Gehrig J., Dong X., Akalin A., Suzuki A.M., et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. 2014;507:381–385. doi: 10.1038/nature12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid C.D., Praz V., Delorenzi M., Perier R., Bucher P. The eukaryotic promoter database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]