Abstract
NCBI's Conserved Domain Database (CDD) aims at annotating biomolecular sequences with the location of evolutionarily conserved protein domain footprints, and functional sites inferred from such footprints. An archive of pre-computed domain annotation is maintained for proteins tracked by NCBI's Entrez database, and live search services are offered as well. CDD curation staff supplements a comprehensive collection of protein domain and protein family models, which have been imported from external providers, with representations of selected domain families that are curated in-house and organized into hierarchical classifications of functionally distinct families and sub-families. CDD also supports comparative analyses of protein families via conserved domain architectures, and a recent curation effort focuses on providing functional characterizations of distinct subfamily architectures using SPARCLE: Subfamily Protein Architecture Labeling Engine. CDD can be accessed at https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
CDD SCOPE AND COVERAGE
The current live CDD version, v3.15, contains 48 963 protein- and protein domain-models, with content obtained from Pfam (1), SMART (2), the COGs collection (3), TIGRFAMS (4), the NCBI Protein Clusters collection (5), and NCBI's in-house data curation effort (6). CDD version v3.16 is scheduled to be released in late 2016, it will include the most recent release of Pfam, version 30 and a total of 50 369 protein and protein-domain models. For CDD v3.16, the fixed assumed size of the domain model database has been increased to match the current size of the model collection, resulting in slightly higher E-values reported by RPS-BLAST. CDD maintains a fixed model database size for E-value computation so that it becomes possible to incrementally update domain annotation without the need to re-compute existing annotation. The increase in the database size parameter will suppress previously reported annotation at the borderline of significance, as the default E-value threshold has not been adjusted accordingly.
Several classifications for large, common, and functionally diverse domain families have recently been updated or added to CDD, such as for the G-protein-coupled receptors (cd14964*), EF-hand domains (cd15900*), MBL-like metallo-hydrolases (cd06262), haloacid-dehalogenases (cd01427*), RING-finger domains (cd00162*), SPRY domains (cd11709), alkaline phosphatases and sulfatases (cd00016), pleckstrin homology domains (cd00900), myosin and kinesin motor domains (cd01363) or metallophosphatases (cd00838) (* available in CDD v3.16).
CDD is part of NCBI's Entrez search and retrieval system and is cross-linked with other databases such as Entrez/protein, Entrez/Gene, 3D-structure (Molecular Modeling Database, MMDB), NCBI BioSystems, PubMed, and PubChem. Domain and site annotation generated via CDD is visible in flat-file and graphical views of protein sequences in Entrez. Currently, CDD annotates ∼250 million sequences in Entrez/protein, about 80% of the proteins excluding sequences from environmental sampling. CDD annotates 96% of structure-derived protein sequences in Entrez that are over 30 residues long.
CDD curators annotate functional sites on NCBI-curated models, such as active sites and binding sites, which are mapped onto protein (query) sequences. Currently, a total of 29 991 site annotations have been created on 10 605 out of 12 805 NCBI-curated domain models*. Conserved sequence patterns have been recorded for 2123 of these site annotations, and their mapping onto query sequences is contingent on pattern matches.
AVAILABILITY AND DATA SHARING
Table 1 lists a set of URLs for services provided by CDD. The RPS-BLAST program is included in NCBI's BLAST software distribution, and pre-computed search databases are available via CDD's FTP site, so that conserved domain searches can be run locally. We have developed a utility, called ‘rpsbproc’, also distributed via CDD's FTP-site, which can be installed locally as a wrapper for RPS-BLAST in order to provide results that match those computed by NCBI's on-line search services (7), including site annotation and the location of conserved domain superfamily footprints.
Table 1. URLs and other resources associated with the CDD project.
| URL | Description |
|---|---|
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation |
| https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi | BATCH CD-Search interface utilizing the RPS-BLAST algorithm and the model database, and to the CDART database of pre-computed domain annotation. Up to 4,000 protein queries may be submitted per request |
| https://www.ncbi.nlm.nih.gov/cdd | Entrez interface to CDD |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml | CDD project home page |
| https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi | CDART domain architecture viewer |
| ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd | CDD FTP site, see README file for content |
| https://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml | Domain hierarchy editor/viewer and protein structure/alignment viewer |
| ftp://ftp.ncbi.nlm.nih.gov/toolbox; executables can be obtained from: https://www.ncbi.nlm.nih.gov/BLAST/download.shtml | RPS-BLAST stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution |
| https://www.ncbi.nlm.nih.gov/sparcle | Entrez interface to SPARCLE (Subfamily Protein Architecture Labeling Engine) |
The CDD group has started a collaboration with the InterPro group at the European Bioinformatics Institute to supplement sequence annotation provided by InterPro with data that are uniquely provided by NCBI's CDD curation effort, including protein domain models for very specific subfamilies and the annotation of functional sites. To date, >1000 domain signatures provided by CDD have been integrated by InterPro (8).
SUBFAMILY DOMAIN ARCHITECTURES AND SPARCLE
Since its inception, the facility of the Conserved Domain Database was enhanced by the CDART (Conserved Domain Architecture Retrieval Tool) service (9), which groups proteins in the Entrez database by common domain architecture. As CDD is a redundant collection, protein domain models are collapsed into domain superfamilies for the sake of defining domain architectures in CDART. The CDART domain architecture viewer is a tool for the comparative analysis of domain architectures by listing architectures that are most similar to that of a query. It also provides powerful options for filtering these similarity results by domain content/composition and taxonomy. Architectures defined on the basis of their domain superfamily footprints can summarize sets of proteins that are functionally very diverse, however. In CDD, a protein domain superfamily is defined as a set of protein domain models that annotate overlapping footprints on protein sequences. An automated clustering method groups models into these superfamily clusters, and the superfamily clusters undergo a subsequent manual review in order to prevent false clustering. In many cases superfamily clusters contain more than one model from a given data contributor, such as Pfam or COG, and clustering may not just reduce redundancy introduced by including multiple data sources, but also collapse information from one data source that would otherwise be useful in distinguishing between functionally distinct protein families. This becomes particularly evident in cases where a superfamily cluster contains one or more NCBI-curated protein domain hierarchies, which were deliberately constructed so that CDD could distinguish between domain and protein sub-families that have a distinct evolutionary history and distinct function.
In order to make use of the richness of information collected in CDD, we have investigated alternative methods for grouping proteins, namely by their Subfamily Domain Architecture (SDA). Here we present a first and simple implementation of that idea, which defines a Subfamily Domain Architecture as the string of the domain model accessions that provide the most concise annotation on a protein. We find that the proteins grouped under such SDAs can often be named accurately, and curators record brief functional characterizations (called functional labels) and supporting evidence. The curation interface used to associate domain architectures with functional descriptions has been named SPARCLE for ‘Subfamily Protein Architecture Labeling Engine’, and that name has been adopted for the public service as well.
To date, CDD curators have assigned names and functional labels to more than 6,500 SDAs. A publicly accessible Entrez database has been made available to support querying and to provide summary information for SDAs as well as links to other databases, most importantly the NCBI protein collection.
For user protein queries submitted to CD-Search, and in the display of pre-computed domain annotation, the content of the functional label assigned to the corresponding SDA is now shown on the results page, if available (see Figure 1). The functional labels are linked to summary pages, which display additional information about a subfamily domain architecture, including evidence for the name and functional label (see Figure 2).
Figure 1.
CD-Search reporting pre-computed domain annotation for the protein with GenBank accession KUG45846, a hypothetical protein from Pseudomonas savastanoi pv. Fraxini. The section circled in red provides the functional label that has been assigned to the subfamily domain architecture characterized by the string ‘cd00714 cd05008 cd05009’, which is shared by over 70 000 sequences in Entrez/protein.
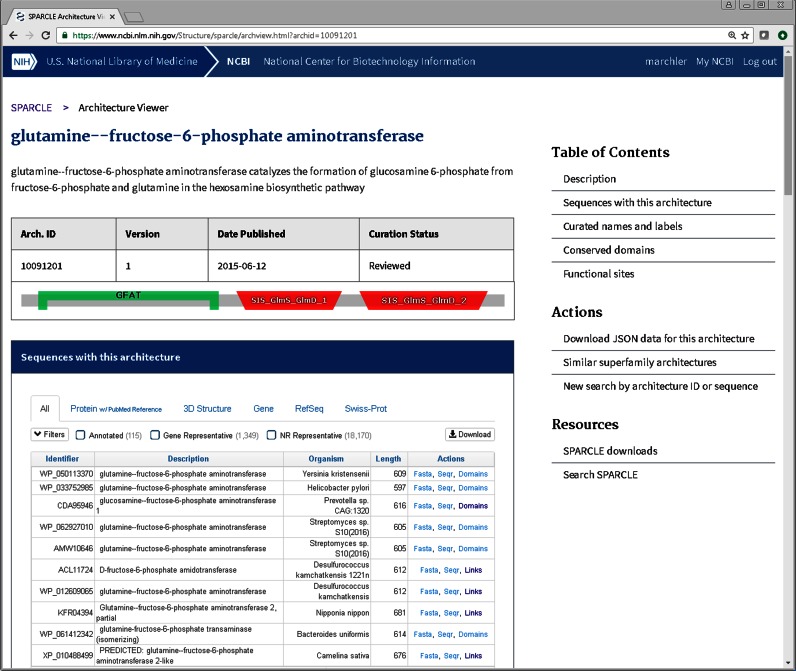
Figure 2.
Subfamily domain architecture summary page. The summary pages include a browser that provides options for retrieving sub-sets of the sequences sharing the same subfamily architecture, such as those from particular sources, a particular organism, or those that are linked to papers in PubMed.
Subfamily domain architectures as defined by the SPARCLE resource vary widely in their coverage and functional diversity. The resolution of this protein classification with respect to specific function depends directly on the availability of specific reagents in the CDD domain model collection, and future curation work in CDD will also be aimed at providing more fine-grained classifications where they may help to better resolve functionally distinct SDAs.
The current set of curated SDA names and labels focuses on architectures common in bacterial genomes. We are investigating methods for automating name and label assignments for architectures. Automatically assigned names and labels will be flagged as not validated, should they be displayed publicly as annotation on query sequences.
Acknowledgments
We thank the NCBI Information Engineering Branch for assistance with software development. We are grateful to the authors of Pfam, SMART, COGs, TIGRFAMs and NCBI's Protein Clusters database for providing access to their resources and data, and the users of CDD for their acknowledgements and invaluable feedback.
Comments, suggestions, and questions are welcome and should be directed to: info@ncbi.nlm.nih.gov.
FUNDING
Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I., Doerks T., Bork P. SMART: recent updates, new developments, and status in 2015. Nucleic Acids Res. 2014;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haft D.H., Selengut J.D., Richter A.R., Harkins D., Basu M.K., Beck E. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013;41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klimke W., Agarwala R., Badretdin A., Chetvernin S., Ciufo S., Fedorov B., Kiryutin B., O'Neill K., Resch W., Resenchuk S., et al. The National Center for Biotechnology Information's protein clusters database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchler-Bauer A., Bryant S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2005;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn R., Attwood T., Babbitt P., Bateman A., Bork P., Bridge A., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M., et al. InterPro in 2017: beyond protein family and domain annotations. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkw1107. doi:10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geer L.Y., Domrachev M., Lipman D.J., Bryant S.H. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]