Abstract
The NHGRI-EBI GWAS Catalog has provided data from published genome-wide association studies since 2008. In 2015, the database was redesigned and relocated to EMBL-EBI. The new infrastructure includes a new graphical user interface (www.ebi.ac.uk/gwas/), ontology supported search functionality and an improved curation interface. These developments have improved the data release frequency by increasing automation of curation and providing scaling improvements. The range of available Catalog data has also been extended with structured ancestry and recruitment information added for all studies. The infrastructure improvements also support scaling for larger arrays, exome and sequencing studies, allowing the Catalog to adapt to the needs of evolving study design, genotyping technologies and user needs in the future.
INTRODUCTION
Genome-wide association studies (GWAS) are a well-established and effective method of identifying genetic loci associated with common diseases or traits (1). GWAS involve the analysis of at least hundreds of thousands of variants across the genome in large cohorts of individuals, often split into cases and controls, to identify variants associated with the trait of interest. The majority of variants identified by GWAS are assumed not to be causal but to tag a region of linkage disequilibrium containing one or more functional variants. GWAS have identified reproducible genomic loci associated with many common human diseases, including cardiovascular disease (2), inflammatory bowel disease (3), type 2 diabetes (4) and breast cancer (5).
The GWAS Catalog (www.ebi.ac.uk/gwas; 6) is a publicly available, manually curated resource of all published GWAS and association results, collaboratively produced and developed by the NHGRI and EMBL-EBI. It includes all eligible GWAS studies since the first published GWAS on age-related macular degeneration in 2005 (7). As of 1st September 2016 it contains 24,218 unique SNP-trait associations from 2,518 publications in 337 different journals. The Catalog summarizes a large body of diverse and unstructured data from the literature in an accessible, expertly curated and quality controlled resource. Catalog data are used by biologists, bioinformaticians and clinical/translational researchers as a starting point for further investigations to identify causal variants, understand disease mechanisms, and establish targets for novel therapies. Examples of such work include the analysis of Catalog data for identification of other traits associated with type 1 diabetes loci by Onengut-Gumuscu et al. (8); for evidence of pleiotropy by Sivakumaran et al. (9); for loci associated with seven common diseases to identify possible causal missense variants by Pal and Moult, 2015 (10); and for identification of new targets for known drugs by Mullen et al. (11). GWAS Catalog data are also integrated into many bioinformatics resources including Ensembl (12), the UCSC Genome Browser (13), PheGenI (14), HuGE Navigator (15) and GWASdb (16).
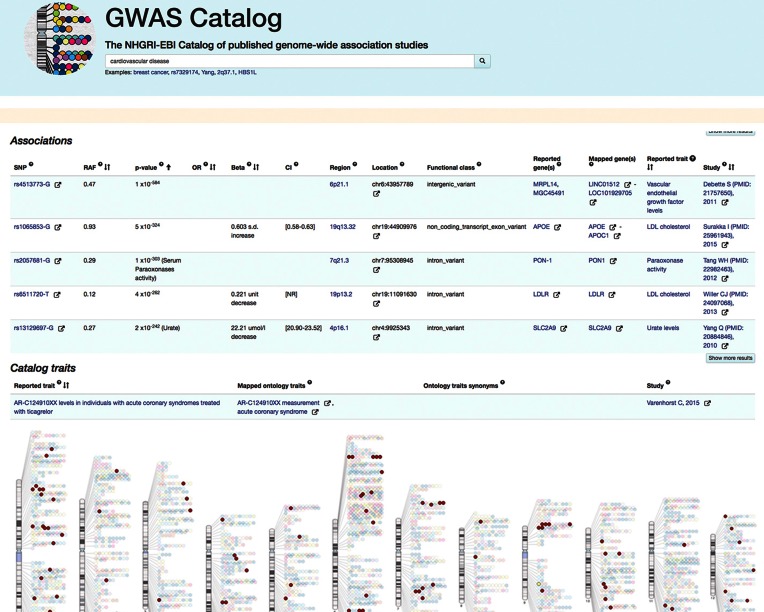
For inclusion in the Catalog, studies and associations must meet strict criteria (www.ebi.ac.uk/gwas/docs/methods); studies must include an array-based GWAS and analysis of >100 000 SNPs with genome-wide coverage, while SNP-trait associations must have a P-value <1 × 10−5. Studies identified through an automated PubMed literature search are reviewed to select those matching the inclusion criteria. In order to summarize the GWAS literature in a consistent manner in the Catalog, each GWAS analysis is entered separately into the curation system using standards for describing ancestry and traits, and following extensive extraction guidelines at the study and SNP levels. Trained curators, with expertise in deciphering study design, read each paper to determine how to represent the GWAS analyses in the most scientifically accurate and accessible manner. The curators assess whether any appropriate replication analyses were performed; the number and ancestry of samples included in the analysis; and the traits analyzed. All traits are mapped to terms from the Experimental Factor Ontology (EFO; 17). This enables structured querying and visualization of data in the GWAS diagram (Figure 1) e.g. searching for ‘cardiovascular disease’ displays all associations both with this specific trait and its sub-traits, including for example ‘myocardial infarction’ and ‘coronary artery disease’.
Figure 1.
A composite diagram showing association and trait search results for ‘cardiovascular disease’ visualized in the user interface and on the GWAS Diagram. http://www.ebi.ac.uk/gwas/search?query=cardiovascular%20disease. The diagram can be reached from http://www.ebi.ac.uk/gwas/diagram.
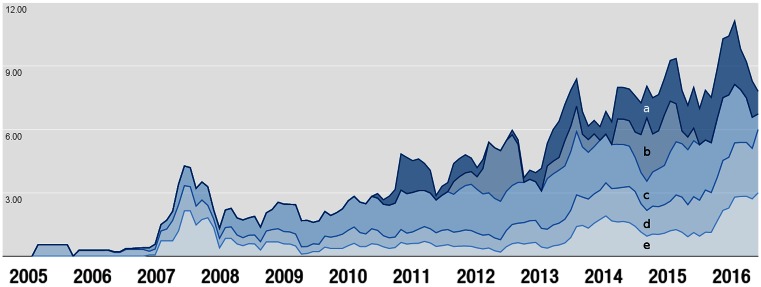
GWAS study design has evolved and complexity has increased since the inception of the Catalog, rendering it more challenging to understand and represent the study design, and to extract greater volumes of data. Specifically, there has been significant growth in the number of publications describing multiple GWAS, SNP-by-SNP and SNP-by-environment interaction studies, and in the number of traits and ancestry categories analyzed per publication (Figure 2), along with an average 50% increase in eligible SNP-trait associations year on year over the last 4 years.
Figure 2.
Increasing complexity of GWAS studies over time (A) number of SNP-by-environment interaction studies, (B) number of SNP-by-SNP interaction publications, (C) number of traits per publication, (D) number of ancestry categories each GWAS publication analyzed and (E) number of GWAS analyses per publication. Values were normalized to provide equal weighting to each category.
Since our last publication (6), we have made substantial changes to the infrastructure that improve the GWAS Catalog and ensure sustainability into the future. In parallel we have curated over 600 publications and 10,000 SNP-trait associations since November 2014. We developed a new Catalog infrastructure, launched in March 2015 with a redesigned database, improved data model, curation interface, user interface and download files. In addition, the new website (www.ebi.ac.uk/gwas/) has ontology supported search functionality and an interactive search results interface. All software for the Catalog's infrastructure is open source and made available via a public GitHub repository (https://github.com/EBISPOT/goci) using an Apache 2.0 licence. All Catalog data are freely and publicly available facilitating integration with bioinformatics resources.
IMPROVED DATA REPRESENTATION AND ANNOTATION
Accurate data capture is essential to make data correctly available for searching, visualization and integration with other resources. To ensure the GWAS Catalog is able to accurately capture study design and association results of increasing complexity we reviewed and redesigned the Catalog database schema. The representation of effect sizes has been improved with odds ratio and beta coefficient information now captured and displayed in separate fields, improving the accessibility of this data for users. We also mandated the use of structured, rather than free text, terms for key concepts including beta unit and direction, platform manufacturer, number of SNPs analyzed and whether imputation was used. This will improve consistency and integration potential with other complementary resources. Changes were made to the Catalog infrastructure to improve the representation of composite genomic elements, including haplotypes and SNP-by-SNP associations. Each individual variant is now captured separately in the database allowing variant-specific mapping, searches, links and visualization. The new schema design is also more flexible and will support future extensions to the scope of the GWAS Catalog to meet evolving study designs and users needs, as discussed in future work.
To improve the quality and accuracy of the SNP and associated genomic data in the Catalog, we have redeveloped the variant mapping pipeline. The new pipeline accesses Ensembl's REST API (https://rest.ensembl.org/) (18) enabling live validation within the curation system for: SNP ID validation; reported gene ID validation; checking that the SNP and reported gene are on the same chromosome. This delivers more accurate data in the Catalog as errors are reported and corrected immediately, decreasing the need for post-hoc curation. The new pipeline has also increased the proportion of variants that map to the genome, from 92% to 96%, improving the completeness of genetic location, mapped gene and cytogenetic data. In future, the flexibility of this pipeline will allow integration of additional information from Ensembl to improve functional annotation, for example with all genes within a specified genomic region from both the RefSeq (19) and GENCODE (20) gene sets. These future enhancements are supported in the redesigned database with the model now capturing mapping of multiple genes to a single variant, and the distance to each gene. In addition the new pipeline is used to update the current dataset to the most recent genome build.
IMPROVED DATA CURATION
GWAS Catalog data are manually extracted from the literature and entered via a curation interface. The curation interface supports all aspects of the process, including progress reporting, tracking of studies, data entry and quality control. We have re-developed and deployed this interface at the EMBL-EBI focusing on ease of navigation, usability and scaling of curation processes. The curation homepage provides summary information and tracking for all studies within the GWAS Catalog, while the study-specific data entry pages now have separate tabs for study, sample, association and curator information. Additional tabs have been added to allow curators to attach and directly access files relating to the study (e.g. PDF of publication and supplementary materials), print all study-specific information and view provenance information for study-specific data entry. Direct mapping of traits to the Experimental Factor Ontology (EFO) at both the study and association level, is available within the curation interface, facilitating mapping within the curation workflow. These improvements will support future scaling of curatorial activities through author deposition, as discussed in future work.
IMPROVED DATA ACCESS AND VISUALIZATION
We have launched a new GWAS Catalog website (www.ebi.ac.uk/gwas), redesigned based on feedback with improved querying and navigation. All GWAS Catalog data continue to be publicly available from the website via a search interface (Figure 1; www.ebi.ac.uk/gwas/search), download (www.ebi.ac.uk/gwas/docs/downloads) and diagram (www.ebi.ac.uk/gwas/diagram), however, each of these methods has been developed and extended to provide an easier and more intuitive user experience. The new website also includes improved documentation with specific pages describing details of the Catalog's eligibility criteria and methods (www.ebi.ac.uk/gwas/docs/methods), and ontology representation of traits (www.ebi.ac.uk/gwas/docs/ontology).
We have harmonized the GWAS Catalog data release strategy, with all available elements (searchable data, spreadsheets and diagram) now being released weekly. This has allowed us to introduce date-based versioning and provide all weekly data releases since March 2016 on the project's FTP site (ftp://ftp.ebi.ac.uk/pub/databases/gwas/releases/).
Searching of Catalog content is now via a single search bar, with more searchable fields and improved functionality. The new search is driven by a Solr index (http://lucene.apache.org/solr/), which allows simultaneous searching across a wide range of data fields, including title, author, journal, reported trait, mapped (ontology) trait, sample descriptions, genes and SNPs. In addition, it also provides ontology expansion support, returning results for mapped trait's synonyms and child terms. For example, a search of ‘inflammatory bowel disease’ will also return results for children of this trait such as ‘Crohn's disease’ and ‘ulcerative colitis’. Equally, search results for ‘cancer’ will include all sub-types of cancer, whether they contain a lexical match to ‘cancer’ (e.g. breast cancer, esophageal cancer) or not (e.g. Ewing sarcoma, acute myeloid leukemia).
Search results are displayed in an interactive graphical user interface with improved data visualization, interrogation and integration with data from external resources. Results are displayed in tabular format, organized into separate facets for studies, associations and traits. Search results can be filtered, by P-value, odds ratio, beta coefficient, study date, reported trait, are sortable and can be downloaded in tab-delimited format. Links are available to relevant data in external resources, including the publication entry in Europe PMC (21), variant, genomic location and gene in Ensembl (12), and mapped ontology trait in EFO (17) enabling users to access additional information easily.
The new GWAS Catalog interface also provides additional data not previously available via the now retired NHGRI hosted search interface, for example ancestry data is now available for all studies (released October 2016). This work included the annotation of study samples to an ancestry ontology (www.ebi.ac.uk/ols/ontologies/ancestro), with sample sizes and country of recruitment for each ancestral group. These data are fully searchable enabling new searches for studies performed on samples from a specific ancestral background. We also plan to provide additional filtering functionality based on ancestry to allow inclusion or exclusion of results based on the ancestry of study samples.
The GWAS Catalog's complete data in tab-delimited format (www.ebi.ac.uk/gwas/docs/downloads) is primarily used by bioinformaticians for bulk data access, allowing large-scale analysis of the data, annotation with additional data or integration into resources, e.g. Ensembl. We have expanded the range of download files available, with separate files for all associations and all studies, both with and without ontology mappings, and for ancestry data. We retained the original file format for association data, including all column headings, in order to provide backwards compatibility for programmatic parsing of the data. Studies are now included in a separate spreadsheet which provides a full list of all studies with a count of how many associations were identified for each study. We also provide a dedicated mapping spreadsheet between all reported GWAS Catalog traits and ontology terms, along with the GWAS diagram trait category. This allows users to identify the child terms included under each higher-level trait category on the GWAS diagram for example congenital heart disease is a cardiovascular disease.
The interactive GWAS diagram uses the Web Ontology Language/Resource Description Framework (OWL/RDF, https://www.w3.org/OWL/) for representation of the Catalog data and was previously hosted separately from the main GWAS Catalog. The new diagram portal (www.ebi.ac.uk/gwas/diagram) is now integrated with the new public Catalog infrastructure providing a uniform look and feel and improving transition for users between search and visualization. It is now possible to search the Catalog for a trait or SNP of interest directly from the pop-up information window for each trait. The redevelopment of the diagram generation software using a Virtuoso triplestore to represent the underlying data, has led to more than 1000-fold improvement in diagram generation time, and diagram and data releases are now synchronized weekly. Current and older versions of the GWAS diagram are downloadable providing users with high quality diagram images for publications and presentations and enabling the construction of a diagram time series. The full GWAS Catalog is also available in OWL/RDF format, allowing users to install their own triple store and run their own version of the diagram generation software as well as query the GWAS Catalog in SPARQL or another graph querying language.
IMPROVED USER SUPPORT
The GWAS Catalog is committed to providing support for users. We have improved the documentation on our website, extended the process documentation, and FAQ have been added (www.ebi.ac.uk/gwas/docs/faq). The helpdesk provides rapid responses (48 h) to emails and support requests via gwas-info@ebi.ac.uk, announces major updates from the announce list, and tweets from the GWAS Catalog Twitter account (@GWASCatalog). During the past year we have also delivered our first face-to-face training workshops (training materials available at ftp://ftp.ebi.ac.uk/pub/databases/gwas/training_materials/).
FUTURE WORK
We will continue to improve the data access, scientific value and user experience of the GWAS Catalog and we invite users to request new features. In order to allow full programmatic access to all GWAS Catalog data, we will release a REST API in 2017. We will also develop more advanced searching capabilities such as limiting searches to only a single field (e.g. genes or traits), batch searching and combinatorial searches. New search parameters such as searching across genomic regions and filtering by LD blocks will also be included. The presentation of Catalog data will be improved with the introduction of dedicated SNP-specific pages, followed by study- and gene- specific pages, to supplement the high-level overview presented by the current multi-faceted search results page. These pages will link to other data sources, such as Ensembl, to allow views of functional annotation and pathway information. We will provide improved user support, with dedicated resources for user groups with different needs, including publishing online training at EMBL-EBI's Train online (www.ebi.ac.uk/training/online/).
GWAS study design, genotyping technologies and user needs have advanced since the initial design of the Catalog. It is essential that the Catalog adapts to contain the most relevant and up-to-date studies and association results. Studies using large-scale targeted/non-genome-wide arrays, including the Metabochip, Immunochip and Exome arrays, and genotyping by sequencing are currently not included in the Catalog. Following feedback from our users we have plans to extend the scope of the Catalog to include all large-scale association studies and all SNP-trait associations, regardless of P-value. This, coupled with programmatic access, will vastly improve the utility of the Catalog for large-scale meta-analyses of published GWAS data with increased power to identify causal loci.
To meet the needs of increasing data volumes, study complexity and allow us to extend the scope of data included, we will develop scalable methods of data acquisition. As described above the curation interface has been re-designed with a view to opening it up to authors of eligible papers in the future to facilitate data deposition directly to the GWAS Catalog.
Acknowledgments
The authors wish to thank all of their users and authors of studies included in the GWAS Catalog, the systems teams who maintain their computational infrastructure, the members of their scientific advisory board for their feedback, and their colleagues at Ensembl for distribution of data via their resources and feedback.
FUNDING
National Human Genome Research Institute, National Heart, Lung and Blood Institute; National Institutes of Health Common Fund [U54-HG004028, U41-HG006104, U41-HG007823]; European Molecular Biology Laboratory; L.H., P.H. and H.J. are employees of the National Human Genome Research Institute. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikpay M., Goel A., Won H.-H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium. Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–362. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein R.J., Zeiss C., Chew E.Y., Tsai J.-Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Onengut-Gumuscu S., Chen W.-M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E., et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sivakumaran S., Agakov F., Theodoratou E., Prendergast J.G., Zgaga L., Manolio T., Rudan I., McKeigue P., Wilson J.F., Campbell H. Abundant pleiotropy in human complex diseases and traits. Am. J. Hum. Genet. 2011;89:607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal L.R., Moult J. Genetic basis of common human disease: insight into the role of missense SNPs from genome-wide association studies. J. Mol. Biol. 2015;427:2271–2289. doi: 10.1016/j.jmb.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen J., Cockell S.J., Woollard P., Wipat A. An integrated data driven approach to drug repositioning using gene-disease associations. PLoS One. 2016;11:e0155811. doi: 10.1371/journal.pone.0155811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos E.M., Hoffman D., Junkins H.A., Maglott D., Phan L., Sherry S.T., Feolo M., Hindorff L.A. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet. EJHG. 2014;22:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu W., Gwinn M., Clyne M., Yesupriya A., Khoury M.J. A navigator for human genome epidemiology. Nat. Genet. 2008;40:124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 16.Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.-P.A., Yeager M., Sham P.C., Chanock S.J., Xia Z., et al. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016;44:D869–D876. doi: 10.1093/nar/gkv1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malone J., Holloway E., Adamusiak T., Kapushesky M., Zheng J., Kolesnikov N., Zhukova A., Brazma A., Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinforma. Oxf. Engl. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yates A., Beal K., Keenan S., McLaren W., Pignatelli M., Ritchie G.R.S., Ruffier M., Taylor K., Vullo A., Flicek P. The Ensembl REST API: Ensembl Data for Any Language. Bioinforma. Oxf. Engl. 2015;31:143–145. doi: 10.1093/bioinformatics/btu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Europe PMC Consortium. Europe PMC: a full-text literature database for the life sciences and platform for innovation. Nucleic Acids Res. 2015;43:D1042–D1048. doi: 10.1093/nar/gku1061. [DOI] [PMC free article] [PubMed] [Google Scholar]