Abstract
The Biological General Repository for Interaction Datasets (BioGRID: https://thebiogrid.org) is an open access database dedicated to the annotation and archival of protein, genetic and chemical interactions for all major model organism species and humans. As of September 2016 (build 3.4.140), the BioGRID contains 1 072 173 genetic and protein interactions, and 38 559 post-translational modifications, as manually annotated from 48 114 publications. This dataset represents interaction records for 66 model organisms and represents a 30% increase compared to the previous 2015 BioGRID update. BioGRID curates the biomedical literature for major model organism species, including humans, with a recent emphasis on central biological processes and specific human diseases. To facilitate network-based approaches to drug discovery, BioGRID now incorporates 27 501 chemical–protein interactions for human drug targets, as drawn from the DrugBank database. A new dynamic interaction network viewer allows the easy navigation and filtering of all genetic and protein interaction data, as well as for bioactive compounds and their established targets. BioGRID data are directly downloadable without restriction in a variety of standardized formats and are freely distributed through partner model organism databases and meta-databases.
INTRODUCTION
Biological interactions, whether functional interactions between genes or physical interactions between proteins and other biomolecules, provide the framework to understand how the cell is structured and controlled. Interaction networks reflect the organization of cellular pathways and are hence essential for the interpretation of genomic, functional genomic, phenotypic and chemical screen data. Since the advent of high-throughput approaches to detect protein (1–3) and genetic interactions (4), the depth of coverage and accuracy of biological interaction networks has continued to improve (5,6), with increased resolution at the level of protein isoforms (7) and metabolites (8,9). Extensive network contexts now provide a basis for the rationalization of perturbations caused by disease-associated mutations (10–12) and have helped deconvolve complex mutational profiles generated by genome-wide association studies (GWAS) and next-generation sequencing-based approaches for analysis of the genome (13), transcriptome, and epigenome (14). The network paradigm thus holds the promise of predictive and precision medicine, as illustrated for example by the synthetic lethal interaction networks between cancer driver mutations and established drug targets (15).
The Biological General Repository for Interaction Datasets (BioGRID: https://thebiogrid.org) was originally implemented in 2003 (16) to provide open access to high-throughput (HTP) interaction datasets (1–3), and subsequently to augment and benchmark HTP data with interactions drawn from focused low-throughput (LTP) studies in the biomedical literature (17). Since its inception as a yeast-specific database (16), BioGRID has grown to cover interactions from 66 different species, including all major model organisms and humans. Correspondingly, the number of annotated interactions in BioGRID has increased from 30 000 protein interactions in the original version to more than one million protein, genetic and chemical interactions in the current release (Figure 1).
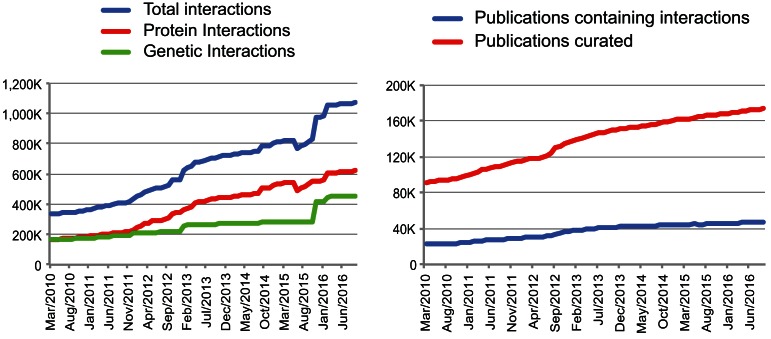
Figure 1.
Increase in data content of BioGRID. Increments in interaction records and source publications reported in BioGRID from March 2010 (release 2.0.62) to September 2016 (release 3.4.140). Left panel shows the increase of annotated protein interactions (red), genetic interactions (green) and total interactions (blue). Right panel shows the number of publications that actually reported protein or genetic interactions (blue) as a function of the total number of publications examined by BioGRID curators (red).
The primary focus of BioGRID is the manual curation of experimentally validated genetic and protein interactions that are reported in peer-reviewed biomedical publications. Text-mining approaches are now routinely used to accelerate and prioritize expert manual curation in BioGRID (18–20). All interactions in BioGRID are annotated by curators according to a structured set of experimental evidence codes. In addition BioGRID captures post-translational modification (PTM) data, such as sites of phosphorylation and ubiquitination, from both LTP and HTP studies. Recently, BioGRID has extended its data content to include the protein and/or genetic interactions of drugs, metabolites and other bioactive small molecules. The current version of BioGRID includes a newly implemented network viewer that allows visualization of all search results in an interactive graphical format. Additional new custom page views also allow the interrogation of PTM sites and chemical interaction data. BioGRID content is updated on a monthly basis and is made freely accessible via the web interface, through downloadable files in standardized formats, and through dissemination by model organism database (MOD) partners (21–26) and other biological resources (27–32).
DATABASE GROWTH AND STATISTICS
Since our 2015 NAR Database report (33), the number of curated interactions housed in BioGRID has increased by 30%. As of September 2016 (version 3.4.140), BioGRID contains 1 072 173 protein and genetic interactions, of which 836 212 are non-redundant interactions. These interactions correspond to 621 639 (470 810 non-redundant) protein interactions and 450 534 (373 762 non-redundant) genetic interactions (Table 1). These data were directly extracted from 47 223 manually annotated peer-reviewed publications, which were identified from the biomedical literature by keyword searches and text-mining approaches. Extensive manual inspection of candidate abstracts and/or full text papers reveals that approximately one in four candidate publications actually contains experimentally documented interaction data, such that many more publications are parsed by curators than are entered into BioGRID as sources of interaction data (Figure 1). All BioGRID interaction records are directly mapped to experimental evidence in the supporting publication, as classified by a structured set of evidence codes that map to the PSI-MI 2.5 standard (34). The BioGRID also currently contains data on 38 559 protein PTMs as curated from 4317 publications. These PTM data are now drawn mainly from high-throughput mass spectrometry studies, which are able to routinely survey many thousands of modification sites for any given PTM type (35). All yeast phosphorylation site data in BioGRID are also currently housed in the PhosphoGRID database (36), but this older database has been essentially subsumed by the new PTM functionalities of BioGRID (see below). BioGRID curation has recently focused on improved coverage for PTMs. For example, a recent themed project on the ubiquitin proteasome system has documented 130 184 sites of ubiquitin modification on proteins encoded by 8681 human genes and 20 019 sites on 2549 yeast proteins, all which will be released as a consolidated dataset by the end of 2016. A new aspect of BioGRID is the coverage of chemical interactions, typically for drug-protein targets and/or drug-gene interactions. BioGRID release 3.3.123 (April 2015) included 27 034 chemical–target interaction records drawn from DrugBank, a database of manually curated drug-target relationships (37). At present, BioGRID contains 2519 unique genes/proteins from 21 organisms that are linked to 4999 total unique chemicals as curated from 8989 publications. 2129 of these interactions are between drugs or other bioactive agents and human genes or proteins.
Table 1. Increase in BioGRID data content since previous update.
| August 2014 (3.2.115) | September 2016 (3.4.140) | ||||||
|---|---|---|---|---|---|---|---|
| Organism | Type | Nodes | Edges | Publications | Nodes | Edges | Publications |
| Arabidopsis thaliana | PI | 7200 | 21 536 | 1414 | 9479 | 41 918 | 2168 |
| GI | 112 | 192 | 66 | 246 | 298 | 125 | |
| Caenorhabditis elegans | PI | 3288 | 6345 | 178 | 3277 | 6341 | 190 |
| GI | 1129 | 2344 | 30 | 1123 | 2330 | 31 | |
| Drosophila melanogaster | PI | 8076 | 37 606 | 416 | 8236 | 38 638 | 454 |
| GI | 1042 | 9980 | 1483 | 1042 | 9979 | 1482 | |
| Escherichia coli | PI | 105 | 102 | 14 | 108 | 109 | 17 |
| GI | 34 | 25 | 11 | 4000 | 166 137 | 15 | |
| Homo sapiens | PI | 18 435 | 237 498 | 23 388 | 20 914 | 365 547 | 25 383 |
| GI | 1364 | 1678 | 273 | 1577 | 1663 | 283 | |
| Mus musculus | PI | 8276 | 22 563 | 3207 | 11 892 | 38 163 | 3529 |
| GI | 259 | 290 | 167 | 275 | 309 | 176 | |
| Saccharomyces cerevisiae | PI | 6410 | 135 690 | 7402 | 6299 | 131 659 | 8074 |
| GI | 5674 | 207 188 | 7257 | 5719 | 212 092 | 7880 | |
| Schizosaccharomyces pombe | PI | 2694 | 11 270 | 1146 | 2946 | 12 817 | 1247 |
| GI | 3158 | 56 745 | 1359 | 3208 | 57 847 | 1459 | |
| Other organisms | ALL | 7999 | 12 367 | 1920 | 9688 | 14 814 | 2250 |
| Total | ALL | 55 528 | 749 912 | 43 149 | 65 031 | 1 072 173 | 47 223 |
Data drawn from monthly release 3.2.115 and 3.4.140 of BioGRID. Nodes refer to genes or proteins, edges refer to interactions. PI, protein (physical) interactions; GI, genetic interactions. All numbers represent total interactions curated.
In 2016, Google Analytics reported that the BioGRID received on average 124 232 page views and 14 444 unique visitors per month, versus 88 080 page views and 12 399 unique visitors per month in 2014. We estimate that these page views correspond to perusal of ∼24 million interactions by BioGRID users in 2016. BioGRID data files were downloaded on average 10 135 times per month in 2016, compared with 9256 downloads per month in 2014. These statistics do not include the widespread dissemination of BioGRID records by various partner databases, which include the MODs Saccharomyces Genome Database (25), PomBase (23), Candida Genome Database (38), WormBase (26), FlyBase (24), TAIR (39), ZFIN (21) and MGD (22) and the meta-database resources NCBI (29), UniProt (28), Pathway Commons (30), BeagleDB (40) and others. In 2016, the BioGRID user base was located primarily in the USA (29%), followed by China (10%), United Kingdom (7%), Germany (6%), Canada (5%), Japan (4%), India (4%), France (4%) and all other countries (31%).
OVERALL CURATION STRATEGY
All interactions in BioGRID must be directly supported by experimental evidence in the source publications as identified by BioGRID curators. All curation activity is controlled by a dedicated Interaction Management System (IMS) that serves as the primary curator interface (33). The IMS is used to build publication lists and standardize all aspects of curation, including controlled vocabularies for experimental evidence, interaction types and gene names. BioGRID interaction annotation is based on Entrez Gene identifiers for genes and proteins. RefSeq protein identifiers are used for the annotation of PTMs, which are typically mapped to RefSeq by mass spectrometry search engines (see the BioGRID WikiPage for further details; URL: https://wiki.thebiogrid.org/doku.php/identifiers). The IMS also tracks all curator contributions for dispute resolution and curation consistency. BioGRID currently contains interaction data for 66 model organisms at varying depths of coverage. BioGRID continues to maintain complete curation of the primary literature for genetic and protein interactions in the model yeasts Saccharomyces cerevisiae (343 751 total interactions, 231 326 non-redundant interactions) and Schizosaccharomyces pombe (70 664 total interactions, 57 699 non-redundant interactions). These datasets are updated on a monthly basis and released for redistribution through the Saccharomyces Genome Database (25) and PomBase (23). Comprehensive curation of protein interactions is also maintained for the model plant Arabidopsis thaliana (39). It is not possible to manually curate the vast and ever-expanding human biomedical literature, which currently exceeds 16 million publications reported in PubMed with an average growth rate of >600 000 papers per year over the past 5 years. Instead, to achieve meaningful depth of coverage in key areas of human biology and disease, BioGRID has established a number of ongoing themed projects centered on cellular functions relevant for central cellular processes and/or major human diseases. Current themed curation projects on particular biological processes include inflammation, chromatin modification, autophagy, the ubiquitin proteasome system, the DNA damage response, phosphorylation-based signaling and stem cell regulators. Curation projects focused on particular diseases include cardiovascular disease and hypertension, brain cancer, and prevalent infectious diseases, such as tuberculosis and HIV.
A recent example of a themed curation project on a central biological process is the conserved autophagy network that targets damaged organelles, cytoplasmic material, and pathogens to the lysosome for degradation. This process is mediated by a core set of 18 autophagy (ATG) proteins that drive membrane formation to mediate both selective and non-selective degradation (41). An additional 116 human genes associated with autophagy were identified through Gene Ontology (GO) annotations and ongoing literature review. This set of 134 genes was used to build a candidate publication list of over 7603 papers for review by BioGRID curators, of which 1277 publications yielded 7888 interactions that were entered into BioGRID. A recent example of a themed curation project with a disease focus is on glioblastoma (GBM) in collaboration with the Stand Up to Cancer (SU2C) team on brain cancer (see www.standup2cancer.ca). GBM has amongst the worst long term survival rates of all human cancers, in part because the brain tumour stem cells (BTSCs) that drive tumour growth readily acquire drug resistance (42). BioGRID curators have coordinated with scientists and clinicians in the SU2C team to identify a core set of 31 genes implicated in GBM, as drawn from cancer genome analyses for genes either mutated or of altered copy number in patient-derived tumour samples (43,44). From this GBM-associated gene list, 2443 papers have been curated to date to yield 8781 interactions. Once completed, this literature-derived GBM interaction network will serve as a resource for the interpretation of genome-scale sequence, transcriptional, epigenetic, proteomic and genetic datasets in GBM, with the goal of identifying new drug targets and drug combinations that are effective against this deadly cancer (42).
Important curation contributions are also made by model organism and other database partners, and are prominently attributed through a ‘curated by’ icon that hyperlinks the record to the original source database. These attributions are listed directly in all search results for the entire BioGRID website and are also provided in download files. The BioGRID also works in close conjunction with the GO consortium (45), both to guide BioGRID curation efforts based on relevant GO terms, and on occasion to help elaborate branches of the GO, for example as pertains to the ubiquitin proteasome system. Finally, BioGRID supports pre-publication deposition of experimental results to facilitate rapid dissemination of HTP datasets generated by resource centers and other groups. For example, BioGRID curators have assisted with the formatting and upload of large-scale genetic interaction datasets for immediate release upon publication (46). In another instance, the biophysical interactions of ORFeome-based complexes (BioPlex) network generated by high-throughput affinity-purification mass spectrometry (6), was uploaded in BioGRID well in advance of publication in order to provide immediate open access to the dataset. Pre-publication data records are fully archived on a monthly basis along with all other BioGRID interactions but are excluded from BioGRID downloads until conversion into full BioGRID records upon publication of the dataset.
TEXT MINING
The vast amount of free form text that is used to report essentially all biomedical knowledge in journals precludes the manual distillation and annotation of biological interactions. Unfortunately, despite recent improvements in natural language processing based on artificial intelligence approaches (47), fully automated text mining systems (TMS) are unable to match the annotation accuracy of expert manual curation. Nonetheless, TMS can be of great utility in supporting biocuration tasks through the proficient triaging of non-pertinent literature and the provision of customized annotation interfaces that can both increase the rate of curation and track text statements that support curator inferences. Text-mining approaches to develop publication queues for curation have now largely superseded simple PubMed queries and are carried out in collaboration with leading text-mining groups. For example, a Support Vector Machine (SVM) method developed by the Textpresso (18,48,49) group was used to select pertinent full-text articles for the HIV, Wnt, arachidonic acid pathway (AAP) interaction networks and the A. thaliana project. In another example, a text-mining system developed by the RLIMS-P group (50) was used to help identify publications containing yeast phosphorylation site data. We have also recently implemented an in-house system that incorporates additional machine learning technologies (Chatr-aryamontri et al., in preparation) (51).
The BioGRID team has strongly supported the biomedical text-mining community through the BioCreative initiative (52) by providing curation expertise and manually annotated gold standard reference datasets (53,54). For example, in the most recent BioCreative competition in 2015 one particular task focused on the development of a curation interface designed specifically for BioGRID curators (55), based on the BioC standard, an extensible mark-up language created to allow interoperability between biomedical text processing systems (56). This BioCreative task also produced an innovative text-based dataset that tracked all statements used by curators to infer interaction data and phenotypes (20). This dataset will allow the refinement of text-mining algorithms to more closely mimic the intuitive sparse coverage approaches used by human curators. The initial version of the BioC-based curation interface devised by the BioCreative consortium will support annotation for both genetic and protein interactions, and will allow both faster extraction of interaction data and better curation quality control.
GENETIC INTERACTION CURATION
The accurate representation of genetic interactions is a challenge due to the complexity of both phenotypes and the precise genetic context of an interaction. In an effort to more precisely describe genetic interactions, and reconcile the different terminologies used within the various model organism research communities, BioGRID has collaborated with WormBase (26) to develop a new Genetic Interactions Structured Terminology (GIST) (Grove et al., in preparation). This effort to delineate well-defined, standardized genetic interaction (GI) terms has been supported by various MODs, including ZFIN (21), FlyBase (24), SGD (25), CGD (38), PomBase (23) and TAIR (39). Importantly, the GIST will not only facilitate the interpretation of genetic interactions, but also the integration of large volumes of genetic interaction data across different species. The acute need for a structured terminology was recognized by both WormBase and BioGRID curators while attempting to coordinate the curation of worm genetic interactions between the two databases. For historical reasons, GI terms in BioGRID were biased towards yeast genetic interaction descriptors that were unduly restrictive because the phenotype was often implicit within the GI term. For example, the common term ‘synthetic lethality’ represents a greater than multiplicative genetic interaction and the implicit phenotype of cell growth. Since there is currently no separate ‘synthetic’ GI term in BioGRID for curating interactions with other phenotypes, the existing GI terms could not be used to effectively curate more complicated phenotypes that arise in yeast and even more frequently in more complex metazoans, including humans. To resolve this issue in a general form, that is to cover all possible GI scenarios, the new GIST was organized according to a structured set of genetic terms that are completely separated from the myriad of possible phenotypes that might be linked to the interaction. The GIST is thus intended to be used in conjunction with all relevant species- or tissue-specific phenotype ontologies such that the type of genetic interaction is curated as a separate entity from the specific phenotype that is scored. This approach allows BioGRID to take full advantage of rigorous phenotype ontologies across model systems and humans, including Uberon, the Monarch Initiative, the Human Phenotype Ontology, and others (57,58). For yeast genetic interactions, 11 of the current BioGRID GI terms map to seven of the new GIST terms that will be used for curation going forward in 2017. This mapping will allow automated back-curation of more than 270 000 yeast genetic interactions associated with over 600 unique phenotypes (25) to be fully automated. BioGRID will also implement GIST for the curation of genetic interactions in human and key model organisms, including yeast, worm, fly, zebrafish and mouse. The use of standardized GI terms will facilitate the cross-species integration of genetic interaction datasets produced by large-scale CRISPR/Cas9-based screens in human cells and other organisms (59,60).
CHEMICAL INTERACTIONS
Since our previous update, the BioGRID has initiated a new pilot project to incorporate curated chemical interaction data and to combine this data with other biological interaction types curated from the literature. Initially, this focus has been on chemical–protein interactions because many biochemical relationships between drugs, toxins and other bioactive compounds and their targets are documented in the literature. A number of drug-discovery associated databases have captured either direct or inferred evidence for drug-target interactions. In order to incorporate previously annotated chemical–protein interaction data into BioGRID, a minimal unified record structure compatible with the diverse annotation systems used across multiple chemical databases was required. We surveyed the content of the major specialized chemical interaction databases, including DrugBank (37), HMdb (61), T3DB (62), BindingDB (63), CTD (64), Therapeutic Target DB (65), ChemBank (66), PharmGKB (67), DGIdb (68), PubChem (69) and ChEMBL (70) to determine the shared fields housed in each of these databases. Based on this survey, a minimal interoperable record structure was designed that contains: the target protein based on UniProt or GeneID identifiers; generic chemical name, synonyms and/or brand name for the chemical agent; the class of agent, such as small molecule, natural product, or biologic; the structural formula of the agent; CAS and ATC identifiers for the agent; the molecular action or effect of the agent; associated citations; and the original database source. This minimal set of fields allows the facile import of data records into BioGRID and effective interoperability between multiple chemical databases. Relevant database sources for all of the associated records are explicitly acknowledged with reciprocal links to the parent database, thereby allowing users the option of direct access to the original source of data in a transparent fashion. As the first test case, BioGRID has recently imported manually curated chemical–target data records from DrugBank (37), which contains >12 800 experimental and approved drugs and >4200 proteins. The downloadable DrugBank files were parsed and drug-target interactions re-mapped to the minimal chemical record structure in BioGRID. The automated mapping was validated by extensive manual review to resolve any issues and ensure data integrity. To display chemical interaction information, the BioGRID interface was modified to include new tabs on the result summary page to show chemical associations for the protein of interest. These chemical associations have been incorporated into the BioGRID network viewer to allow users to visualize chemical, genetic and protein interactions as a single network if desired. BioGRID chemical association data are also made available for download in a standardized tab-delimited file format. These infrastructure developments now allow the straightforward incorporation of chemical–protein and chemical–genetic interaction data from any source into BioGRID.
ENHANCED INTERACTION NETWORK VIEWER
The BioGRID Network Viewer has recently undergone substantial revisions in order to improve its functionality for visualizing complex interaction data (Figure 2). Each search result page now has an embedded Javascript-based viewer that leverages the powerful Cytoscape.js platform (71) to display interactive graph-based data representations. A default network layout provides an intuitive overview of the overall topology of the network. In this view, individual nodes represent each protein, gene, or chemical, with the distance from the center of the network proportional to connectivity for each node. Node size is also proportional to the number of interaction partners for that node. Edge colours depict the type of relationship between entities, namely protein–protein, gene–gene, chemical–protein, chemical–gene or chemical–chemical interactions. Edge thickness represents the quantity of evidence in support of the connection, such that the thicker the edge, the more types of evidence support the interaction. All nodes and edges in the network viewer can be dragged by the user to any desired position and can be clicked (or right-clicked or two-finger clicked on a Macintosh computer) to show additional details such as experimental evidence or additional annotation. Individual nodes can be right-clicked (or two-finger clicked) and the option ‘display network’ chosen to generate a new network with the selected node in the center. Each embedded BioGRID network provides several additional built-in layout options that include grid, concentric circles, single circle and arbor views. Users can apply on-the-fly filtering to show or hide specific types of edges and use toggles to increase or decrease experimental evidence thresholds for edge and node visualization. All networks generated with the viewer can be saved as high-resolution PNG images for use in figures for presentation or publication.
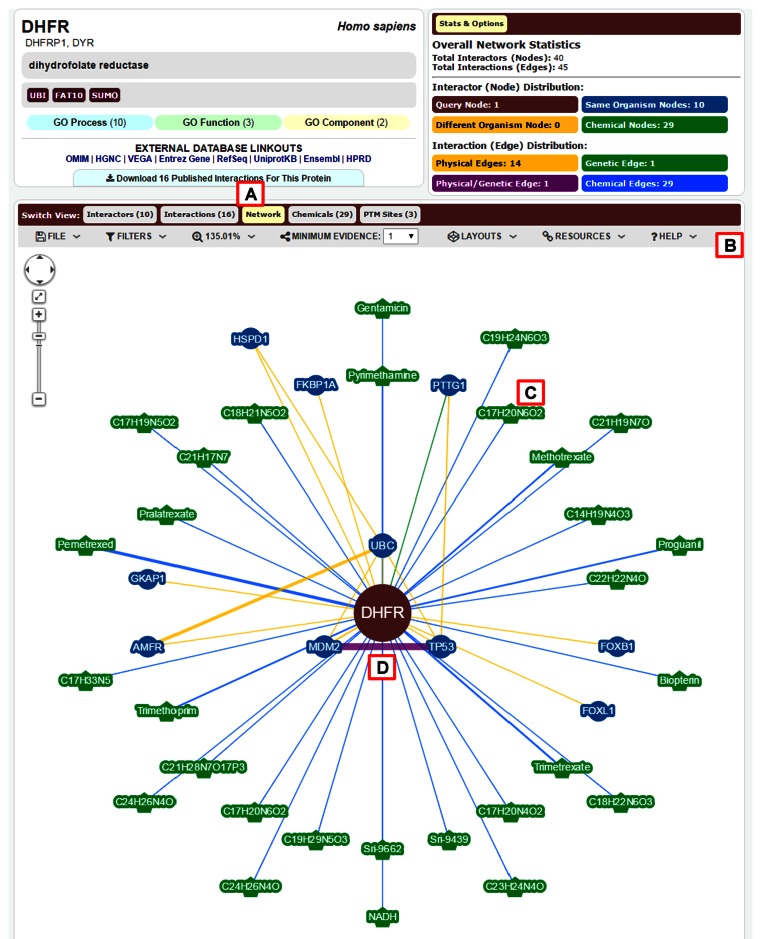
Figure 2.
New BioGRID network viewer. (A) The network tab in the ‘Switch View’ menu opens a network view for the selected query gene, as shown for human DHFR. (B) Users may export the network view as a figure file in PNG format, set filters that show or hide interactions, set thresholds for experimental evidence, and select from a number of layout formats. Explanatory text is provided under the help menu. (C) Node and edge colour indicates the interaction type and node size is proportional to its connectivity. In this example, green nodes represent chemicals and blue nodes represent proteins. When common names are not available, compounds are abbreviated by the chemical formula. (D) Yellow edges represent protein interactions, green edges represent genetic interactions, blue edges represent chemical interactions and purple edges represent both protein and genetic interactions.
VISUALIZATION OF POST-TRANSLATIONAL MODIFICATIONS
We have made several improvements to the BioGRID PTM viewer, which displays PTM sites on the protein sequence of interest (Figure 3). A comprehensive new layout for PTM views indicates all linked protein records, including splice isoforms, and provides links to the curated evidence for the PTM. In contrast to the original PTM viewer, which displayed only phosphorylation sites in budding yeast proteins (36), the new PTM viewer enables visualization of PTMs for all species for all PTMs in BioGRID, including phosphorylation, ubiquitination, acetylation, methylation, sumoylation, fat10ylation, and neddylation. All PTMs for any given query protein can now be viewed on the associated PTM summary page. PTMs that have not been mapped to a specific residue in the protein of interest are now also displayed in addition to site specific PTMs. Proteins annotated with PTMs in the BioGRID are marked by icons on the search list and result summary pages, and clicking on any icon opens the PTM viewer for the entire protein sequence.
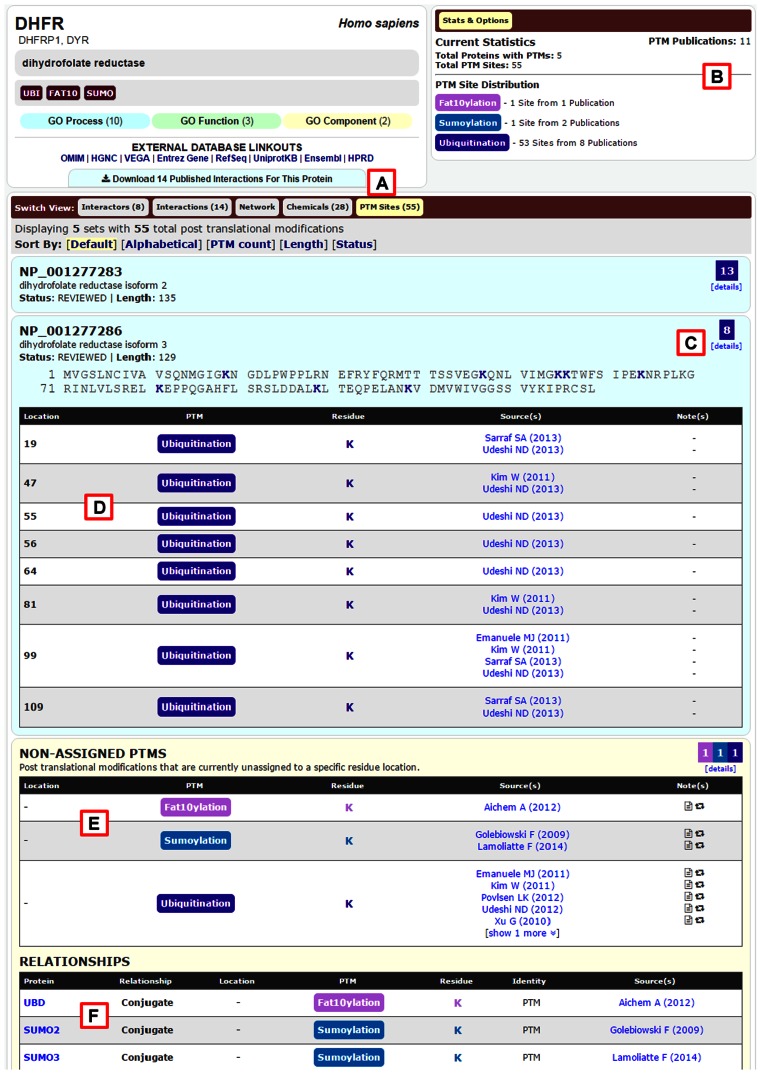
Figure 3.
New BioGRID post-translational modification (PTM) viewer. (A) Users can select the ‘PTM Sites’ tab from the ‘Switch View’ menu to view PTM data when available. (B) The ‘Stats & Options’ box indicates the number of PTM sites and defines the colours assigned to each PTM type. (C) PTM locations are displayed on the protein sequence with modified residues highlighted. (D) Assigned PTM sites are displayed in tabular format with supporting evidence and citations. (E and F) Non-assigned PTMs are displayed at the bottom of the page.
To accompany these improvements to the PTM viewer, we have recently completed an extensive project to migrate 57 819 PTMs that were originally housed in relatively obscure form within interaction records into the PTM viewer. Most notably, for the covalent protein modifier ubiquitin, we reassigned 49 425 annotations previously recorded in BioGRID as covalent protein interactions and demarcated in the free text notes as ‘likely ubiquitin conjugate’. The segregation of covalent ubiquitin modifications from non-covalent ubiquitin interactions properly delineates these two distinct types of interaction, and reduces the previous artificial dominance of ubiquitin as a super-hub in protein interaction networks. Non-covalent interactions between ubiquitin and recognition components of the ubiquitin-proteasome system are still retained as interaction records. As a consequence of the reassignment of ubiquitin and other small protein modifiers as PTMs, the number of protein interaction records for Homo sapiens decreased by 46 946 interactions and for S. cerevisiae decreased by 10 466 interactions in BioGRID release 3.4.125 (June 2015). However, these reductions were more than offset by curation of an even greater number protein interactions for each species since the previous update (Table 1). We anticipate the imminent addition of hundreds of thousands of new ubiquitin PTM sites with the release of a themed ubiquitin curation project in the immediate future.
DATABASE IMPROVEMENTS
In 2013, we completed the deployment of the BioGRID database, tools and web applications to a suite of six virtual machines (VMs) hosted by a commercial provider (Linode, NJ, USA). The VMs provide state-of-the-art processors, scalable memory, and native SSD high performance storage that can be expanded as needed. Each system has a fully redundant backup that runs daily and weekly, and is situated on a 40 Gigabit network for fast access by BioGRID developers and curators in different countries, as well as by BioGRID web interface and REST service users. Since deployment to cloud-based servers, the BioGRID software suite has maintained > 99.9% uptime. Since 2013, we have increased the processor speed and memory available on each VM in order to satisfy increased user demand. The number of VMs has been increased to eight in total in order to support additional new projects. Major improvements have been made to the size, speed, and storage capabilities of the MySQL database that underpins the BioGRID in order to incorporate the various new features described above. Finally, we have made continuous improvements to the extensive BioGRID annotation system that supports all BioGRID operations, including public-facing websites, download files, REST/PSICQUIC services, text-mining algorithms, and internal curation toolsets. The current annotation platform provides references for >100 million unique aliases, identifiers, systematic names and MOD references for >200 organisms, compared to the previous annotation system that supported ∼48 million identifiers for ∼100 supported organisms.
DATA DISSEMINATION
BioGRID data can be searched via the web search page or downloaded in a number of tabular (tab, tab 2 and mitab) and XML (PSI-MI 1.0, PSI-MI 2.5) formats. The BioGRID REST web service supports over 660 active projects worldwide that perform over 100 000 queries per month with an average return of ∼2 million interactions per month. The IMEx consortium PSICQUIC API interface (72) also fielded over 140 000 queries per month to BioGRID from a wide variety of third party plugins. For example, the REST service enables the direct comparison of all data in BioGRID to real time experimental data in the ProHits mass spectrometry LIMS (73). With the release of BioGRID 3.4, we introduced a new search result page that allows the relationship between any two entities to be viewed and linked to independently. This feature enables external resources such as NCBI (29), Uniprot (28) and others (23–25,27,30,31,39,74–76) to link to individual entity relationships described within a publication rather than simply to an entire search page result as previously. BioGRID search results now include more details, including the number of interaction partners, interactions, PTMs and chemical interactions. Furthermore, when applicable each result page also indicates whether a particular interaction was curated as part of one or more themed projects. We have continued to update our online Wiki documentation with detailed information on all aspects of BioGRID tools and resources (see https://wiki.thebiogrid.org). In early 2016, we released two protocol papers that outline key functions in step-by-step processes to aid new users in using the platform (77,78). The BioGRID also maintains an active e-mail help desk to assist users and facilitate the direct deposition of large datasets (biogridadmin@gmail.com). We continue to update and post all new related source code repositories at our GitHub organizational page (https://github.com/BioGRID) and we continue to update both our Twitter (https://twitter.com/biogrid) and YouTube Channel (https://www.youtube.com/user/TheBioGRID) with the latest BioGRID news and feature updates.
FUTURE DEVELOPMENTS
The BioGRID will continue to annotate high quality protein, genetic and chemical interaction data, with increasing attention to human datasets as focused on themes of central biological processes and specific human diseases. The BioGRID curation pipeline will be enhanced through the integration of ever more sophisticated text-mining tools, which will be implemented in collaboration with text-mining groups and the BioCreative consortium (19,48,49,55,79,80). These efforts will be augmented by collaborations with diverse database partners, including MODs, phenotype databases, and chemical databases. In particular, chemical–protein interaction datasets will be prioritized for elaboration with specific attention to drugs, metabolites, toxins and bioactive small molecules with the goal of facilitating network-based approaches to drug-discovery (37,63,70,81). We will continue to improve search and visualization tools to expedite the analysis of interaction datasets and to provide additional resources and support for the propagation of BioGRID interaction data through partner databases. An imminent new update to the BioGRID database architecture will allow the seamless acquisition and integration of human genetic and chemical–genetic interaction data generated in human cell lines and various model organisms by CRISPR/Cas9 gene editing technology (59,60). These improvements will also allow the precise capture of more complex interaction contexts, for example higher order genetic interactions, splice isoform dependent protein interactions, and tissue-specific interactions (82). The BioGRID will thus continue to evolve as a biological interaction data resource for the biomedical research community.
Acknowledgments
The authors thank Chris Grove, Paul Sternberg and Anastasia Baryshnikova for ongoing collaborative development of the Genetic Interaction Structured Terminology. We also thank John Aitchison, Brenda Andrews, Gary Bader, Andre Bernards, Judy Blake, Charlie Boone, Stephen Burley, Peter Dirks, Andrew Emili, Russ Finley, Michael Gilson, Anne-Claude Gingras, Steve Gygi,Melissa Haendel, Wade Harper, Eva Huala, Trey Ideker, Igor Jurisica, Thom Kaufman, Craig Knox, Chris Mungal, Chad Myers, Ivan Sadowski, Paul Sternberg, Olga Troyanskaya, Monte Westerfield, John Wilbur, David Wishart, Val Wood, Helen Yu and Cathy Wu for support, discussions and/or access to pre-publication datasets. We wish to dedicate this work to the memory of William Gelbart of Harvard University who, as the Principal Investigator of FlyBase, always provided his unreserved support for BioGRID.
FUNDING
National Institutes of Health Office of Research Infrastructure Programs [R01OD010929 and R24OD011194 to M.T. and K.D.]; National Institutes of Health National Heart, Lung and Blood Institute [U54HL117798 Curation Core to K.D., Garret FitzGerald overall P.I.]; Genome Canada Largescale Applied Proteomics/Ontario Genomics Institute OGI-069 [to M.T. and Anne Claude Gingras]; Genome Québec International Recruitment Award [to M.T.]; Canada Research Chair in Systems and Synthetic Biology [to M.T.]. Funding for open access charge: National Institute of Health [R01OD010929].
Conflict of interest statement. None declared.
REFERENCES
- 1.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 3.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 5.Rolland T., Tasan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X., Coulombe-Huntington J., Kang S., Sheynkman G.M., Hao T., Richardson A., Sun S., Yang F., Shen Y.A., Murray R.R. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., Gianoulis T.A., Yip K.Y., Gerstein M., Snyder M. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143:639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego O., Betts M.J., Gvozdenovic-Jeremic J., Maeda K., Matetzki C., Aguilar-Gurrieri C., Beltran-Alvarez P., Bonn S., Fernandez-Tornero C., Jensen L.J., et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahni N., Yi S., Taipale M., Fuxman Bass J.I., Coulombe-Huntington J., Yang F., Peng J., Weile J., Karras G.I., Wang Y., et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofree M., Shen J.P., Carter H., Gross A., Ideker T. Network-based stratification of tumor mutations. Nat. Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun S., Yang F., Tan G., Costanzo M., Oughtred R., Hirschman J., Theesfeld C.L., Bansal P., Sahni N., Yi S., et al. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 2016;26:670–680. doi: 10.1101/gr.192526.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium. Roadmap Epigenomics, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivas R., Shen J.P., Yang C.C., Sun S.M., Li J., Gross A.M., Jensen J., Licon K., Bojorquez-Gomez A., Klepper K., et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell. 2016;63:514–525. doi: 10.1016/j.molcel.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breitkreutz B.J., Stark C., Tyers M. The GRID: the general repository for interaction datasets. Genome Biol. 2003;4:R23. doi: 10.1186/gb-2003-4-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reguly T., Breitkreutz A., Boucher L., Breitkreutz B.J., Hon G.C., Myers C.L., Parsons A., Friesen H., Oughtred R., Tong A., et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller H.M., Kenny E.E., Sternberg P.W. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S., Kwon D., Shin S.Y., Wilbur W.J. PIE the search: searching PubMed literature for protein interaction information. Bioinformatics. 2012;28:597–598. doi: 10.1093/bioinformatics/btr702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islamaj Doğan R., Kim S., Chatr-Aryamontri A., Chang C.S., Oughtred R., Rust J., Wilbur W.J., Comeau D.C., Dolinski K., Tyers M. The BioC-BioGRID corpus: full text articles annotated for curation of protein–protein and genetic interactions. Database (Oxford) 2016 doi: 10.1093/database/baw147. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruzicka L., Bradford Y.M., Frazer K., Howe D.G., Paddock H., Ramachandran S., Singer A., Toro S., Van Slyke C.E., Eagle A.E., et al. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015;53:498–509. doi: 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bult C.J., Eppig J.T., Blake J.A., Kadin J.A., Richardson J.E., Mouse Genome Database Group Mouse genome database 2016. Nucleic Acids Res. 2016;44:D840–D847. doi: 10.1093/nar/gkv1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDowall M.D., Harris M.A., Lock A., Rutherford K., Staines D.M., Bahler J., Kersey P.J., Oliver S.G., Wood V. PomBase 2015: updates to the fission yeast database. Nucleic Acids Res. 2015;43:D656–D661. doi: 10.1093/nar/gku1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Attrill H., Falls K., Goodman J.L., Millburn G.H., Antonazzo G., Rey A.J., Marygold S.J., FlyBase Consortium FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016;44:D786–D792. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard T.K., Hitz B.C., Engel S.R., Song G., Balakrishnan R., Binkley G., Costanzo M.C., Dalusag K.S., Demeter J., Hellerstedt S.T., et al. The Saccharomyces Genome Database Variant Viewer. Nucleic Acids Res. 2016;44:D698–D702. doi: 10.1093/nar/gkv1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C., et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur O., Anwar N., Schultz N., Bader G.D., Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F.S., Cesareni G., et al. Protein interaction data curation: the International Molecular Exchange (IMEx) Consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerrien S., Orchard S., Montecchi-Palazzi L., Aranda B., Quinn A.F., Vinod N., Bader G.D., Xenarios I., Wojcik J., Sherman D., et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roux P.P., Thibault P. The coming of age of phosphoproteomics–from large data sets to inference of protein functions. Mol. Cell Proteomics. 2013;12:3453–3464. doi: 10.1074/mcp.R113.032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadowski I., Breitkreutz B.J., Stark C., Su T.C., Dahabieh M., Raithatha S., Bernhard W., Oughtred R., Dolinski K., Barreto K., et al. The PhosphoGRID Saccharomyces cerevisiae protein phosphorylation site database: version 2.0 update. Database (Oxford) 2013;2013:bat026. doi: 10.1093/database/bat026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skrzypek M.S., Binkley J., Binkley G., Miyasato S.R., Simison M., Sherlock G. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw924. doi:10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E. The Arabidopsis Information Resource: Making and mining the “gold standard" annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernards A. Ras superfamily and interacting proteins database. Methods Enzymol. 2006;407:1–9. doi: 10.1016/S0076-6879(05)07001-1. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 42.Dirks P.B. Brain tumor stem cells: the cancer stem cell hypothesis writ large. Mol. Oncol. 2010;4:420–430. doi: 10.1016/j.molonc.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costanzo M., Van der Sluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D., et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pastur-Romay L.A., Cedron F., Pazos A., Porto-Pazos A.B. Deep artificial neural networks and neuromorphic chips for big data analysis: pharmaceutical and bioinformatics applications. Int J Mol. Sci. 2016;17:E1313. doi: 10.3390/ijms17081313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang R., Schindelman G., Van Auken K., Fernandes J., Chen W., Wang X., Davis P., Tuli M.A., Marygold S.J., Millburn G., et al. Automatic categorization of diverse experimental information in the bioscience literature. BMC Bioinformatics. 2012;13:16. doi: 10.1186/1471-2105-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Auken K., Fey P., Berardini T.Z., Dodson R., Cooper L., Li D., Chan J., Li Y., Basu S., Muller H.M., et al. Text mining in the biocuration workflow: applications for literature curation at WormBase, DictyBase and TAIR. Database (Oxford) 2012;2012:bas040. doi: 10.1093/database/bas040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torii M., Arighi C.N., Li G., Wang Q., Wu C.H., Vijay-Shanker K. RLIMS-P 2.0: A generalizable rule-based information extraction system for literature mining of protein phosphorylation information. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;12:17–29. doi: 10.1109/TCBB.2014.2372765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S., Shin S.Y., Lee I.H., Kim S.J., Sriram R., Zhang B.T. PIE: an online prediction system for protein–protein interactions from text. Nucleic Acids Res. 2008;36:W411–W415. doi: 10.1093/nar/gkn281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q., Abdul S.S., Almeida L., Ananiadou S., Balderas-Martinez Y.I., Batista-Navarro R., Campos D., Chilton L., Chou H.J., Contreras G., et al. Overview of the interactive task in BioCreative V. Database (Oxford) 2016;2016:baw119. doi: 10.1093/database/baw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatr-Aryamontri A., Winter A., Perfetto L., Briganti L., Licata L., Iannuccelli M., Castagnoli L., Cesareni G., Tyers M. Benchmarking of the 2010 BioCreative Challenge III text-mining competition by the BioGRID and MINT interaction databases. BMC Bioinformatics. 2011;12(Suppl 8):S8. doi: 10.1186/1471-2105-12-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krallinger M., Vazquez M., Leitner F., Salgado D., Chatr-Aryamontri A., Winter A., Perfetto L., Briganti L., Licata L., Iannuccelli M., et al. The protein–protein interaction tasks of BioCreative III: classification/ranking of articles and linking bio-ontology concepts to full text. BMC Bioinformatics. 2011;12(Suppl 8):S3. doi: 10.1186/1471-2105-12-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S., Islamaj Dogan R., Chatr-Aryamontri A., Chang C.S., Oughtred R., Rust J., Batista-Navarro R., Carter J., Ananiadou S., Matos S., et al. BioCreative V BioC track overview: collaborative biocurator assistant task for BioGRID. Database (Oxford) 2016;2016:baw121. doi: 10.1093/database/baw121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Comeau D.C., Islamaj Dogan R., Ciccarese P., Cohen K.B., Krallinger M., Leitner F., Lu Z., Peng Y., Rinaldi F., Torii M., et al. BioC: a minimalist approach to interoperability for biomedical text processing. Database (Oxford) 2013;2013:bat064. doi: 10.1093/database/bat064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groza T., Kohler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T., et al. The human phenotype ontology: semantic unification of common and rare disease. Am. J. Hum. Genet. 2015;97:111–124. doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurry J.A., Kohler S., Washington N.L., Balhoff J.P., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M., et al. Navigating the phenotype frontier: The Monarch Initiative. Genetics. 2016;203:1491–1495. doi: 10.1534/genetics.116.188870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 61.Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wishart D., Arndt D., Pon A., Sajed T., Guo A.C., Djoumbou Y., Knox C., Wilson M., Liang Y., Grant J., et al. T3DB: the toxic exposome database. Nucleic Acids Res. 2015;43:D928–D934. doi: 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis A.P., Grondin C.J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Wiegers T.C., Mattingly C.J. The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin C., Zhang C., Zhu F., Xu F., Chen S.Y., Zhang P., Li Y.H., Yang S.Y., Wei Y.Q., Tao L., et al. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res. 2014;42:D1118–D1123. doi: 10.1093/nar/gkt1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seiler K.P., George G.A., Happ M.P., Bodycombe N.E., Carrinski H.A., Norton S., Brudz S., Sullivan J.P., Muhlich J., Serrano M., et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whirl-Carrillo M., McDonagh E.M., Hebert J.M., Gong L., Sangkuhl K., Thorn C.F., Altman R.B., Klein T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffith M., Griffith O.L., Coffman A.C., Weible J.V., McMichael J.F., Spies N.C., Koval J., Das I., Callaway M.B., Eldred J.M., et al. DGIdb: mining the druggable genome. Nat. Methods. 2013;10:1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Suzek T., Zhang J., Wang J., He S., Cheng T., Shoemaker B.A., Gindulyte A., Bryant S.H. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–D1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies M., Nowotka M., Papadatos G., Dedman N., Gaulton A., Atkinson F., Bellis L., Overington J.P. ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res. 2015;43:W612–W620. doi: 10.1093/nar/gkv352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.del-Toro N., Dumousseau M., Orchard S., Jimenez R.C., Galeota E., Launay G., Goll J., Breuer K., Ono K., Salwinski L., et al. A new reference implementation of the PSICQUIC web service. Nucleic Acids Res. 2013;41:W601–W606. doi: 10.1093/nar/gkt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu G., Zhang J., Choi H., Lambert J.P., Srikumar T., Larsen B., Nesvizhskii A.I., Raught B., Tyers M., Gingras A.C. Using ProHits to store, annotate, and analyze affinity purification-mass spectrometry (AP-MS) data. Curr. Protoc. Bioinformatics. 2012 doi: 10.1002/0471250953.bi0816s39. doi:10.1002/0471250953.bi0816s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Razick S., Magklaras G., Donaldson I.M. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murali T., Pacifico S., Yu J., Guest S., Roberts G.G., 3rd, Finley R.L., Jr DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–D743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lardenois A., Gattiker A., Collin O., Chalmel F., Primig M. GermOnline 4.0 is a genomics gateway for germline development, meiosis and the mitotic cell cycle. Database (Oxford) 2010;2010:baq030. doi: 10.1093/database/baq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oughtred R., Chatr-aryamontri A., Breitkreutz B.J., Chang C.S., Rust J.M., Theesfeld C.L., Heinicke S., Breitkreutz A., Chen D., Hirschman J., et al. Use of the BioGRID database for analysis of yeast protein and genetic interactions. Cold Spring Harb. Protoc. 2016;2016 doi: 10.1101/pdb.prot088880. pdb prot088880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oughtred R., Chatr-aryamontri A., Breitkreutz B.J., Chang C.S., Rust J.M., Theesfeld C.L., Heinicke S., Breitkreutz A., Chen D., Hirschman J., et al. BioGRID: a resource for studying biological interactions in yeast. Cold Spring Harb. Protoc. 2016;2016 doi: 10.1101/pdb.top080754. pdb top080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwon D., Kim S., Shin S.Y., Chatr-aryamontri A., Wilbur W.J. Assisting manual literature curation for protein–protein interactions using BioQRator. Database (Oxford) 2014;2014:bau067. doi: 10.1093/database/bau067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shin S.Y., Kim S., Wilbur W.J., Kwon D. BioC viewer: a web-based tool for displaying and merging annotations in BioC. Database (Oxford) 2016;2016:baw106. doi: 10.1093/database/baw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rose P.W., Prlic A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C., et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]