Abstract
The National Center for Biotechnology Information (NCBI) provides a large suite of online resources for biological information and data, including the GenBank® nucleic acid sequence database and the PubMed database of citations and abstracts for published life science journals. The Entrez system provides search and retrieval operations for most of these data from 37 distinct databases. The E-utilities serve as the programming interface for the Entrez system. Augmenting many of the Web applications are custom implementations of the BLAST program optimized to search specialized data sets. New resources released in the past year include iCn3D, MutaBind, and the Antimicrobial Resistance Gene Reference Database; and resources that were updated in the past year include My Bibliography, SciENcv, the Pathogen Detection Project, Assembly, Genome, the Genome Data Viewer, BLAST and PubChem. All of these resources can be accessed through the NCBI home page at www.ncbi.nlm.nih.gov.
INTRODUCTION
NCBI overview
The National Center for Biotechnology Information (NCBI), a center within the National Library of Medicine at the National Institutes of Health, was created in 1988 to develop information systems for molecular biology. Since the beginning the foundation of these systems has been molecular sequence data, such as the nucleic acid sequence data in GenBank® (1), which NCBI continues to maintain and which continues to receive data through the international collaboration with DDBJ and ENA as well as from the scientific community. Over the years the amount and variety of data that NCBI maintains has expanded enormously, and can be generally divided into six categories: Literature, Health, Genomes, Genes, Proteins and Chemicals (Table 1). Each of these six categories has a corresponding web page that lists the relevant databases and tools, along with links to tutorials and other information. Links to these pages are also provided in Table 1. NCBI also provides a variety of services to support the research enterprise: (i) facilities that allow submission of scientific data and open-access publications, (ii) facilities for downloading large and/or customized datasets, (iii) educational events and materials about NCBI products, (iv) software and services to support an expanding developer community, (v) software tools to analyze and/or display NCBI data and (vi) direct involvement in research in computational biology. These services, along with all other data resources, are available through the NCBI home page at www.ncbi.nlm.nih.gov (2). In most cases, the data underlying these resources and executables for the software described are available for download at ftp.ncbi.nlm.nih.gov.
Table 1. The Entrez databases (as of 3 September 2016).
| Database | Records | Annual growth | Description |
|---|---|---|---|
| Literature | www.ncbi.nlm.nih.gov/home/literature.shtml | ||
| Books | 528 176 | 18.2% | Books and reports |
| PubMed Central | 4 066 155 | 11.9% | Full-text journal articles |
| PubMed | 26 413 966 | 4.7% | Scientific and medical abstracts/citations |
| MeSH | 265 382 | 2.4% | Ontology used for PubMed indexing |
| NLM Catalog | 1 551 801 | 1.4% | Index of NLM collections |
| Health | www.ncbi.nlm.nih.gov/home/health.shtml | ||
| GTR | 48 612 | 52.0% | Genetic testing registry |
| ClinVar | 159 184 | 27.4% | Human variations of clinical significance |
| PubMed Health | 62 991 | 14.0% | Clinical effectiveness, disease and drug reports |
| dbGaP | 223 662 | 7.6% | Genotype/phenotype interaction studies |
| MedGen | 292 341 | 7.1% | Medical genetics literature and links |
| Genomes | www.ncbi.nlm.nih.gov/home/literature.shtml | ||
| SRA | 3 092 408 | 82.2% | High-throughput DNA and RNA sequence read archive |
| Assembly | 90 727 | 52.3% | Genome assembly information |
| BioSample | 5 224 211 | 43.2% | Descriptions of biological source materials |
| dbVar | 6 147 903 | 37.2% | Genome structural variation studies |
| BioProject | 193 972 | 27.4% | Biological projects providing data to NCBI |
| Genome | 16 962 | 25.3% | Genome sequencing projects by organism |
| SNP | 819 309 474 | 16.1% | Short genetic variations |
| Taxonomy | 1 617 350 | 13.3% | Taxonomic classification and nomenclature catalog |
| Nucleotide | 210 148 411 | 5.2% | DNA and RNA sequences |
| Clone | 38 083 613 | 2.0% | Genomic and cDNA clones |
| GSS | 39 614 616 | 0.6% | Genome survey sequences |
| Probe | 32 405 018 | 0.1% | Sequence-based probes and primers |
| Genes | www.ncbi.nlm.nih.gov/home/genes.shtml | ||
| GEO DataSets | 2 008 226 | 22.1% | Functional genomics studies |
| GEO Profiles | 128 414 055 | 18.1% | Gene expression and molecular abundance profiles |
| Gene | 24 351 351 | 13.8% | Collected information about gene loci |
| PopSet | 257 306 | 11.0% | Sequence sets from phylogenetic and population studies |
| EST | 76 257 001 | 0.3% | Expressed sequence tag sequences |
| UniGene | 6 473 284 | 0.0% | Clusters of expressed transcripts |
| HomoloGene | 141 268 | 0.0% | Homologous gene sets for selected organisms |
| Proteins | www.ncbi.nlm.nih.gov/home/proteins.shtml | ||
| Protein | 307 799 547 | 37.7% | Protein sequences |
| Structure | 121 463 | 9.2% | Experimentally-determined biomolecular structures |
| Conserved Domains | 52 411 | 3.5% | Conserved protein domains |
| Protein Clusters | 820 546 | 0.0% | Sequence similarity-based protein clusters |
| Chemicals | www.ncbi.nlm.nih.gov/home/chemicals.shtml | ||
| PubChem Compound | 91 679 397 | 50.9% | Chemical information with structures, information and links |
| PubChem Substance | 223 159 019 | 41.8% | Deposited substance and chemical information |
| BioSystems | 879 994 | 9.3% | Molecular pathways with links to genes, proteins and chemicals |
| PubChem BioAssay | 1 218 668 | 5.6% | Bioactivity screening studies |
This article provides a brief overview of the NCBI Entrez system of databases, followed by a summary of resources that were either introduced or significantly updated in the past year. A more complete discussion of NCBI resources can be found elsewhere (2), and is also available on the home pages of individual databases, on the NCBI Learn page (www.ncbi.nlm.nih.gov/learn/) or in the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/).
The Entrez system
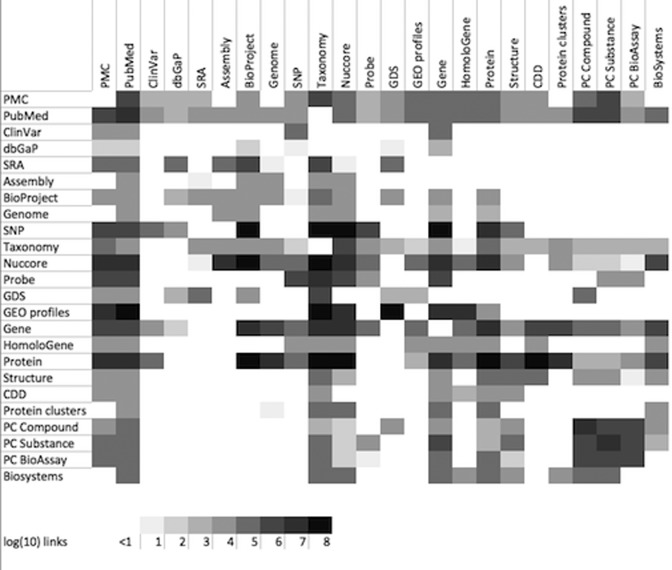
Entrez (2,3) is an integrated database retrieval system that provides access to a diverse set of 37 databases that together contain 2.1 billion records (Table 1). Links to the web portal for each of these databases are provided on the Entrez GQuery page (www.ncbi.nlm.nih.gov/gquery/). Entrez supports text searching using simple Boolean queries, downloading of data in various formats, and linking records between databases based on asserted relationships. In their simplest form, these links may be cross-references between a sequence and the abstract of the paper in which it is reported, or between a protein sequence and either its coding DNA sequence or its 3D-structure. Computationally derived links between neighboring records, such as those based on computed similarities among PubMed abstracts, allow rapid access to groups of related records. A summary of available links for selected databases is shown in Figure 1. The LinkOut service expands the range of links to include external resources, such as organism-specific genome databases. The records retrieved in Entrez can be displayed in many formats and downloaded singly or in batches. An Application Programming Interface for Entrez functions (the E-utilities) is available, and detailed documentation is provided at eutils.ncbi.nlm.nih.gov.
Figure 1.
Graphical Depiction of Selected Entrez Links. Each cell in the matrix is shaded according to the log (base 10) of the number of records in the source database (rows) that have an Entrez link to the destination database (columns). Diagonal cells represent computational links (e.g. pubmed related articles) and off-diagonal cells assert biological relationships (e.g. nuccore to taxonomy). The matrix is not diagonal because an individual record in a source database may have many links to a destination database (e.g. genome to protein).
Data sources and collaborations
NCBI receives data from three sources: direct submissions from external investigators, national and international collaborations or agreements with data providers and research consortia, and internal curation efforts. Details about direct submission processes are available from the NCBI Submit page (www.ncbi.nlm.nih.gov/home/submit.shtml) and from the resource home pages (e.g. the GenBank page, www.ncbi.nlm.nih.gov/genbank/). NCBI staff provide identifiers to submitters for their data generally within 2–5 business days, depending on the destination database and the complexity of the submission. More information about the various collaborations, agreements, and curation efforts are also available through the home pages of the individual resources.
RECENT DEVELOPMENTS
Literature updates
My Bibliography
My Bibliography is a tool that allows researchers to manage and share an online collection of their citations and, where relevant, associate these citations with grants (www.ncbi.nlm.nih.gov/books/NBK53595/). The My Bibliography interface now offers new features that ease the process of adding new citations. The main bibliography page now contains an ‘Add from PubMed’ button that opens a PubMed search window in place so that users can add citations without leaving their bibliography. In addition, an ‘Upload a file’ button allows users to upload a large set of citations in RIS (Research Information Systems) format.
SciENcv
SciENcv is an online system that allows researchers to maintain biosketches that are submitted with grant applications (www.ncbi.nlm.nih.gov/sciencv/). Similar to My Bibliography, SciENcv now allows users to search PubMed from within SciENcv and add citations. This can be done in the ‘Personal Statement’ or ‘Contributions’ section. In addition, SciENcv now supports the Biosketch format used by the Institute of Educational Sciences (IES) at the Department of Education.
Health updates
NCBI pathogen detection
The NCBI Pathogen Detection Project (www.ncbi.nlm.nih.gov/pathogens/) integrates bacterial pathogen genomic sequences originating in food, environmental sources, and patients. It quickly clusters and identifies related sequences to uncover potential food contamination sources, helping public health scientists investigate foodborne disease outbreaks. This pipeline has been in operation since 2013 when FDA, CDC and now USDA agreed to sequence all Listeria isolates collected in the United States and submit the data in real time to NCBI as part of a pilot project. The success of the project, with a reduction in outbreak cluster size along with more clusters being identified, means that all major foodborne bacterial pathogens (Campylobacter, Escherichia coli and Shigella spp., Listeria and Salmonella) are slated to be sequenced in real time in the United States within the next year. A number of additional pathogens have been added since 2013 as part of several other projects including efforts to identify antimicrobial resistance organisms (see a full listing on the Pathogen Detection home page). Once public health agencies submit raw sequencing data for each isolate to the Sequence Read Archive (SRA), NCBI assembles the sequences and clusters the resulting genomes by single-linkage clustering using SNP distances and a threshold of 50 SNPs. Then, within each cluster, a phylogenetic tree is constructed. The set of pathogens can be searched and browsed using the Pathogen Isolates Browser (www.ncbi.nlm.nih.gov/pathogens/isolates/#/search/). This tool allows users to browse isolates by their metadata, such as whether the isolate comes from a clinical or environmental source, its geographical location, or its collection date. For those isolates that are within 50 SNPs of any other isolate, the tool integrates these metadata with links to the SNP tree for each isolate. As of 1 September 2016, this project has processed 88 170 pathogens. This system allows public health officials to quickly determine the genetic relatedness of isolates to aid in traceback investigations of outbreaks, thereby improving the safety of the food supply.
Antimicrobial resistance gene reference database
The NCBI Pathogen Detection team has constructed a reference database of acquired resistance genes and proteins (www.ncbi.nlm.nih.gov/bioproject/PRJNA313047/). This dataset integrates curated resources from several collaborators with novel sequences that are submitted for allele registration. NCBI now hosts the beta lactamase allele registry previously hosted at the Lahey Clinic and is responsible for the assignment of many beta lactamase families (www.ncbi.nlm.nih.gov/pathogens/submit_beta_lactamase/). To provide the most comprehensive dataset of acquired resistance mechanisms, newly released alleles are integrated into the reference dataset along with sequences involved in other antimicrobial resistance mechanisms that have been curated by RefSeq or by several external collaborators. Currently, this dataset does not include resistance arising from point mutations in housekeeping genes or from promoters that increase expression. As of 1 September 2016, the database consisted of 3423 curated records, and that number is expected to grow by several hundred by the end of the year. The NCBI Prokaryotic Genome Annotation Pipeline uses curated hidden Markov models (HMMs) from the antimicrobial reference set to identify antimicrobial resistance genes and proteins to the most specific level possible (exact allele or general protein family) and aids in the annotation and identification of resistant pathogens as part of the NCBI Pathogen Detection pipeline.
Genome updates
Sequence identifiers
As described elsewhere (1), NCBI is in the process of phasing out the practice of assigning GI numbers as identifiers for records in the sequence databases (Nucleotide, EST, GSS, Popset and Protein). This decision was reached in response to several factors, including the rapidly growing size of the sequence databases and the fact that many WGS and TSA records have never had GI numbers. Moving to accession.version as the primary identifier thus provides a consistent method for identifying and versioning all sequence records at NCBI. Over the coming months, new sequence records will be assigned these accession.version identifiers only. It should be emphasized that existing GI numbers will not be removed from the data: records that have both an accession.version and GI identifier will retain both identifiers indefinitely and will be retrievable by both identifiers, although GI numbers will no longer appear in GenBank and FASTA format displays. NCBI is adding support for accession.version identifiers to all services that do not yet support them. As this process develops, NCBI will post announcements on our news and social media platforms, as well as in GenBank release notes.
Assembly
The Assembly database (4) is a collection of genome assemblies from both prokaryotes and eukaryotes, and now also includes data from metagenomes. Assembly records also have richer displays that include, where applicable, common names of organisms, indications that a prokaryotic dataset was derived from type material, and reports of anomalies or other reasons that a given dataset was not included in the Reference Sequences (RefSeq) database. Many eukaryotic assembly records for annotated genomes provide links to the NCBI Genome Data Viewer (see below). Users searching Assembly will now find helpful sidebar filters that can restrict search results by organism group, status, assembly level, and more.
Genome data viewer
The NCBI Genome Data Viewer (GDV) (www.ncbi.nlm.nih.gov/genome/gdv/help/) is a powerful and flexible tool for visualizing data in the context of an annotated eukaryotic genome. GDV employs several ‘widgets’ that offer particular functions, such as viewing a genome's ideogram and selecting an individual chromosome to view, searching for locations or annotations, uploading external data to user-defined tracks, adding or removing NCBI data tracks, viewing and navigating to features of interest, and finally, viewing a given portion of the selected chromosome along with the currently selected set of data tracks. Access to GDV is provided both from Assembly and GEO (Gene Expression Omnibus), and the access point determines the details of the views. Assembly records that support GDV displays have a ‘View the Genome’ link in the right sidebar.
Genome
The Genome database now offers a revised organism browser that provides pre-computed distance trees for all prokaryotic genomes (www.ncbi.nlm.nih.gov/genome/browse/). Moreover, NCBI continues to update the Genome FTP site (ftp.ncbi.nlm.nih.gov/genomes/) by improving the organization of the data and expanding the types of data available. For assemblies with annotation, files are now provided that contain all annotated coding sequence (CDS) and RNA features, along with files containing annotation hashes that allow users to easily monitor annotation changes. To reflect the metagenomes now available in the Assembly database, a new FTP directory contains these data (ftp.ncbi.nlm.nih.gov/genomes/genbank/metagenomes/). These and several additional changes are fully described in the Genome FTP README file (ftp.ncbi.nlm.nih.gov/genomes/all/README.txt).
BLAST updates
In 2016, NCBI released a new home page for BLAST (Basic Local Alignment Search Tool, blast.ncbi.nlm.nih.gov) with a cleaner design that emphasizes commonly used tools and provides easier access to a variety of specialized sequence analysis tools. A ‘BLAST Genomes’ search box allows users to find appropriate genomic BLAST pages and databases for an organism of interest, and clearly labeled links provide access to BLAST software and databases for download, the BLAST API and BLAST instances at cloud providers. Specialized tools, such as Primer-BLAST, the Needleman-Wunsch global aligner, IgBLAST (5), and COBALT (6) (produces protein multiple sequence alignments), now have more prominent and readable links on the page.
The BLAST taxonomy report has also been revised to improve navigation. The overall report page still contains three individual reports, but each report is now collapsible. The Lineage Report shows a taxonomic view of the BLAST results that emphasizes the organism with the strongest match as measured by the BLAST score. The Organism Report groups the BLAST matches by organism and provides links to the individual alignments for each matching sequence. Finally, the Taxonomy Report presents a traditional taxonomic tree classification of the BLAST results, emphasizing those taxonomic nodes that contain the most matches.
The scoring of web BLAST searches has also been updated. Web BLAST searches now score a selenocysteine residue the same as a cysteine residue instead of as an ambiguous residue. The Conserved Domain (CDD) results presented with a blastx search using RPS-tblastn now use composition-based statistics to reduce the number of false positives (7).
Protein updates
iCn3D
In April 2016 NCBI released iCn3D, a WebGL-based application that provides interactive displays of three-dimensional structures of macromolecules and chemicals (www.ncbi.nlm.nih.gov/Structure/icn3d/docs/icn3d_about.html). This new application provides functionality similar to that of Cn3D (8), NCBI's standalone structure viewer; however, iCn3D runs directly in web browsers and so does not require users to install an application. Now both Cn3D and iCn3D are linked from the record pages of the NCBI Molecular Modeling Database (MMDB) as well as from VAST (Vector Alignment Search Tool) result pages. Users can incorporate iCn3D views into their own web pages, and can also download the source code (github.com/ncbi/icn3d).
HistoneDB 2.0
‘HistoneDB 2.0–with variants’ is a new database of histone protein sequences classified by histone types and variants (9). The database contains a manually curated set of histone sequences grouped into 30 different variant subsets with variant-specific annotations. This set is supplemented by an automatically extracted set of histone sequences from NCBI protein sequence databases. The interactive web site supports various searching strategies: browsing of phylogenetic trees; on-demand generation of multiple sequence alignments with feature annotations; classification of histone-like sequences, and browsing of the taxonomic diversity for every histone variant.
MutaBind
MutaBind is a new computational resource that evaluates the effects of sequence variants and disease mutations on protein interactions and calculates the quantitative changes in binding affinity (10). The MutaBind method uses molecular mechanics force fields, statistical potentials, and fast side-chain optimization algorithms. The MutaBind server maps and visualizes mutations on a structural protein complex, calculates the associated changes in binding affinity, estimates the confidence of the prediction, and produces a model of the mutant for download.
Chemical updates
PubChem (11), a resource that focuses on small molecules and their roles as diagnostic and therapeutic agents, introduced several improvements in the past year. PubChem now provides views of Laboratory Chemical Safety Summaries that contain health and safety data for PubChem Compound records with a GHS hazard classification (Globally Harmonized System of Classification and Labeling of Chemicals). These reports are linked next to the ‘Safety Summary’ field near the top of a Compound's record page. The BioAssay record pages were also redesigned in a mobile-friendly style similar to the current Compound and Substance pages. These pages have numerous enhancements including improved data tables and extended download capabilities. Additional details about these and other PubChem developments are available on the PubChem blog (pubchemblog.ncbi.nlm.nih.gov).
FOR FURTHER INFORMATION
The resources described here include documentation, other explanatory material and references to collaborators and data sources on their respective web sites. An alphabetical list of NCBI resources is available from a link above the category list on the left side of the NCBI home page. The NCBI Help Manual and the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/), both available as links in the common page footer, describe the principal NCBI resources in detail. The NCBI Learn page (www.ncbi.nlm.nih.gov/learn/) provides links to documentation, tutorials, webinars, courses, and upcoming conference exhibits. A variety of video tutorials are available on the NCBI YouTube channel that can be accessed through links in the standard NCBI page footer. A user-support staff is available to answer questions at info@ncbi.nlm.nih.gov. Updates on NCBI resources and database enhancements are described on the NCBI News site (www.ncbi.nlm.nih.gov/news/), NCBI social media sites (FaceBook, Twitter, and LinkedIn), the ‘NCBI Insights’ blog, and the several mailing lists and RSS feeds that provide updates on services and databases. Links to these resources are in the NCBI page footer and on the NCBI News site.
APPENDIX
NCBI Resource Coordinators: Richa Agarwala, Tanya Barrett, Jeff Beck, Dennis A. Benson, Colleen Bollin, Evan Bolton, Devon Bourexis, J. Rodney Brister, Stephen H. Bryant, Kathi Canese, Chad Charowhas, Karen Clark, Michael DiCuccio, Ilya Dondoshansky, Michael Feolo, Kathryn Funk, Lewis Y. Geer, Viatcheslav Gorelenkov, Wratko Hlavina, Marilu Hoeppner, Brad Holmes, Mark Johnson, Viatcheslav Khotomlianski, Avi Kimchi, Michael Kimelman, Paul Kitts, William Klimke, Sergey Krasnov, Anatoliy Kuznetsov, Melissa J. Landrum, David Landsman, Jennifer M. Lee, David J Lipman, Zhiyong Lu, Thomas L. Madden, Tom Madej, Aron Marchler-Bauer, Ilene Karsch-Mizrachi, Terence Murphy, Rebecca Orris, James Ostell, Christopher O'Sullivan, Vasuki Palanigobu, Anna R. Panchenko, Lon Phan, Kim D. Pruitt, Kurt Rodarmer, Wendy Rubinstein, Eric W. Sayers, Valerie Schneider, Conrad L Schoch, Gregory D. Schuler, Stephen T. Sherry, Karl Sirotkin, Karanjit Siyan, Douglas Slotta, Alexandra Soboleva, Vladimir Soussov, Grigory Starchenko, Tatiana A. Tatusova, Kamen Todorov, Bart W. Trawick, Denis Vakatov, Yanli Wang, Minghong Ward, W. John Wilbur, Eugene Yaschenko, Kerry Zbicz.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 4.Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A., et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016;44:D73–D80. doi: 10.1093/nar/gkv1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papadopoulos J.S., Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 7.Schaffer A.A., Aravind L., Madden T.L., Shavirin S., Spouge J.L., Wolf Y.I., Koonin E.V., Altschul S.F. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Geer L.Y., Chappey C., Kans J.A., Bryant S.H. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 9.Draizen E.J., Shaytan A.K., Marino-Ramirez L., Talbert P.B., Landsman D., Panchenko A.R. HistoneDB 2.0: a histone database with variants–an integrated resource to explore histones and their variants. Database (Oxford) 2016;2016:baw014. doi: 10.1093/database/baw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M., Simonetti F.L., Goncearenco A., Panchenko A.R. MutaBind estimates and interprets the effects of sequence variants on protein-protein interactions. Nucleic Acids Res. 2016;44:W494–W501. doi: 10.1093/nar/gkw374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]