Abstract
The first ever cyanobacterial genome sequence was determined two decades ago and CyanoBase (http://genome.microbedb.jp/cyanobase), the first database for cyanobacteria was simultaneously developed to allow this genomic information to be used more efficiently. Since then, CyanoBase has constantly been extended and has received several updates. Here, we describe a new large-scale update of the database, which coincides with its 20th anniversary. We have expanded the number of cyanobacterial genomic sequences from 39 to 376 species, which consists of 86 complete and 290 draft genomes. We have also optimized the user interface for large genomic data to include the use of semantic web technologies and JBrowse and have extended community-based reannotation resources through the re-annotation of Synechocystis sp. PCC 6803 by the cyanobacterial research community. These updates have markedly improved CyanoBase, providing cyanobacterial genome annotations as references for cyanobacterial research.
INTRODUCTION
Bacteria belong to several phyla, such as the Proteobacteria and Firmicutes, and exhibit an extremely high level of diversity. Because of this heterogeneity, no database currently covers all of the genome sequences within a particular phylum, with the exception of one, the Cyanobacteria. Cyanobacteria is a very diverse phylum (1) from which >300 species have been sequenced, and the genome annotation database CyanoBase (http://genome.microbedb.jp/cyanobase) includes these multi-genus genomes. CyanoBase was originally established two decades ago as a genome database for Synechocystis sp. PCC 6803, which was the first cyanobacterial genome to be sequenced (2). Since then, it has been continuously extended to include the genomes of additional cyanobacteria and related species (3–6), covering 39 genera.
More than 300 cyanobacterial genome sequences are currently available and the rapid advances that are being made in sequencing technologies will result in many more in the near future. However, prior to this update, CyanoBase only included 39 sequences (6). Therefore, the development of a high-quality automated annotation system was required to rectify this situation. Synechocystis sp. PCC 6803 represents an appropriate reference genome for automated annotation, as it is one of the best characterized cyanobacteria with many mutants and omics data. However, its annotation data have not been updated since 2003. Therefore, high-quality annotation was required, which ideally should be performed manually by experimental researchers covering multifarious aspects of cyanobacterial biology.
In this report, we outline how we have newly incorporated 337 cyanobacterial genomes into CyanoBase. We also describe the development of a web-based annotation system that is easily accessible for experimental researchers. Using this system, we were able to perform high-quality re-annotation of Synechocystis sp. PCC 6803 through the use of a cyanobacterial research community covering various biological aspects. This re-annotated data will contribute to the automated and high-quality annotation of newly sequenced cyanobacterial genomes.
DATA RESOURCES
Reference genomes
CyanoBase integrates reference genomes from original genome projects conducted by Kazusa DNA Research Institute and from public sequence databases (2–6). We added 337 new genome entries into CyanoBase based on recent genome sequencing projects. As a result, CyanoBase has been extended to currently include 376 completely sequenced genomes, of which 86 are complete genomes and 290 are draft genomes, including unannotated contigs and scaffolds. In traditional classification, morphological characters are used to divide them into five subsections (7,8). In this update, species from subsection II and V were newly added to the list and now it is covering all the subsections of cyanobacteria (Supplementary Table S1).
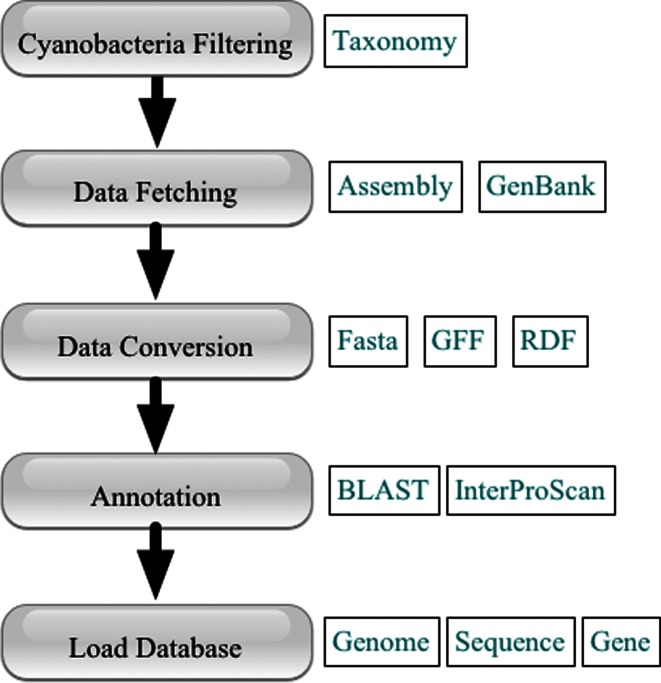
To deal with the rapidly increasing number of sequenced genomes, we also developed a processing pipeline for CyanoBase records, which are gathered from cyanobacterial genomic records in the International Nucleotide Sequence Database Collaboration (INSDC) based on the complete taxonomy subtree descended from the cyanobacteria taxon (taxonomy id: 1117) in the taxonomy database. We used a resource description framework (RDF) format for all of the assembly records for cyanobacteria. This pipeline is almost fully automated in converting, annotating, and importing the cyanobacterial genome datasets into CyanoBase (Figure 1).
Figure 1.
Flowchart representing data processing pipeline for CyanoBase.
Metadata and cross references
CyanoBase also contains metadata associated with cyanobacterial genomes that have been collected from multiple sources, such as the National Center for Biotechnology Information's (NCBI's) assembly database and GenBank sequence records. Also, we reviewed other databases and web resources of cyanobacteria with different aspects and contents. For example, CyanoExpress (9) is a database for gene expression, CyanoClust (10) is a database for clustering of homologous proteins, and CyanoLyase (11) is a database specialized for phycobilin lyases. CyanoBase represents one of the most comprehensive databases for cyanobacteria in the comparison of genome number of resources (Supplementary Table S2).
USER INTERFACE
Genome project
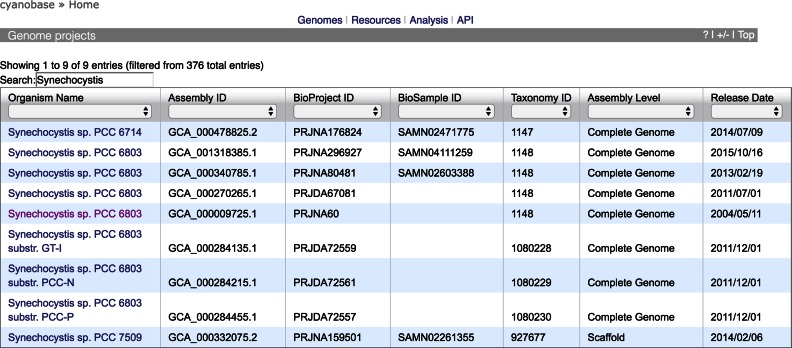
CyanoBase is derived from genome project data that have been submitted to the public nucleic acid databases DNA Data Bank of Japan, European Molecular Biology Laboratory, and GenBank. INSDC assembly records (12,13) contain information about a particular genome assembly and are supported by unique assembly-level identifiers. In these records, all of the pieces of a genome are collected together in ways that are much more flexible and powerful for indexing and retrieval purposes. In this update of CyanoBase, the number of genome projects included was increased from 39 to 376, and draft genomes were also introduced as database entries for the first time. We also provided a new user interface for genome projects (Figure 2) and applied TogoStanza (http://togostanza.org) for visualization of the list of genome projects—a generic web framework that enables reusable web components to be developed to assist the development of Semantic Web components such as querying SPARQL endpoints and visualizing the retuned results.
Figure 2.
A sample Genome Project View page resulting from the keyword search ‘Synechocystis’. The table is filterable and sortable for each column.
Genome browser
CyanoBase originally used GBrowse as a genomic browser and to graphically illustrate the genomic context, indicating the length, direction and function of the gene and the surrounding genes. To provide an equivalent service on a larger scale, we replaced GBrowse (14) with JBrowse (15), which is very lightweight and has few server resource requirements. JBrowse allows display features or quantitative data to be obtained directly from a SPARQL endpoint, which the CyanoBase annotation resource stores in RDF.
COMMUNITY ANNOTATION
Among the hundreds of sequenced cyanobacteria, the unicellular cyanobacterium Synechocystis sp. PCC 6803 has reigned as a cyanobacterial model for two decades, being the first cyanobacterium and fourth living organism to have its entire genome sequenced. Therefore, cyanobacterial researchers searched the newly reported genes from peer-reviewed research literature and annotated these in Synechocystis sp. PCC 6803. This effort resulted in the re-annotation of ∼30% of the genes (1096 of 3725 genes) in the genome of Synechocystis sp. PCC 6803, and decreased the rate of ‘function unknown genes’ to less than half (46.3%). This type of high-quality gene annotation is becoming increasingly important, as such information can be used as a platform for newly sequenced cyanobacterial genomes, particularly for the automated annotation of draft genome sequences. However, some genes had been annotated with an ambiguous function, such as probable/putative glycosyltransferase. We newly annotated 46 of these genes, but if we include them in the function unknown category, the proportion of function unknown genes remains at more than half (55.1%). To increase the accuracy of the annotation, we need more experiment-based information about these function unknown genes to allow the complete annotation of all genes in cyanobacterial genomes.
Acknowledgments
We thank Eli Kaminuma for suggestions and comments on the database development, Yasuhiro Tanizawa for the operation of the CyanoBase Web services. We also thank members of MicrobeDB.jp and TogoGenome projects for the collaboration on the Semantic Web-based developments. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics. We are also grateful to Hitoshi Nakamoto for information on chaperone proteins, and Norio Murata and Masahiko Ikeuchi for letting us conduct high-quality gene annotation of Synechocystis sp. PCC 6803.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Integrated Database Project, Ministry of Education, Culture, Sports, Science and Technology of Japan; National Bioscience Database Center (NBDC) of the Japan Science and Technology Agency (JST). Funding for open access charge: National Bioscience Database Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shih P.M., Wu D., Latifi A., Axen S.D., Fewer D.P., Talla E., Calteau A., Cai F., Tandeau de Marsac N., Rippka R., et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.KanekoT. SatoS., KotaniH. TanakaA., AsamizuE. NakamuraY., MiyajimaN. HirosawaM., SugiuraM. SasamotoS., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura Y., Kaneko T., Hirosawa M., Miyajima N., Tabata S. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 1998;26:63–67. doi: 10.1093/nar/26.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura Y., Kaneko T., Tabata S. CyanoBase, the genome database for Synechocystis sp. strain PCC6803: status for the year 2000. Nucleic Acids Res. 2000;28:72. doi: 10.1093/nar/28.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakao M., Okamoto S., Kohara M., Fujishiro T., Fujisawa T., Sato S., Tabata S., Kaneko T., Nakamura Y. CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res. 2010;38:D379–D381. doi: 10.1093/nar/gkp915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujisawa T., Okamoto S., Katayama T., Nakao M., Yoshimura H., Kajiya-Kanegae H., Yamamoto S., Yano C., Yanaka Y., Maita H., et al. CyanoBase and RhizoBase: Databases of manually curated annotations for cyanobacterial and rhizobial genomes. Nucleic Acids Res. 2014;42:D666–670. doi: 10.1093/nar/gkt1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rippka R.J., Deruelles J.B., Waterbury J.B., Herdman M., Stanier R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979;111:1–61. [Google Scholar]
- 8.Castenholz R.W. General characteristics of the cyanobacteria. In: Boone DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. 2nd edn. Vol. 1. NY: Springer-Verlag; 2001. pp. 474–487. [Google Scholar]
- 9.Hernández-Prieto M.A., Semeniuk T.A., Giner-Lamia J., Futschik M.E. The transcriptional landscape of the photosynthetic model cyanobacterium Synechocystis sp. PCC6803. Sci Rep. 2016;6:22168. doi: 10.1038/srep22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki N.V., Sato N. CyanoClust: comparative genome resources of cyanobacteria and plastids. Database. 2010;2010:bap025. doi: 10.1093/database/bap025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bretaudeau A., Coste F., Humily F., Garczarek L., Le Corguillé G., Six C., Ratin M., Collin O., Schluchter W.M., Partensky F. CyanoLyase: a database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res. 2013;41:D396–D401. doi: 10.1093/nar/gks1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitts P.A., Church D.M., Thibaud-Nissen F., Choi J., Hem V., Sapojnikov V., Smith R.G., Tatusova T., Xiang C., Zherikov A., et al. Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res. 2016;44:D73–D80. doi: 10.1093/nar/gkv1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pakseresht N., Alako B., Amid C., Cerdeño-Tárraga A., Cleland I., Gibson R., Goodgame N., Gur T., Jang M., Kay S., et al. Assembly information services in the European Nucleotide Archive. Nucleic Acids Res. 2014;42:D38–D43. doi: 10.1093/nar/gkt1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L., et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:1–12. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]