Abstract
PubChem's BioAssay database (https://pubchem.ncbi.nlm.nih.gov) has served as a public repository for small-molecule and RNAi screening data since 2004 providing open access of its data content to the community. PubChem accepts data submission from worldwide researchers at academia, industry and government agencies. PubChem also collaborates with other chemical biology database stakeholders with data exchange. With over a decade's development effort, it becomes an important information resource supporting drug discovery and chemical biology research. To facilitate data discovery, PubChem is integrated with all other databases at NCBI. In this work, we provide an update for the PubChem BioAssay database describing several recent development including added sources of research data, redesigned BioAssay record page, new BioAssay classification browser and new features in the Upload system facilitating data sharing.
INTRODUCTION
PubChem BioAssay (1–4) is an open access database hosted by the National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH). It started in 2004 serving as a public repository for information generated from chemogenomic, medicinal chemistry and functional genomics research. All data in the database are freely accessible to the public for searching and download. Recent reviews on the community's use of the PubChem resource (5–7) highlighted that the collection of bioactivity and toxicity data in PubChem BioAssay has greatly supported research in several fields such as medicinal chemistry, drug discovery, pharmaceutical genomics and informatics research. Small molecule data in PubChem BioAssay are cross-linked to chemical structures via the referenced samples in the assay. The PubChem BioAssay database is also linked to other biomedical and literature databases hosted at NCBI such as PubMed, Protein, Gene, Taxonomy etc. Metadata in the database are integrated with the NCBI's search engine, Entrez, making the PubChem BioAssay database accessible by interactive keyword search using the web interface and by programmatic retrieval via E-Utilities. Assay data can also be retrieved and analyzed via web-based and programmatic tools provided by PubChem. An update for the services and their URLs for accessing, searching, downloading and analyzing PubChem BioAssay data is provided in Table 1. Most of the web based services can also be accessed at https://pubchem.ncbi.nlm.nih.gov/assay/.
Table 1. A list of PubChem BioAssay services.
| Service | Description | URL example |
|---|---|---|
| BioAssay Record Page | Access and download a bioassay record | https://pubchem.ncbi.nlm.nih.gov/bioassay/805 |
| BioAssay Search | Search BioAssay Database with Entrez | https://www.ncbi.nlm.nih.gov/pcassay/ |
| BioAssay Search, Advanced page | An interface for searching multiple search fields | https://www.ncbi.nlm.nih.gov/pcassay/limits |
| An interface for reviewing search history and refining search results with Boolean operation | https://www.ncbi.nlm.nih.gov/pcassay/advanced | |
| PubChem Upload | Substance and BioAssay submission system | https://pubchem.ncbi.nlm.nih.gov/upload/ |
| BioAssay FTP | FTP for all PubChem BioAssay records and related information | ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/ |
| BioAssay Data Standard | XML Data specification for PubChem BioAssay data model | ftp://ftp.ncbi.nlm.nih.gov/pubchem/data_spec/ |
| BioAssay Service Home | BioAssay Service Home | https://pubchem.ncbi.nlm.nih.gov/assay/ |
| BioAssay Classification | Browse BioAssay classification tree | https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?p=classification |
| Bioactivity Data Tool | Retrieve a full data table from a single bioassay record | https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?aid=1811 |
| Retrieve and download cross-assay bioactivity data for a single substance sample (SID), chemical structure (CID), protein target (GI, UniProt or GenBank accession), gene target (GeneID) or publication (PMID) | https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?sid=103164874 | |
| https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?cid=2244 | ||
| https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?gi=29725609 | ||
| https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?uniprot=P00533 | ||
| https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?ncbiacc=NP_005219 | ||
| https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?geneid=1956 | ||
| https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?pmid=25728019 | ||
| Bioassay Download Tool | A flexible download interface | https://pubchem.ncbi.nlm.nih.gov/assay/assaydownload.cgi |
| PubChem PUG/REST/SOAP | Programmatic tool and REST api for data retrieval | https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html |
| https://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html | ||
| PubChem Widget Help | PubChem widgets enable you to display PubChem data in your pages | https://pubchem.ncbi.nlm.nih.gov/widget/docs/widget_help.html |
| Structure-Activity Analysis (SAR) | Analyze and visualize Structure-Activity relationship with clustering tools and a heatmap-style display | https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?p=heat |
| Dose-response Curve Tool | Analyze bioassay test results and visualize dose-response curve | https://pubchem.ncbi.nlm.nih.gov/assay/plot.cgi?plottype=1 |
| Scatter Plot/Histogram | Analyze bioassay test results with histogram or scatter plot | https://pubchem.ncbi.nlm.nih.gov/assay/plot.cgi?plottype=2 |
| Related BioAssays | Summarize bioassay relationship by: same assay project, overlap of active compounds, overlap of active gene, target sequence similarity, deposited annotation, same publication and gene interaction | https://pubchem.ncbi.nlm.nih.gov/bioassay/1510#section=Same-Project-BioAssays |
| https://pubchem.ncbi.nlm.nih.gov/bioassay/1510#section=Related-BioAssays | ||
| BioActivity Summary - Compound-centric | Summarize and analyze bioactivity data for a set of records, presented from the compound point of view | https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.cgi?tab=1 |
| BioActivity Summary - Assay-centric | Summarize and analyze bioactivity data for a set of records, presented from the assay point of view | https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.cgi?tab=2 |
| BioActivity Summary - Target-centric | Summarize and analyze bioactivity data for a set of records, presented from the target point of view | https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.cgi?tab=3 |
With continuous development towards supporting open data during the past 12 years, the PubChem BioAssay database is committed to meet the increasing need from the community for information archival, retrieval and mining. PubChem BioAssay stays as a leading repository of research data pertaining to drug discovery by: (i) supporting broad types of bioactivity information with an optimized and flexible data model; (ii) maintaining steady enhancement of database infrastructure and scalability; (iii) utilizing new technology for data archival, viewing, indexing, search and download; (iv) enhancing data upload system; (v) integrating with other biomedical resources. In this work, we provide an update on several aspects of the information resource, including data content and data sources growth, database infrastructure consolidation, the redesigned and widgetized BioAssay record page, new BioAssay classification browser and added features for previously provided web services. Enhanced management for assay data embargo, release and sharing of on-hold data by the PubChem Upload system is also described.
BioAssay DATA
The PubChem BioAssay database currently contains over one million records holding 230 000 000 bioactivity outcomes deposited by over 80 organizations (data sources) across the world. The data content for the time period of 2004–2013 and 2014–2016 is given in Table 2. High-throughput screening (HTS) data were provided by laboratories and screening centers from academic institutes, universities, government organizations as well as pharmaceutical companies including those participated the HTS campaigns for the development of chemical probes funded by the NIH Molecular Libraries Program (8,9). The database also contains literature-based data including both publication driven depositions submitted by journal authors and those contributed by PDBbind (10), IUPHAR (11), BindingDB (12), ChEMBL (13) and other literature curation projects. Additionally, third-party annotations with community adopted vocabulary and ontology (13–15) for the assay data in PubChem, such as assay format, assay type, detection method and cell line are collected and linked to the pertinent data sets, which are shown in the BioAssay record page and used for searching and filtering assay data in Entrez and data analysis tools.
Table 2. PubChem BioAssay statistics.
| Total | Chemical assays | RNAi assays | |||
|---|---|---|---|---|---|
| 2004–2013 | 2014–present | 2004–2013 | 2014–present | ||
| assay records (AID) | 1 218 687 | 737 994 | 480 616 | 57 | 48 |
| substance samples (SID) | 3 576 066 | 2 755 032 | 1 396 693 | 213 030 | 293 499 |
| chemical structures (CID) | 2 283 533 | 1 956 998 | 986 237 | - | - |
| bioactivity outcomes | 231 303 607 | 222 198 148 | 8 764 075 | 701 993 | 419 081 |
| data points | 1 514 223 504 | 1 403 289 248 | 100 451 032 | 9 404 999 | 7 473 465 |
| species | 3543 | 2730 | 1895 | 6 | 2 |
| protein targets | 10 636 | 7450 | 6972 | - | - |
| protein targets (human) | 4771 | 3378 | 3495 | - | - |
| gene targets | 55 714 | - | - | 38 694 | 52 986 |
| gene targets (human) | 24 888 | - | - | 24 460 | 22 656 |
| gene targets (phenotype) | 15 866 | - | - | 12 816 | 4524 |
Nearly one-third of the assay data sources were added in the past three years. The majority of these data sources deposited RNAi data to PubChem along with manuscript submission to journals supporting open access for functional genomic research. Nature Cell Biology led this effort by recommending RNAi data deposition to public repository, which recently also started to call small molecule data sharing. Other open access journals, such as PLoS One, joined the force lately for recommending data sharing via public repository and brought in the deposition of several small molecule data sets in PubChem (16,17). Assay data contributed by these sources are linked to the respective publications indexed in PubMed (16–36), allowing PubChem users to access the articles for additional information, and vice versa, PubMed users also gain access to the research data in the BioAssay archive supporting machine readable format. All BioAssay depositors and the associated information, such as affiliations, summary of data submissions may be viewed at the PubChem Data Source page at: https://pubchem.ncbi.nlm.nih.gov/sources/, whereas data sources are grouped by geographic location and various other categories. One may access the submissions of a specific depositor by following the Substance or BioAssay record counts presented under the ‘Data Counts by Type’ field.
The significant growth of the BioAssay database requires a robust and scalable database system. A set of relational databases and tables are set up to: (i) archive bioassay submissions, track update and provide version control; (ii) maintain data embargo and release status; (iii) record and derive links and relationships among assays and other biomedical information; (iv) store third-party annotations and link to the respective data sets; (v) provide search indexes; (vi) support data retrieval for web display, rest API and data analysis tools; (vi) facilitate daily update for BioAssay FTP. Many efforts have been invested to enhance the database infrastructure. Additional mechanisms were implemented for tracking information about BioAssay protein and gene targets. Mappings between NCBI protein GI number, GenBank accessions and UniProt ID were created to facilitate data retrieval and integration. These efforts were made to facilitate: (i) bioassay submission; (ii) integration of deposited small-molecule and RNAi data; (iii) integration of biological annotations for proteins and genes from public biomedical databases; (iv) access to the bioactivity data in PubChem using non-NCBI sequence identifier of the assay target; (v) and development of new PubChem services to enhance BioAssay target search; (vi) improve discoverability of the biological data in the database.
NEW WEB FEATURES AND SERVICES
PubChem BioAssay provides web-based and programmatic tools for data search, access, analysis and download. Several recently developed web services are described here for improving assay data search and navigation by classifying deposited metadata and third-party annotations.
BioAssay record page
PubChem provides a full access to each deposited BioAssay record. The PubChem BioAssay Record page, replacing the legacy Summary page, has been revamped to streamline data flow, support data and service reutilization and unify the web page presence across the PubChem resource. Taking the advantage of new web technology, the data-driven interface was designed and optimized for both touch- and mouse-based devices with a similar approach for the recent revamp of the PubChem Compound Summary page and the Substance Record page (37). The widgetized web page consisting of multiple sections automatically adapts to the available screen size with a responsive design, making it friendly for navigating page content and reviewing information with desktops, tablets and mobile phones. Furthermore, the new design provides an ability to embed any section or subsection of the page as a widget in another web page without the need of separate codebase eliminating the burden of maintenance by a third-party, which is greatly beneficial to non-PubChem resources that are interested in integrating PubChem BioAssay data. Information and instruction about embedding the PubChem widget are available at https://pubchem.ncbi.nlm.nih.gov/widget/docs/widget_help.html.
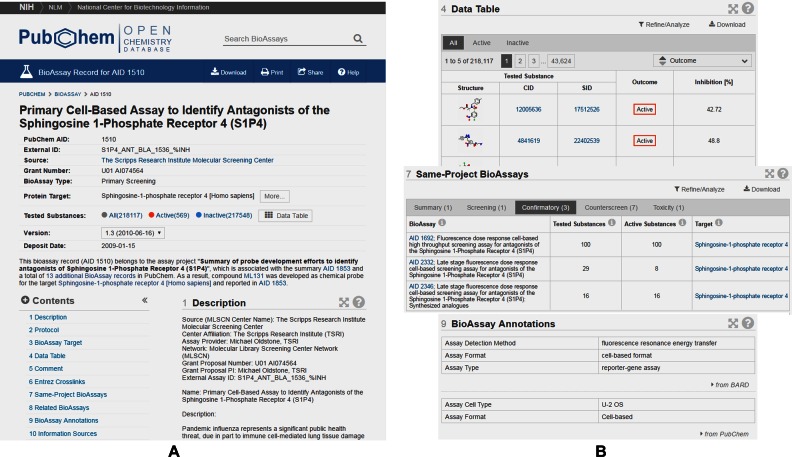
A deposited BioAssay record can be accessed by an AID number, the primary accession. An example for a data set reporting an assay for identifying antagonists of the Sphingosine 1-Phosphate Receptor 4 (AID: 1510; https://pubchem.ncbi.nlm.nih.gov/bioassay/1510) is shown in Figure 1. The BioAssay Record page provides a primary access with version control to the initial submission and all subsequent updates of an assay. It also provides third-party annotations and links to tools supporting data analysis and download. The ‘Download’ button on top allows one to download depositor-provided metadata and assay result, as well as chemical structures for tested small-molecule samples. The table of contents can be expanded for quick navigation to each individual section. Full data set is retrieved by default at the Data Table section. Additionally, the assay data table may be partitioned according to activity outcomes (e.g. active, inactive, or subset with micromolar activity or nanomolar activity), allowing users to quickly filter, select and download the results of interest. To facilitate hit evaluation, data comparison and target identification, the structure image of a small molecule sample links to the specific bioactivity analysis tool that shows all available across-assay data for the compound through CID (PubChem Compound accession for unique chemical structure). Similarly, for RNAi assay (e.g. https://pubchem.ncbi.nlm.nih.gov/bioassay/1904), the gene ID under the gene target column links to a specific bioactivity analysis tool that summarizes all RNAi data as well as small molecule targeting the gene, allowing one to discovery small-molecule tools, compare results with other research and identify biological functionalities suggested by other RNAi screens. The display of depositor submitted information is followed by sections presenting related BioAssay data sets from the same or multiple projects. The presentation of depositor provided related assay data sets helps one to track the development of an assay project, and facilitate data validation and interpretation, across-assay comparison when combined with the target and publication based related assay data sets derived by PubChem. An excerpt at the top of the record page prompts the availability of such related data highlighting the importance of data integration for assay data interpretation and reutilization. Third-party annotations are presented as the last section of the page together with indication of the sources.
Figure 1.
A bioassay record (AID 1510, https://pubchem.ncbi.nlm.nih.gov/bioassay/1510). (A) The overview of the record page. The table of contents provides quick navigation to a list of sections shown on the page. Each section has an anchor and its URL can be used for widget embedding. (B) Selected sections: Data Table, Same-Project BioAssays and BioAssay Annotations.
BioAssay classification tool
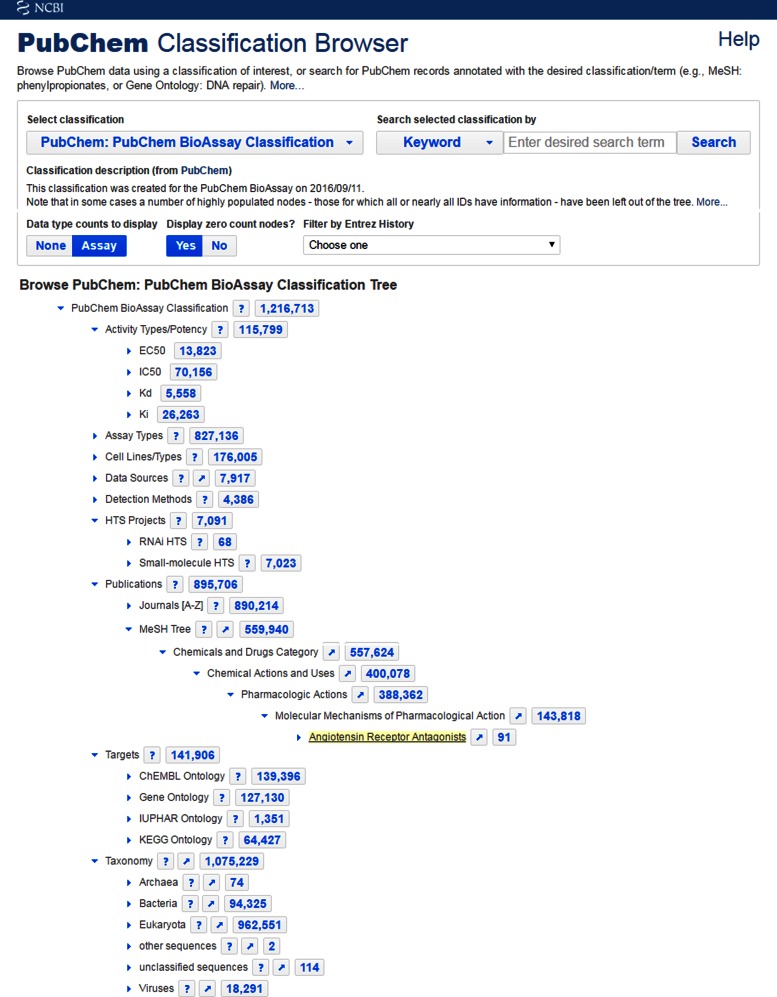
A hierarchical tree view is developed upon the software frame of the PubChem Classification Browser providing an additional approach to browse, search and access the BioAssay data. The tool (available at https://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?p=classification and https://pubchem.ncbi.nlm.nih.gov/classification/#hid=80) offers an overview of the assay data sets with attributes of common interest by putting together assay metadata, third-party annotations, classifications of assay target, taxonomy and associated publications. It organizes the BioAssay records with nodes in the hierarchy presenting various classifications and annotations. This tool allows one to browse the distribution of BioAssay data among nodes in the hierarchy of interest, aggregate information for a particular sub-class and perform specific search by a data source, assay type etc. (Figure 2). The counts shown with nodes in the hierarchical display link to assay data entries indexed in Entrez upon clicking. Individual assay lists sent from the classification browser can be combined using the Entrez's query refining functionality, providing a powerful way to drill down to the desired data sets satisfying multiple search criteria in one's mind. As examples of usage, users can drill down to IC50 or Kd data under ‘Activity Types/Potency’, or to cell-based or biochemical data under ‘Assay Types’. Users can also browse the over 7000 data sets from HTS projects under ‘HTS Projects’, including both genomic wide RNAi screenings and those chemical probe development projects funded by the NIH Molecular Libraries Program.
Figure 2.
The PubChem BioAssay Classification Tree (https://pubchem.ncbi.nlm.nih.gov/classification/#hid=80). A hierarchical display is provided, which can be navigated and explored by expanding to the sub-trees upon clicking on the triangle icon  . A click on the numbers on a node (showing the count of BioAssay records with that annotation) leads to a report in Entrez for the associated assay records.
. A click on the numbers on a node (showing the count of BioAssay records with that annotation) leads to a report in Entrez for the associated assay records.
The ‘Publications’ branch allows one to browse assay data sets by journal name and publication year of the associated primary citation. More importantly, the tool allows one to search data in BioAssay by research subject based on the classification of the PubMed citations with controlled vocabulary of biomedical terms provided in MeSH. As an example shown in Figure 2, one can follow the branch ‘Chemical and Drugs Category’ under the ‘MeSH Tree’ to retrieve the assay data entries reporting bioactivity for angiotensin receptor antagonists, which link to the abstracts of the publications in PubMed. For the ‘Targets’ branch in the tool, four types of protein ontologies and classifications have been incorporated for assay targets including ChEMBL, GO, IUPHAR and KEGG. Recording taxonomy ID, one of the key metadata fields in the BioAssay data model, enables the organization of the BioAssay data sets by biological organisms to provide a hierarchical display of the assay data sets based on the taxonomy classifications maintained at NCBI, whereas the ‘Taxonomy’ tree in the classification tool resembles a subset of the NCBI Taxonomy tree with taxonomy nodes having assay data.
New features for bioactivity analysis tool
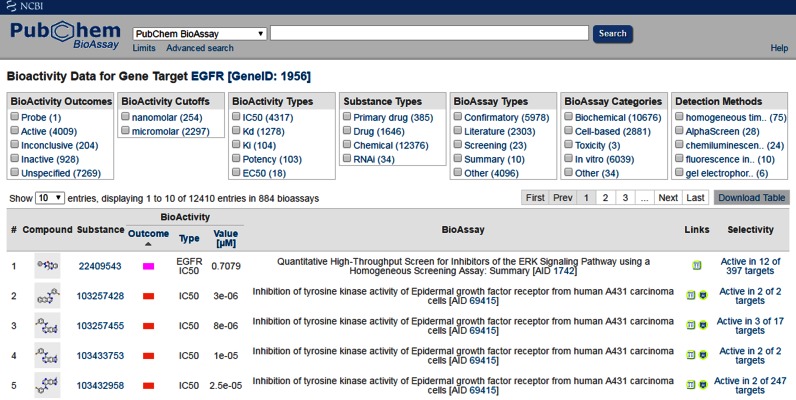
PubChem BioAssay allows several ways to reference protein and gene targets. An assay target is often linked to many small molecule samples tested in hundreds of assays making it important to combine the information for identifying chemical tools with proper selectivity assessment. Bioactivity analysis tools were previously developed for retrieving and aggregating dynamically across-assay bioactivity data for a protein or gene target. With the growth of BioAssay submissions, it is critical to provide selection functionality to slice and dice the information. In addition, for therapeutic targets, it is essential to indicate the drugs that were primarily developed for treating disease via the query target. Moreover, it is important to emphasize all drug molecules tested against the query target to facilitate the identification of alternative therapeutic effects of these drugs toward drug repositioning. Several new selection functionality for filtering bioactivity data were added and upgraded to these tools by incorporating recently obtained assay annotations, which enables the retrieval of assay data from a particular detection method, the selection of toxicity result or subsetting bioactivity data from drug molecules (Figure 3, https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?geneid=1956). As another feature, the ‘Selectivity’ column is added to assist target selectivity assessment for the tested chemical sample, which counts the total unique targets that a chemical sample was tested against and the number of targets it was active for. In addition to taking gene ID, this bioactivity analysis tool also accepts GenBank accession or UniProt ID for specifying the query target, SID or CID for specifying the tested samples (RNAi reagents or small molecules), PMID for specifying the literature that contains assay data. The service is also optimized to support programmatic download of bioactivity data with filtering functionality.
Figure 3.
Collective and cross-assay bioactivity data for a specific gene target in the PubChem BioAssay database (https://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.html?geneid=1956). The filters at the top of the web page may be used to drill down to a subset of interest. For example, using the filters provided under the ‘Substance Types’ section, one may retrieve the RNAi data by clicking on ‘RNAi (34)’. The ‘Primary drug (385)’ filter in the section allows the retrieval of the bioactivity data for drugs that were developed to specifically target the query protein/gene, while the ‘Drug (1646)’ filter retrieves bioactivity for any drugs in general which were tested in the assays. This latter filter allows one to identify drug molecules that show experimental evidence (based on PubChem BioAssay data) for binding or affecting the query protein/gene target so that their potential for drug repositioning (against the query protein/gene target) may be further explored. Drug and target information supporting these two filters were obtained from annotations in the DrugBank.
PUBLIC ACCESS, SEARCH, DOWNLOAD
PubChem provides multiple means to access, search and download the BioAssay data including the BioAssay Record page and classification tool described above. PubChem BioAssay is indexed in Entrez (https://www.ncbi.nlm.nih.gov/pcassay/) under numerous fields to support keyword search. It is cross-linked with several other biomedical databases such as with PubMed via the provided citation in the assay submission, or to NCBI Gene via the assay target specification. As a result, users of genomic information in Entrez may retrieve biological test result in PubChem relating to the gene target, and PubMed users can go to BioAssay to retrieve data as discussed in a publication.
BioAssay data can be downloaded using: (i) download functions in the BioAssay Record page supporting ASN, XML, JSON and CSV formats; (ii) A web-based service for bulk download at https://pubchem.ncbi.nlm.nih.gov/assay/assaydownload.cgi taking AID list and optionally SID (PubChem Substance accession) list; (iii) programmatic tools provided by NCBI's E-Utilities (https://www.ncbi.nlm.nih.gov/books/NBK25497/), PubChem PUG/SOAP (https://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html) and PUG/REST (https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html) (38), which provide great flexibility for retrieving AID specific metadata and assay result, database links and bioactivity data across multiple assays for a compound or target; (iv) Daily updated PubChem BioAssay FTP at ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/, primarily providing open access to all deposited BioAssay records via ASN, XML, JSON and CSV formats. New information is added under the directory ‘Extras’ at BioAssay FTP including: (i) the file ‘Aid2Annotation’ providing third party annotations in the tag/value structure; (ii) the file ‘Aid2GiGeneidAccessionUniprot’ containing AID and the mapping of target identifiers between protein GI, GenBank accession, UniProt ID and Gene ID); (iii) a subfolder ‘VendorCatalogs’ containing files for several RNAi product vendors with mapping between vendor catalog ID and the assigned PubChem SID for the RNAi sample.
PubChem UPLOAD FOR BioAssay SUBMISSION
As a public repository accepting submission of complex research data, a robust and user friendly deposition system plays a key role. User interface for the PubChem Upload system (https://pubchem.ncbi.nlm.nih.gov/upload/) has been continuously optimized. Several features are now added for managing data embargo, release and sharing on-hold data among depositor maintained user group. PubChem allows data embargo to provide depositors the needed time for getting research published or completing patent application. Only Upload account holders could have a full access to a BioAssay record under embargo previously. A newly developed mechanism now provides a full access privilege to collaborators, journal reviewers or editors via a specifically requested URL. Upload now supports bulk release of embargoed BioAssay records by taking a list of BioAssay accessions (AID). In addition, it streamlines the release of the BioAssay records and related Substance records so that once depositors request a list of AID to be released, the associated on-hold Substance records are looked up and released automatically by the Upload system. A FAQ section (https://pubchem.ncbi.nlm.nih.gov/upload/docs/upload_faq.html) is added to the Upload help page (https://pubchem.ncbi.nlm.nih.gov/upload/docs/upload_help.html) providing quick tips for common questions and update operations.
SUMMARY
PubChem started in 2004 as a public repository for biological data from small molecule and RNAi screenings. As of today over 80 organizations and laboratories across the world have shared research data via PubChem BioAssay. There are many challenges for developing and maintaining public repositories. The community have put force together for critical thoughts to define guidance for desirable data management. Recently, the FAIR (Findability, Accessibility, Interoperability and Reusability) principle has been proposed to provide guidance for managing public data, maintaining data flow and sharing analysis tools and pipelines. This effort is to bring clarity and encourage public data stakeholders to work toward the simple guidance together with funding agency, researcher and publisher to harmonize research data and maximize the value of scholarly digital publishing.
The PubChem BioAssay repository has been designed and developed largely complying with the FAIR principle when reviewed retrospectively. The BioAssay data model was designed with machine readability and all data in the database is freely available to the community. The assay data can be searched using the NCBI Entrez system: https://www.ncbi.nlm.nih.gov/pcassay/. PubChem provides additional tools to support data search, access and analysis. Many add-on services and tools are developed by the community to extend and complement the functionality of the PubChem resource and to provide additional annotations to the data content in PubChem (5). The interplay between PubChem and the community's efforts are mutual beneficial. The information platform at PubChem is under continuous development to encourage re-use of the cheminformatics, chemical biology and functional genomics research data in PubChem, and to enable and ease the integration by community's effort. PubChem will continue to improve services and tools as technology advances, integrate with third-party annotations and other public biomedical data, and work with funding agencies and publishers supporting research data archiving and reutilization. PubChem welcomes the community to share the resource and contribute to the repository.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM). The authors thank all submitters who have contributed data to PubChem.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH); National Library of Medicine (NLM). Funding for open access charge: National Center for Biotechnology Information (NCBI).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang Y., Suzek T., Zhang J., Wang J., He S., Cheng T., Shoemaker B.A., Gindulyte A., Bryant S.H. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–D1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Zhou Z., Han L., Karapetyan K., Dracheva S., Shoemaker B.A., et al. PubChem's BioAssay database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Bolton E., Dracheva S., Karapetyan K., Shoemaker B.A., Suzek T.O., Wang J., Xiao J., Zhang J., Bryant S.H. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng T., Pan Y., Hao M., Wang Y., Bryant S.H. PubChem applications in drug discovery: a bibliometric analysis. Drug Discov. Today. 2014;19:1751–1756. doi: 10.1016/j.drudis.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qader A.A., Urraca J., Torsetnes S.B., Tonnesen F., Reubsaet L., Sellergren B. Peptide imprinted receptors for the determination of the small cell lung cancer associated biomarker progastrin releasing peptide. J. Chromatogr. A. 2014;1370:56–62. doi: 10.1016/j.chroma.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H., Zhang J., Kim M.T., Boison A., Sedykh A., Moran K. Big data in chemical toxicity research: the use of high-throughput screening assays to identify potential toxicants. Chem. Res. Toxicol. 2014;27:1643–1651. doi: 10.1021/tx500145h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Austin C.P., Brady L.S., Insel T.R., Collins F.S. NIH molecular libraries initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 9.Schreiber S.L., Kotz J.D., Li M., Aube J., Austin C.P., Reed J.C., Rosen H., White E.L., Sklar L.A., Lindsley C.W., et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell. 2015;161:1252–1265. doi: 10.1016/j.cell.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z., Li Y., Han L., Li J., Liu J., Zhao Z., Nie W., Liu Y., Wang R. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics. 2015;31:405–412. doi: 10.1093/bioinformatics/btu626. [DOI] [PubMed] [Google Scholar]
- 11.Southan C., Sharman J.L., Benson H.E., Faccenda E., Pawson A.J., Alexander S.P., Buneman O.P., Davenport A.P., McGrath J.C., Peters J.A., et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016;44:D1054–D1068. doi: 10.1093/nar/gkv1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bento A.P., Gaulton A., Hersey A., Bellis L.J., Chambers J., Davies M., Kruger F.A., Light Y., Mak L., McGlinchey S., et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visser U., Abeyruwan S., Vempati U., Smith R.P., Lemmon V., Schurer S.C. BioAssay Ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinformatics. 2011;12:257. doi: 10.1186/1471-2105-12-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howe E.A., de Souza A., Lahr D.L., Chatwin S., Montgomery P., Alexander B.R., Nguyen D.T., Cruz Y., Stonich D.A., Walzer G., et al. BioAssay Research Database (BARD): chemical biology and probe-development enabled by structured metadata and result types. Nucleic Acids Res. 2015;43:D1163–D1170. doi: 10.1093/nar/gku1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crowther G.J., Hillesland H.K., Keyloun K.R., Reid M.C., Lafuente-Monasterio M.J., Ghidelli-Disse S., Leonard S.E., He P., Jones J.C., Krahn M.M., et al. Biochemical screening of five protein kinases from Plasmodium falciparum against 14 000 cell-active compounds. PloS One. 2016;11:e0149996. doi: 10.1371/journal.pone.0149996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z., Bhattacharya A., Ivanov D.N. Identification of small-molecule inhibitors of the HuR/RNA interaction using a fluorescence polarization screening assay followed by NMR validation. PloS One. 2015;10:e0138780. doi: 10.1371/journal.pone.0138780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swenson J.M., Colmenares S.U., Strom A.R., Costes S.V., Karpen G.H. The composition and organization of Drosophila heterochromatin are heterogeneous and dynamic. eLife. 2016;5:e16096. doi: 10.7554/eLife.16096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voter A.F., Manthei K.A., Keck J.L. A high-throughput screening strategy to identify protein-protein interaction inhibitors that block the Fanconi Anemia DNA repair pathway. J. Biomol. Screen. 2016;21:626–633. doi: 10.1177/1087057116635503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J., Li N., Oh K.S., Dutta B., Vayttaden S.J., Lin B., Ebert T.S., De Nardo D., Davis J., Bagirzadeh R., et al. Comprehensive RNAi-based screening of human and mouse TLR pathways identifies species-specific preferences in signaling protein use. Science Signal. 2016;9:ra3. doi: 10.1126/scisignal.aab2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt C.K., Galanty Y., Sczaniecka-Clift M., Coates J., Jhujh S., Demir M., Cornwell M., Beli P., Jackson S.P. Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat. Cell Biol. 2015;17:1458–1470. doi: 10.1038/ncb3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin R., Elf S., Shan C., Kang H.B., Ji Q., Zhou L., Hitosugi T., Zhang L., Zhang S., Seo J.H., et al. 6-Phosphogluconate dehydrogenase links oxidative PPP, lipogenesis and tumour growth by inhibiting LKB1-AMPK signalling. Nat. Cell Biol. 2015;17:1484–1496. doi: 10.1038/ncb3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmich F., Szczurek E., Kreibich S., Dilling S., Andritschke D., Casanova A., Low S.H., Eicher S., Muntwiler S., Emmenlauer M., et al. gespeR: a statistical model for deconvoluting off-target-confounded RNA interference screens. Genome Biol. 2015;16:220. doi: 10.1186/s13059-015-0783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lang L., Ding H.F., Chen X., Sun S.Y., Liu G., Yan C. Internal ribosome entry site-based bicistronic in situ reporter assays for discovery of transcription-targeted lead compounds. Chem. Biol. 2015;22:957–964. doi: 10.1016/j.chembiol.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupte A., Baker E.K., Wan S.S., Stewart E., Loh A., Shelat A.A., Gould C.M., Chalk A.M., Taylor S., Lackovic K., et al. Systematic screening identifies dual PI3K and mTOR inhibition as a conserved therapeutic vulnerability in osteosarcoma. Clin. Cancer Res. 2015;21:3216–3229. doi: 10.1158/1078-0432.CCR-14-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telford B.J., Chen A., Beetham H., Frick J., Brew T.P., Gould C.M., Single A., Godwin T., Simpson K.J., Guilford P. Synthetic lethal screens identify vulnerabilities in GPCR signaling and cytoskeletal organization in E-cadherin-deficient cells. Mol. Cancer Ther. 2015;14:1213–1223. doi: 10.1158/1535-7163.MCT-14-1092. [DOI] [PubMed] [Google Scholar]
- 27.Pasetto M., Antignani A., Ormanoglu P., Buehler E., Guha R., Pastan I., Martin S.E., FitzGerald D.J. Whole-genome RNAi screen highlights components of the endoplasmic reticulum/Golgi as a source of resistance to immunotoxin-mediated cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E1135–E1142. doi: 10.1073/pnas.1501958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pena I., Pilar Manzano M., Cantizani J., Kessler A., Alonso-Padilla J., Bardera A.I., Alvarez E., Colmenarejo G., Cotillo I., Roquero I., et al. New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource. Sci. Rep. 2015;5:8771. doi: 10.1038/srep08771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyati S., Schinske-Sebolt K., Pitchiaya S., Chekhovskiy K., Chator A., Chaudhry N., Dosch J., Van Dort M.E., Varambally S., Kumar-Sinha C., et al. The kinase activity of the Ser/Thr kinase BUB1 promotes TGF-beta signaling. Science Signal. 2015;8:ra1. doi: 10.1126/scisignal.2005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shan C., Elf S., Ji Q., Kang H.B., Zhou L., Hitosugi T., Jin L., Lin R., Zhang L., Seo J.H., et al. Lysine acetylation activates 6-phosphogluconate dehydrogenase to promote tumor growth. Mol. Cell. 2014;55:552–565. doi: 10.1016/j.molcel.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudryavtsev D., Makarieva T., Utkina N., Santalova E., Kryukova E., Methfessel C., Tsetlin V., Stonik V., Kasheverov I. Marine natural products acting on the acetylcholine-binding protein and nicotinic receptors: from computer modeling to binding studies and electrophysiology. Marine Drugs. 2014;12:1859–1875. doi: 10.3390/md12041859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costantino L., Sotiriou S.K., Rantala J.K., Magin S., Mladenov E., Helleday T., Haber J.E., Iliakis G., Kallioniemi O.P., Halazonetis T.D. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowther G.J., Booker M.L., He M., Li T., Raverdy S., Novelli J.F., He P., Dale N.R., Fife A.M., Barker R.H., Jr, et al. Cofactor-independent phosphoglycerate mutase from nematodes has limited druggability, as revealed by two high-throughput screens. PLoS Negl. Trop. Dis. 2014;8:e2628. doi: 10.1371/journal.pntd.0002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falkenberg K.J., Gould C.M., Johnstone R.W., Simpson K.J. Genome-wide functional genomic and transcriptomic analyses for genes regulating sensitivity to vorinostat. Sci. Data. 2014;1:140017. doi: 10.1038/sdata.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasson S.A., Kane L.A., Yamano K., Huang C.H., Sliter D.A., Buehler E., Wang C., Heman-Ackah S.M., Hessa T., Guha R., et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013;504:291–295. doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George A.J., Purdue B.W., Gould C.M., Thomas D.W., Handoko Y., Qian H., Quaife-Ryan G.A., Morgan K.A., Simpson K.J., Thomas W.G., et al. A functional siRNA screen identifies genes modulating angiotensin II-mediated EGFR transactivation. J. Cell Sci. 2013;126:5377–5390. doi: 10.1242/jcs.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S., Thiessen P.A., Bolton E.E., Bryant S.H. PUG-SOAP and PUG-REST: web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015;43:W605–W611. doi: 10.1093/nar/gkv396. [DOI] [PMC free article] [PubMed] [Google Scholar]