Abstract
Secondary metabolites produced by microbes have diverse biological functions, which makes them a great potential source of biotechnologically relevant compounds with antimicrobial, anti-cancer and other activities. The proteins needed to synthesize these natural products are often encoded by clusters of co-located genes called biosynthetic gene clusters (BCs). In order to advance the exploration of microbial secondary metabolism, we developed the largest publically available database of experimentally verified and predicted BCs, the Integrated Microbial Genomes Atlas of Biosynthetic gene Clusters (IMG-ABC) (https://img.jgi.doe.gov/abc/). Here, we describe an update of IMG-ABC, which includes ClusterScout, a tool for targeted identification of custom biosynthetic gene clusters across 40 000 isolate microbial genomes, and a new search capability to query more than 700 000 BCs from isolate genomes for clusters with similar Pfam composition. Additional features enable fast exploration and analysis of BCs through two new interactive visualization features, a BC function heatmap and a BC similarity network graph. These new tools and features add to the value of IMG-ABC's vast body of BC data, facilitating their in-depth analysis and accelerating secondary metabolite discovery.
INTRODUCTION
Microbes produce a variety of compounds, known as secondary metabolites (SMs) or natural products, which play many important physiological roles. Some SMs confer the ability to survive adverse conditions, while others function in bacterial communication (1) or are used as weapons of inter- and intra-species competition (2). These diverse functions make SMs a great source of biotechnologically relevant compounds with antimicrobial, anti-cancer and other activities (3–5). Additionally, biologically synthesized compounds achieve a chemical structure complexity unmatched by synthetic chemistry. Whereas traditionally SMs have been isolated from microbial cultures, the recent advances in DNA sequencing technologies have opened a new avenue to the discovery and characterization of SMs (6). The cornerstone of this capability lies in the observation that SM synthesis is typically encoded by the genes that are clustered together on the chromosomes or plasmids (biosynthetic gene clusters or BCs) (7). This feature of SMs has been exploited by several computational tools, which were used to predict and annotate BCs in thousands of microbial genomes (8). The result of this breakthrough was not only the discovery of previously unknown biosynthetic potential in well-studied organisms (9), but also identification of proteins with novel enzymatic activities and novel classes of SMs (10).
In order to facilitate the discovery and analysis of BCs and SMs in bacterial genomes and metagenomes, a data mart called the Atlas of Biosynthetic gene Clusters within the Integrated Microbial Genomes system (11) (IMG-ABC) was introduced in 2015 (12). BCs were predicted and annotated across all microbial isolate genomes and a set of metagenomes using a combination of Clusterfinder (10) and antiSMASH (13). After the creation of the data mart, annotation with antiSMASH became part of the standard annotation pipeline for newly imported isolate genomes. The result of this integration was the growth of the IMG-ABC database to contain more than 730 000 BCs in 40 034 isolate microbial genomes and >310 000 BCs in 2416 metagenomes. Additionally, a set of experimentally verified BCs and their associated SMs were collected from the NCBI and imported into the database. IMG-ABC also provided the means to search these large datasets based on BC and SM attributes, such as BC type, Pfam (14) and EC content (15), BC length, SM activity etc. Here, we present an update of IMG-ABC, which includes several new features improving data mining and navigation, such as a set of tools for BC visualization and comparison, and targeted BC identification across thousands of isolate bacterial genomes with ClusterScout.
NEW FEATURES AND UPDATES
Open access to browsing
IMG-ABC can be accessed at https://img.jgi.doe.gov/abc/ and can be browsed without a login requirement. However, creating a free IMG account enables users to utilize IMG-ABC's ‘BC Set’ feature, which allows saving groups of BCs on the server side for future retrieval and sharing.
Find similar BCs function
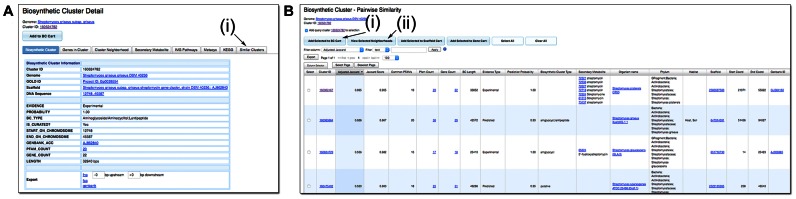
In the initial version of IMG-ABC, users could search the BC database using various attributes, such as BC type, SM produced, or the presence of specific Pfams (14). With this update, we are introducing the ability to search for BCs with Pfam composition similar to that of a query BC. This function is available through the ‘Similar Clusters’ tab on the Biosynthetic Cluster Detail page (Figure 1A). Briefly, in order to find BCs with similar Pfam composition, Pfams assigned to the genes of the query BC are collected and searched against the IMG-ABC database for BCs containing at least three of these Pfams. In order to minimize the running time of the search, the ubiquitous ABC Transporter family (PF00005) is not considered at this step. At least 500 BCs (if possible) are collected in order of descending number of shared Pfams. Next, a pairwise comparison between all collected BCs against the query BC is performed. In the resulting Pairwise Similarity Results table two scores are reported: first is the ‘Jaccard Score’, which is the Jaccard Index (the ratio of the number of shared distinct Pfam domains to the total number of distinct Pfam in both BCs); second is the ‘Adjusted Jaccard’, which is based on the modified Jaccard Index introduced by Cimermančič et al. (10) and scaled by normalizing to 0.64e−1 so that the score reported is between 0 and 1. The result of the similarity search is the 100 most similar BCs, sorted by descending Adjusted Score (Figure 1B). In addition to the two similarity scores and other BC metadata, the table contains the total number of distinct Pfams shared between the query and each reported BC.
Figure 1.
Similarity Search (A) BC similarity search is accessible through the ‘Similar Clusters’ tab (i) in the Biosynthetic Cluster Detail page, in this case the streptomycin BC from Streptomyces griseus. (B) The results are presented as a table which includes the ‘Jaccard Score’ and the ‘Adjusted Jaccard Score’. As expected, two more experimentally verified BCs known to produce streptomycin are retrieved, in addition to multiple predicted BCs. Users can further analyze these BCs by (i) adding them to the BC Cart, or (ii) visualize the neighborhoods of selected BCs to compare them with the query BCs.
ClusterScout: custom identification of BCs in large datasets
The latest version of the popular BC identification tool antiSMASH (version 3) annotates >40 types of BCs based on the presence of certain key protein domains (13). However, this list is not comprehensive as there many BC types not currently included in the annotation logic. It should be noted, however, that the stand-alone version of antiSMASH provides the ability to implement a customized search logic based not only on Pfam annotations but also on custom protein hidden Markov model (HMM) profiles. Similar functionality in IMG-ABC is provided by ClusterScout, a tool that uses gene Pfam content for the identification of BCs across all publicly available isolate bacterial genomes in IMG.
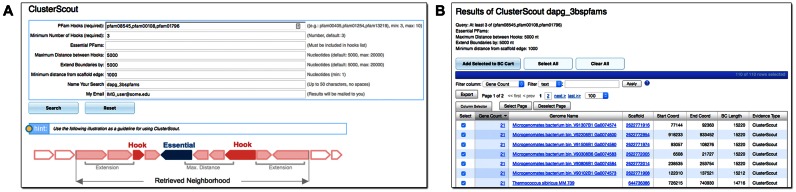
The algorithm used by ClusterScout is summarized in Figure 2A. Briefly, a user must select a set of Pfams (‘hooks’, minimum 3) and specify the maximum distance between them (maximum 20 kb) (Figure 1B). The user must also set the minimum number of hooks required (minimum 3), and may optionally define a subset of specific Pfam hooks that need to be present in the cluster (‘essential hooks’). Additionally, the user can extend the boundaries of the cluster by up to 20 kb. When the extended boundary falls within a coding sequence, the cluster boundary is extended to the start/stop of the overlapping gene(s). Lastly, users can define the minimum distance between the identified cluster and the scaffold ends (default 1 kb), which is useful in eliminating potentially incomplete BCs from the results.
Figure 2.
ClusterScout. (A) User Interface with algorithm logic schematic. (B) The results from a ClusterScout search are presented in a table, and custom BCs can be added to the BC Cart for analysis.
Depending on the complexity of the query and the ubiquity of the selected Pfams, ClusterScout may take a few minutes to several hours to complete its run. For this reason, the process is run in the background and, upon completion, the user is notified by an email with a link to the results (Figure 2B). The number of reported clusters is limited to the top 500 candidates, selected first based on the number of hooks they contain, and then by their sequence length. Custom clusters identified by ClusterScout can be added to a BC Cart for further analysis (Figure 3A). The results of a ClusterScout search are saved for up to 24 hours on the IMG-ABC server, so users are encouraged to download the results or, using a registered account, save them in a BC Set.
Figure 3.
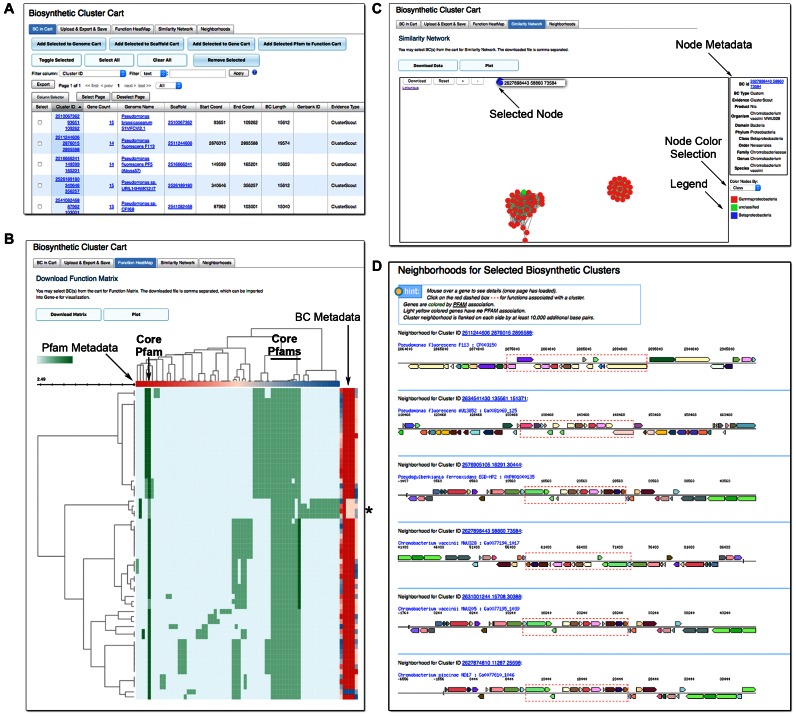
IMG-ABC case study. (A) The BC Cart is the virtual space where BC analysis can be performed. (B) The function heatmap visualization can be used to study the Pfam content of BCs. Cells are colored with hues of green based on the number of copies of the selected Pfam in the BC (darker signifies a higher copy number). Hovering the mouse pointer over a cell provides the number of copies of that Pfam in the BC. Pfams (columns) that occur in all BCs (rows) likely define the core functions of the BCs in view. Column and row metadata are found on top and to the right, respectively, of the heatmap. Hovering the mouse pointer over these metadata cells provides more detail information. These metadata can be used for quick visual inspection and identification of patterns. For example, betaproteobacteria containing the DAPG BC are easily discoverable (asterisk). (C) The similarity network graph provides another way to summarize the data. The BCs in this example fall into three distinct groups; two groups contain gammaproteobacteria (red nodes) while one group consists of betaproteobacteria (purple nodes). The green node represents experimentally verified BC for DAPG (from a gammaproteobacterium Pseudomonas fluorescens) in the IMG-ABC database. Clicking on a node reveals the metadata associated with the BC (table on the right). The color of the nodes can be changed to display different metadata, such as taxonomic classification or evidence. (D) Visualization of the putative DAPG BC neighborhoods from the four betaproteobacterial BCs and one BC from each gammaproteobacterial group shows that although the flanking regions of the BCs differ, the core genes are conserved, thus it is likely that these newly discovered BCs indeed encode the necessary proteins for DAPG production.
BC cart: enabling in-depth analysis of selected BCs
The new version of IMG-ABC includes the BC cart, a feature familiar to IMG users (11), which serves to collect the BCs of interest for further in-depth analysis (Figure 3A). BCs can be added to the cart from several entry points in the User Interface, including the Biosynthetic Cluster Detail page, various summary tables that can be accessed by browsing BCs or SMs, or the results of searches. In the BC Cart, BCs can be exported or imported through the ‘Upload & Export & Save’ tab. The BC Cart also serves as a portal to accessing IMG's rich analysis features through the ability to add data to other types of Carts, such as the Genome, Scaffold and Gene Carts. Additionally, the Pfam contents of selected BCs can be added to a Function Cart for detailed functional analysis.
Most importantly, the BC Cart contains the features specifically designed to facilitate visualization and analysis of BCs. A user can visualize and compare the architectures of selected BCs, and the content of flanking genomic areas, by using the ‘Neighborhoods’ tab. Additionally, Pfam content and modular architecture of BCs can be explored through the ‘Function Heatmap’ tab and BC similarities can be visualized through the ‘Similarity Network’ tab. The latter two features are described in detail below.
Hierarchically clustered heatmap visualization
Calculating the similarity between BCs is an effective way to identify BCs that may encode a similar SM. However, these similarity scores may be misleading due to imprecise prediction of BC boundaries, whereby the similarity scores are influenced by the flanking genes erroneously included in the BC. Additionally, small but important differences (e.g. presence or absence of a single protein domain conferring unique enzymatic activity) between highly similar BCs may not be reflected in the similarity scores. In order to enable more detailed BC comparisons, we are introducing the capability to dynamically build BC heatmaps based on Pfam annotations. This is achieved through an implementation of the InCHlib Javascript library (16). Cells in the heatmap represent counts of Pfams (columns) found in selected BCs (rows). The heatmap display appearance is customizable and allows users to inspect functional similarities and differences between multiple BCs by assessing their Pfam content. The user can also quickly access the Pfam and BC metadata by hovering the mouse pointer, respectively, over the column headers and columns on the right of the map displaying BC taxonomic information (Figure 3B). Additionally, the data are hierarchically clustered on both the Pfam and the BC axis using an InCHlib clustering tool with the Jaccard distance and Ward linkage options. As a result of 2D clustering, BCs with similar Pfam content are clustered together, and Pfams with similar distributions across the BCs of interest are also clustered together. Clustering the data on both axes facilitates the identification of conserved function modules. These may represent the core functions of the BC or auxiliary functions participating in modification of the core SM. The number of BCs and Pfams shown in the user interface is limited to 100 BCs and 50 most frequent Pfams in the selected dataset, with an option to download the data for up to 500 BCs and all Pfams in a format compatible with the Gene-e tool (http://www.broadinstitute.org/cancer/software/GENE-E/).
Dynamic BC similarity network visualization
The construction and visualization of networks based on the similarity between BCs has been a successful approach in the identification of novel BC families (10). Additionally, superimposing experimentally verified BCs on these networks can provide clues for the potential SM encoded by putative BCs. With this update of IMG-ABC, we are introducing the ability to visualize BC similarity networks (Figure 3C) by performing all-versus-all pairwise comparisons of selected BCs to create a similarity matrix. This matrix is filtered to include only the pairs with an Adjusted Jaccard Score equal or >0.5, and BCs without a single score above this threshold are not represented in the network graph. The visualization of this network is based on an implementation of linkurious (http://linkurio.us/), which enables users to quickly navigate and inspect the BC similarity network, and to visualize a set of relevant metadata through a selection menu. The similarity data and metadata can also be downloaded for visualization with Cytoscape (17).
IMG-ABC case study
To showcase the power of the new IMG-ABC features applied to a large database of annotated genomes we present a BC discovery and characterization case study using an example of 2,4-diacetylphloroglucinol (DAPG) biosynthesis, which is annotated by the current version of antiSMASH (13) as type 3 polyketide synthase cluster without SM product prediction. DAPG is a secondary metabolite with important biocontrol properties (18). The aim of our case study is to identify all DAPG clusters in IMG genomes, analyze their structure and elucidate taxonomic and habitat distribution of DAPG biosynthesis pathway. The steps used in this case study are as follows:
We start by identifying the Pfams of enzymes participating in DAPG biosynthesis, which will be then used as the input in a ClusterScout query. These Pfams can be found by searching for experimentally verified DAPG biosynthesis clusters using ‘Search by BC Attributes’ menu and inspecting the Pfam content of each gene in the cluster through the ‘Genes in Cluster’ tab of Biosynthetic Cluster Detail page and IMG's gene detail pages. In cases of genes with multiple Pfams, the Pfam with the longest model can be selected to improve the accuracy of the ClusterScout search. For the DAPG BC these are pfam08545, pfam00108 and pfam00195 for PhlA (hydroxymethylglutaryl-CoA synthase), PhlC (acetyl-CoA acetyltransferase) and PhlD (phloroglucinol synthase), respectively. Next we use the ClusterScout tool, limiting the search to genomic regions in which these Pfams are found within 5000 bp from each other, and extend the boundaries of the discovered locus by 5000 bp. We also set the minimum distance from the scaffold end to 1000 bp. This query returns 62 putative BCs (Supplementary Table SI), which are added to BC cart for analysis (Figure 3A).
Next, we use the function heatmap feature to identify co-occurring Pfams. In the Function Heatmap tab, by selecting ‘Plot’ option a set of Pfams present in all the putative clusters (the ‘core’) can be identified (Figure 3B). By hovering the mouse pointer over the column headers of the ‘core’ Pfams we observe that in addition to the original 3 ‘hook’ Pfams, pfam16859, pfam00440 and pfam07690 appear to be part of the ‘core’ DAPG pathway. This is in agreement with experimental data for DAPG biosynthesis pathway, as the first two of these Pfams are found in the transcription regulator (PhlF), which controls expression of this pathway, while the last one is found in the putative DAPG exporter protein (PhlE) (18).
Taxonomic affiliation of putative DAPG BCs, which can be explored by hovering the mouse pointer over row metadata (far right columns), identifies a distinct group of clusters found in betaproteobacteria (marked with an asterisk in Figure 3B). To the best of our knowledge, this is the first observation of the DAPG pathway in betaproteobacteria.
Alternatively, the diversity of putative DAPG clusters can be investigated by visualizing a BC similarity network by clicking on the ‘Similarity Network’ tab (Figure 3C). In our example, BCs fall into three major groups, and by coloring the nodes based on their taxonomy (Class of the host organism) we observe that two of these groups are found in gammaproteobacteria, in which DAPG biosynthesis pathway has been experimentally elucidated, while the smallest group originates from betaproteobacteria.
Lastly, the chromosomal organization of putative DAPG clusters can be investigated using the ‘Neighborhood’ view (Figure 3D). In order to simplify visualization, we can select only one representative from each of the two large gammaproteobacterial groups, and all four of the newly discovered betaproteobacterial BCs. As expected, the organization of this cluster is very similar between betaproteobacteria and distinct from both gammaproteobacterial clusters. However, organization of the ‘core’ genes is similar between all clusters suggesting that these betaproteobacterial clusters likely encode DAPG biosynthesis pathway.
To summarize the results of our case study, we have used ClusterScout to devise a custom query and search for putative DAPG BCs across >40 000 isolate genomes in the IMG database. This search and subsequent analysis using IMG-ABC tools took less than an hour, and has led to the identification of putative DAPG biosynthesis clusters in four betaproteobacteria: a bacterium identified as Pseudogulbenkiania ferrooxidans EGD-HP2 (19) and three members of Chromobacterium genus, Chromobacterium vaccinii MWU205 (20), C. vaccinii MWU328 (20) and Chromobacterium piscinae ND17 (21). The IMG tools for analysis of biogeography and habitat specificity of these isolates indicate that all of them were collected from freshwater environments across the globe (Northeastern India, Northeastern United States of America and Malaysia). This finding is remarkable considering that so far DAPG production in bacteria has been described mostly in plant-associated bacteria and in a bacterial symbiont of the red-backed salamander (22). Interestingly, C. vaccinii strains, in which DAPG clusters were found, were studied because of their biocontrol activity against mosquito larvae (20). Since DAPG is known to elicit a lethal hyperactive immune response in Drosophila melanogaster larvae (23) and has known toxicity against other organisms (24), this finding raises the possibility that DAPG may be responsible for larvicidal activity of the C. vaccinii strains. This example demonstrates the power of IMG-ABC data and tools in supporting the analysis of genome sequences, enabling biological interpretation of sequence data and formulation of testable hypotheses.
FUTURE DIRECTIONS
The explosion in the number of sequenced isolate microbial genomes and metagenomes provides the opportunity to use bioinformatic methods for the discovery and analysis of biosynthetic gene clusters. IMG-ABC contains the largest publicly available collection of experimentally identified and predicted BCs. The value of this vast body of data increases by the addition of new features and tools expanding the ability of researchers to explore and interpret these data. Users can now design their own criteria to search for putative BCs using ClusterScout and analyze their findings using similarity networks and hierarchically clustered function heatmaps. Although these tools are currently available for isolate genomes only, in future updates we plan to expand their usage to metagenomes, as improved metagenomic assembly methods and sequencing strategies increase the length of metagenomic contigs and scaffolds. Additionally, we plan to introduce the ability to use other types of protein clusters and functional annotations, such as EC numbers (15) and COG assignments (25) as inputs for ClusterScout-powered BC discovery. Lastly, the content of the IMG-ABC database will be updated to include BC predictions from the latest version of antiSMASH, which not only includes more BC types in the identification algorithm, but also implements ClusterFinder to improve BC boundary prediction (13). IMG-ABC will continue to be provided to the users without the need for registration, making the exploration of global microbial secondary metabolism accessible to the entire scientific community.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Director, Office of Science, Office of Biological and Environmental Research, Life Sciences Division, U.S. Department of Energy [DE-AC02-05CH11231]; National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]. Funding for open access charge: University of California.
Conflict of interest statement. None declared.
REFERENCES
- 1.Piel J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 2.Thomashow L.S., Weller D.M. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F., Rodriguez S., Keasling J.D. Metabolic engineering of microbial pathways for advanced biofuels production. Curr. Opin. Biotechnol. 2011;22:775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling L.L., Schneider T., Peoples A.J., Spoering A.L., Engels I., Conlon B.P., Mueller A., Schäberle T.F., Hughes D.E., Epstein S., et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caboche S. Biosynthesis: bioinformatics bolster a renaissance. Nat. Chem. Biol. 2014;10:798–800. doi: 10.1038/nchembio.1634. [DOI] [PubMed] [Google Scholar]
- 7.Jensen P.R. Natural products and the gene cluster revolution. Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.07.006. doi:10.1016/j.tim.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medema M.H., Fischbach M.A. Computational approaches to natural product discovery. Nat. Chem. Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 10.Cimermančič P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J., et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markowitz V.M., Chen I.-M.A., Chu K., Szeto E., Palaniappan K., Pillay M., Ratner A., Huang J., Pagani I., Tringe S., et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014;42:D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadjithomas M., Chen I.-M.A., Chu K., Ratner A., Palaniappan K., Szeto E., Huang J., Reddy T.B.K., Cimermančič P., Fischbach M.A., et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. mBio. 2015;6 doi: 10.1128/mBio.00932-15. doi:10.1128/mBio.00932-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W., et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb E.C. Recommendations of the Nomenclature Committee of the International Unionof Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. 1992. Enzyme nomenclature 1992. [Google Scholar]
- 16.Škuta C., Bartůněk P., Svozil D. InCHlib – interactive cluster heatmap for web applications. J. Cheminformatics. 2014;6:44. doi: 10.1186/s13321-014-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bangera M.G., Thomashow L.S. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 1999;181:3155–3163. doi: 10.1128/jb.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puranik S., Talkal R., Qureshi A., Khardenavis A., Kapley A., Purohit H.J. Genome sequence of the pigment-producing bacterium Pseudogulbenkiania ferrooxidans, isolated from Loktak lake. Genome Announc. 2013;1 doi: 10.1128/genomeA.01115-13. doi:10.1128/genomeA.01115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vöing K., Harrison A., Soby S.D. Draft genome sequence of Chromobacterium vaccinii, a potential biocontrol agent against mosquito (Aedes aegypti) larvae. Genome Announc. 2015;3 doi: 10.1128/genomeA.00477-15. doi:10.1128/genomeA.00477-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan K.-G., Yunos N.Y.M. Whole-genome sequencing analysis of chromobacterium piscinae strain ND17, a quorum-sensing bacterium. Genome Announc. 2016;4 doi: 10.1128/genomeA.00081-16. doi:10.1128/genomeA.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brucker R.M., Baylor C.M., Walters R.L., Lauer A., Harris R.N., Minbiole K.P.C. The identification of 2,4-diacetylphloroglucinol as an antifungal metabolite produced by cutaneous bacteria of the salamander plethodon cinereus. J. Chem. Ecol. 2007;34:39–43. doi: 10.1007/s10886-007-9352-8. [DOI] [PubMed] [Google Scholar]
- 23.Parker R. The effects of the bacteria, Pf-5, and its metabolite, DAPG, on the innate immune response of drosophila melanogaster. Acad. Excell. Showc. Sched. 2013 http://digitalcommons.wou.edu/aes_event/2013/biol/10 [Google Scholar]
- 24.Meyer S.L.F., Halbrendt J.M., Carta L.K., Skantar A.M., Liu T., Abdelnabby H.M.E., Vinyard B.T. Toxicity of 2,4-diacetylphloroglucinol (DAPG) to plant-parasitic and bacterial-feeding nematodes. J. Nematol. 2009;41:274. [PMC free article] [PubMed] [Google Scholar]
- 25.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]