Abstract
Major advancements have recently been made in mass spectrometry-based proteomics, yielding an increasing number of datasets from various proteomics projects worldwide. In order to facilitate the sharing and reuse of promising datasets, it is important to construct appropriate, high-quality public data repositories. jPOSTrepo (https://repository.jpostdb.org/) has successfully implemented several unique features, including high-speed file uploading, flexible file management and easy-to-use interfaces. This repository has been launched as a public repository containing various proteomic datasets and is available for researchers worldwide. In addition, our repository has joined the ProteomeXchange consortium, which includes the most popular public repositories such as PRIDE in Europe for MS/MS datasets and PASSEL for SRM datasets in the USA. Later MassIVE was introduced in the USA and accepted into the ProteomeXchange, as was our repository in July 2016, providing important datasets from Asia/Oceania. Accordingly, this repository thus contributes to a global alliance to share and store all datasets from a wide variety of proteomics experiments. Thus, the repository is expected to become a major repository, particularly for data collected in the Asia/Oceania region.
INTRODUCTION
Recent improvements in mass spectrometry (MS) have yielded large amounts of proteomics data (1). In order to ensure reliability of the data and the capacity for re-analysis in the future, it is necessary to construct a high-quality public data repository for promising datasets (2), similar to the public data repositories for DNA sequences (3,4) and gene expression profiles (5,6). However, such large-scale repositories can be difficult to maintain. Indeed, some public data repositories for proteomes, e.g. Peptidome (7) and Tranche (8), have been closed owing to issues with the sustainability of database maintenance. The databases PRoteomics IDEntifications (PRIDE) (9), mass spectrometry Interactive Virtual Environment (MassIVE, http://massive.ucsd.edu/) and PeptideAtlas SRM Experiment Libraries (PASSEL) (10) are currently available public data repositories.
The ProteomeXchange (PX) consortium was proposed in 2008 and developed over several years, leading to a publication with substantial use information in 2014 (1). PX provides coordinated submission of MS datasets for proteomics to these three proteomics data repositories. However, data transfer to current submission points of ProteomeXchange, such as PRIDE in Europe and MassIVE in the USA for MS/MS datasets and PASSEL in the USA for SRM datasets via the Internet from Asia/Oceania is usually very slow and highly troublesome. Moreover, the size of MS proteomic dataset files to be deposited is likely to be very large, e.g. >100GB, requiring upload times of more than tens of hours from Japan. Such a slow transmission may be due to latency or delays before the actual data transfer. Generally, uploading huge files to a distant file server via the Internet occurs with latency and makes the net transfer speed very slow. To avoid this problem, web services for accelerating transfer speed, such as the Aspera (http://asperasoft.com/) file transfer protocol, are often used; however, these services are expensive and can have disadvantages with regard to the sustainability of data repository maintenance. Additionally, with these types of software, users are required to install specialized software on their computers, imposing a load on users.
Here, we introduce a new public repository, jPOSTrepo (Japan ProteOme STandard Repository), which is an international standard data repository for proteomes. We successfully developed a high-speed file upload system and user-interface with open-source libraries; all the submission operations can be completed within a web-browser. In addition, the repository provides the functions to facilitate data input for details of wet experimental protocols. The repository is expected to have a dramatic increase in the number of deposits. Furthermore, the repository will contribute to improving the sustainability of the PX consortium by receiving some of the increasing data deposits in worldwide, which is currently performed by other PX repositories located only in Europe and the USA (11). Our repository, like other PX repositories, will contribute to proteome data sharing among worldwide researchers.
DATABASE DESCRIPTION
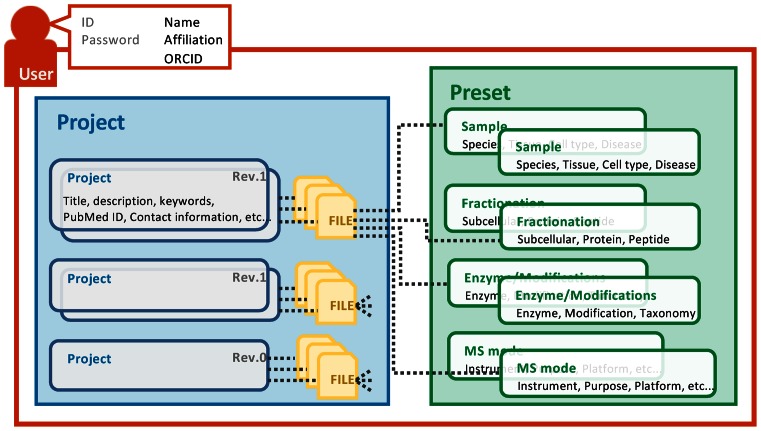
jPOSTrepo is a public data repository for datasets obtained from proteomics experiments. Although users are required to sign up for the repository to upload and manage their datasets, once the depositor makes the dataset public, data are available to be downloaded without requiring a sign up process. Figure 1 shows the management of deposited data in our repository. Generally, researchers use different experimental methods for different experiments, although a single researcher usually uses a limited number of experimental methods. Thus, our repository also manages information related to the experimental procedures as a ‘preset’ and information specific to each experiment as a ‘project’. For the deposit of datasets, users are required to register their experimental details as presets. After the presets are registered, users create a project to deposit datasets and apply the configured presets to proteome data files.
Figure 1.
Diagram of a schematic model of the jPOSTrepo file management system.
Presets for experimental procedures
jPOSTrepo describes proteomics-related biological experimental procedures as four different types of presets: Sample, Fractionation, Enzyme/Modifications and MS mode. Registration of experimental conditions as presets can allow users to easily reuse data when they create other projects using the same experimental conditions. A researcher in one laboratory may use the same cell lines, the same experimental procedures, and the same mass spectrometers for multiple experiments; thus, registration of the experimental procedures as presets could save time when inputting meta-data information.
‘Sample’ preset
The ‘Sample’ preset includes biological sample information, such as species, tissue, cell type, and disease. For the input of such data, the ‘choosing from options’ style-user interface is more preferable than the ‘inputting manually’ style-interface; users can save their inputs and the use of unified terms improves data retrieval. For example, the options of ‘species’ are derived from the NCBI organismal classification (NCBITaxon) (12) ontology. Similarly, ontology/controlled vocabulary-based options are available for almost all items in all four presets, e.g. for the experiment and analysis pipelines of MS, Proteomics Standard Initiative (PSI)-Mass Spectrometry Controlled Vocabulary (CV) (13) is available, and for post-translational modifications (PTMs), the UNIMOD (14) CV is available. When no terms of interest are available in the default term sets, users can search the term from the EMBL-EBI Ontology Lookup Service (15), which is embedded in the input form.
‘Fractionation’ preset
A general shotgun proteome analysis based on liquid chromatography tandem MS (LC-MS/MS) often requires prefractionation of samples at the protein and/or peptide levels. The ‘Fractionation’ preset is a generalized and simplified representation of various methodologies for the prefractionation process, such as the removal of highly expressed proteins by affinity chromatography for isolating low-abundance proteins and the enrichment of specific types of digested peptides, e.g. phosphorylated peptides using chemo-affinity chromatography with metal oxides or metal ions. The contents of this preset are classified into three separation levels: subcellular, protein and peptide. The subcellular level describes mainly subcellular locations of proteins, including whole cells. The protein level describes the prefractionation methods before protease digestion, such as sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoprecipitation. The peptide level describes the peptide prefractionation method, e.g. reversed phase LC at basic pH. The numbers of fractions and replicates are also described.
‘Enzyme/Modifications’ preset
The ‘Enzyme/Modifications’ preset accepts information of parameters specified in the database search. A generic shotgun proteomics workflow contains a digestion step by proteases (or chemicals) to produce peptides. The Enzyme/Modification preset describes the name of the employed enzymes or chemicals. For the enzyme name, combinations of more than one enzyme are also allowed as input. In addition to the enzyme information, this preset addresses supposed PTMs; fixed and variable modifications specified in the database search are required as input. The species name can be described; similarly, the species name specified in database search should be input independently of the description in the ‘Sample’ preset.
‘MS mode’ preset
The ‘MS mode’ preset accepts information regarding the types of MS instruments (mass spectrometers) and the configurations used in the project. For isobaric labelling experiments, such as iTRAQ (16) and Tandem Mass Tag (17), multiple samples labeled by isobaric tags are measured at the same time. Information of such multiplex experiments is also described in this MS mode preset.
Profile consisting of four types of presets
Raw proteomics data files (MS measurements) are linked to a ‘profile,’ i.e. the combination of the four presets. Our repository is designed to reduce user time and load for submission; thus, users can apply a profile to multiple files at one time using a simple drag-and-drop approach on the web browser. The repository accepts both raw mass spectrum files and peak list files generated from spectra, i.e. mzML (18) and Mascot Generic Format (MGF; http://www.matrixscience.com/help/data_file_help.html); database-search result files, i.e. mzIdentML (19) and mzTab (20); commonly used software, such as Mascot (21), X!Tandem (22) and MaxQuant (23) and the settings used in database searches.
File upload
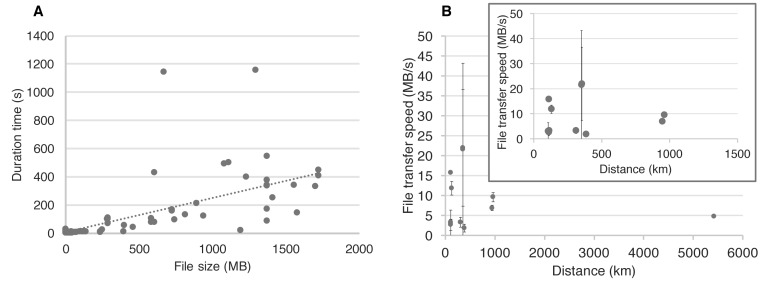
After linkage of the meta-data profile (presets) and deposited data files, users can actually upload the files to the repository. During this upload process, the files are split into fragments of smaller size, called ‘chunks,’ which are subsequently uploaded to the repository in parallel. The latency, i.e. the delay before the actual data transfer, is known to increase during data transfer via the Internet as the length of the data communication path increases. Thus, the data transfer speed between physically distant locations is often extremely slow. The parallel upload of small-sized chunks can improve this latency problem. As shown in Figure 2A, when datasets are uploaded to our repository, a positive correlation is observed between the file size and the transfer duration time; the average transfer speed is quite fast, ∼9MB/s. In addition, the file transfer speed is, in most cases, independent of the distance from where the user deposits the data (Figure 2B). The most distantly located site was more than 5000 km far from the server location, and the file transfer speed was sufficient (∼5MB/s). Thus, the effect of latency would be limited during file upload to the repository, even from more distant locations.
Figure 2.
File transfer speed to jPOSTrepo. (A) Linear relationships between the file size and duration for uploading to the repository. (B) File transfer speed to the repository is independent of the distance between users and the repository server. Even at the most distant location (>5000 km), outside of Japan, where the repository server is established, a high transfer speed of ∼5MB/s was achieved. The inner panel shows magnification of the same figure excluding data for the most distant location. Bars represent standard deviations at each distance.
Partial and complete submission
jPOSTrepo is currently a member of the PX consortium. The PX consortium provides standard criteria of quality for datasets deposited from users. The complete submission recommended by the PX consortium requires meta-data for biological samples, experimental procedures used in the study, and the corresponding relationship between each peptide in the search result files and each peak in the peak list files. The repository performs the validation process for the complete submission. For the complete submission of datasets, a table view of search results is provided, and the corresponding relationships of the mass peaks and the peptides can be visualized. As with the other repositories, our repository functions to make datasets public as a partial submission. The partial submission requires raw data files, search result files, and valid metadata under the standard criteria of the PX consortium for complete submission.
Data publication and download
When the validation process is finished and the deposited datasets are determined to be valid in the context of the PX consortium criteria, users can finish and lock the submission process, yielding both jPOST and PX identifiers. At this stage of the submission of a ‘project’, the dataset deposited to the repository is under an ‘embargo’ as private, and is automatically published at the ‘announcement date’ as public, set by users themselves. During this embargo period, users can issue a specialized URL and password to access the project by anyone who knows them, such as journal editors and reviewers. In addition, users can revise the temporarily locked project according to the comments by reviewers. Revision numbers are provided for the revised data.
The datasets of publicly available projects can be downloaded without any limitations. In addition, public projects can be searched by registered keywords, i.e. ontology terms, CVs and principal investigator names registered as presets and projects.
System implementation
The file upload system in our repository employs an open-source JavaScript library, flow.js (https://github.com/flowjs/). The visualization system of the corresponding relationships of mass peaks and peptides for the complete submission employs an open-source library, Lorikeet (http://uwpr.github.io/Lorikeet/), which is a plugin of the jQuery library (https://jquery.com/) to view MS/MS spectra annotated with fragment ions.
DISCUSSION
We have developed and launched jPOSTrepo to share and store a variety of proteome datasets for researchers worldwide. This repository is now operating in the Asia/Oceania area where no international proteome repository had been established, and accepts MS raw and processed data for proteomics from all over the world as a member of the PX consortium. Users can obtain a global common accession number for their deposited data by both the ‘complete’ submission or the ‘partial’ submission. The repository also stores detailed metadata, such as samples, instruments, analysis software, and settings, as four types of presets with some ontologies and CVs. It also provides an ultra-fast file uploader and user-friendly web interface.
All metadata submitted in our repository, especially all experimental procedures described in the current four types of presets, are expressed with ontologies/vocabularies and are therefore ‘computer-readable.’ When this information is expressed further by some unified framework, such as the Resource Description Framework (RDF) data model, the proteome data could also be linked to a wide variety of other data, such as genomes and transcriptomes, under the ‘linked-open data’ concept. Therefore, our repository could be used to develop a system to enrich RDF expression for proteomic datasets and joint/integrate them with other data resources.
Acknowledgments
The jPOST team would like to thank all the data submitters and collaborators for their contributions and members of the PX consortium for their support. The computational resource was provided in part by SuperComputer System, Institute for Chemical Research, Kyoto University.
FUNDING
Database Integration Coordination Program from National Bioscience Database Center, Japan Science and Technology Agency [15650519]. Funding for open access charge: National Bioscience Database Center, Japan Science and Technology Agency [15650519].
Conflict of interest statement. None declared.
REFERENCES
- 1.Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N., et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prince J.T., Carlson M.W., Wang R., Lu P., Marcotte E.M. The need for a public proteomics repository. Nat. Biotechnol. 2004;22:471–472. doi: 10.1038/nbt0404-471. [DOI] [PubMed] [Google Scholar]
- 3.Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mashima J., Kodama Y., Kosuge T., Fujisawa T., Katayama T., Nagasaki H., Okuda Y., Kaminuma E., Ogasawara O., Okubo K., et al. DNA data bank of Japan (DDBJ) progress report. Nucleic Acids Res. 2016;44:D51–D57. doi: 10.1093/nar/gkv1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T., et al. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji L., Barrett T., Ayanbule O., Troup D.B., Rudnev D., Muertter R.N., Tomashevsky M., Soboleva A., Slotta D.J. NCBI Peptidome: a new repository for mass spectrometry proteomics data. Nucleic Acids Res. 2010;38:D731–D735. doi: 10.1093/nar/gkp1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith B.E., Hill J.A., Gjukich M.A., Andrews P.C. Tranche distributed repository and ProteomeCommons.org. Methods Mol. Biol. 2011;696:123–145. doi: 10.1007/978-1-60761-987-1_8. [DOI] [PubMed] [Google Scholar]
- 9.Vizcaíno J.A., Csordas A., Del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrah T., Deutsch E.W., Kreisberg R., Sun Z., Campbell D.S., Mendoza L., Kusebauch U., Brusniak M.-Y., Hüttenhain R., Schiess R., et al. PASSEL: the PeptideAtlas SRMexperiment library. Proteomics. 2012;12:1170–1175. doi: 10.1002/pmic.201100515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S., et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw936. doi:10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer G., Montecchi-Palazzi L., Ovelleiro D., Jones A.R., Binz P.-A., Deutsch E.W., Chambers M., Kallhardt M., Levander F., Shofstahl J., et al. The HUPO proteomics standards initiative- mass spectrometry controlled vocabulary. Database. 2013;2013:bat009. doi: 10.1093/database/bat009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creasy D.M., Cottrell J.S. Unimod: Protein modifications for mass spectrometry. Proteomics. 2004;4:1534–1536. doi: 10.1002/pmic.200300744. [DOI] [PubMed] [Google Scholar]
- 15.Cote R., Reisinger F., Martens L., Barsnes H., Vizcaino J.A., Hermjakob H. The ontology lookup service: bigger and better. Nucleic Acids Res. 2010;38:W155–W160. doi: 10.1093/nar/gkq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross P.L. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Hamon C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 18.Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W.H., Rompp A., Neumann S., Pizarro A.D., et al. mzML—a Community standard for mass spectrometry data. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.R110.000133. R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones A.R., Eisenacher M., Mayer G., Kohlbacher O., Siepen J., Hubbard S.J., Selley J.N., Searle B.C., Shofstahl J., Seymour S.L., et al. The mzIdentML data standard for mass spectrometry-based proteomics results. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014381. M111.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griss J., Jones A.R., Sachsenberg T., Walzer M., Gatto L., Hartler J., Thallinger G.G., Salek R.M., Steinbeck C., Neuhauser N., et al. The mzTab data exchange format: communicating mass-spectrometry-based proteomics and metabolomics experimental results to a wider audience. Mol. Cell. Proteomics. 2014;13:2765–2775. doi: 10.1074/mcp.O113.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins D.N., Pappin D.J.C., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Craig R., Beavis R.C. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 23.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]