Abstract
The database of Genotypes and Phenotypes (dbGaP) Data Browser (https://www.ncbi.nlm.nih.gov/gap/ddb/) was developed in response to requests from the scientific community for a resource that enable view-only access to summary-level information and individual-level genotype and sequence data associated with phenotypic features maintained in the controlled-access tier of dbGaP. Until now, the dbGaP controlled-access environment required investigators to submit a data access request, wait for Data Access Committee review, download each data set and locally examine them for potentially relevant information. Existing unrestricted-access genomic data browsing resources (e.g. http://evs.gs.washington.edu/EVS/, http://exac.broadinstitute.org/) provide only summary statistics or aggregate allele frequencies. The dbGaP Data Browser serves as a third solution, providing researchers with view-only access to a compilation of individual-level data from general research use (GRU) studies through a simplified controlled-access process. The National Institutes of Health (NIH) will continue to improve the Browser in response to user feedback and believes that this tool may decrease unnecessary download requests, while still facilitating responsible genomic data-sharing.
INTRODUCTION
Broad and responsible sharing of genomic data is fundamental to the mission of the National Institutes of Health (NIH) Genomic Data Sharing (GDS) Policy (https://gds.nih.gov/03policy2.html). The database of Genotypes and Phenotypes (dbGaP) (1,2) was developed to archive and distribute the results of genotype-phenotype studies, and includes genomic data from cohort studies, clinical trials and others studies. dbGaP is a highly utilized tool for sharing individual-level data (Appendix – Glossary) and summary-level data such as allele frequencies. Data submitted to dbGaP include genotype, phenotype, exposure, expression array, epigenomic and pedigree data from genome-wide association studies (GWAS), sequencing studies and other large-scale genomic studies.
In August 2008, in response to the emergence of new statistical approaches that could be used to re-identify research participants using summary-level information and aggregated genotype data (3), NIH moved summary-level dbGaP data from unrestricted-access to controlled-access (Appendix – Glossary) (https://gds.nih.gov/pdf/Data%20Sharing%20Policy%20Modifications.pdf). As a consequence, unrestricted-access resources could thereafter only provide summary study statistics or aggregate allele frequencies. Investigators seeking aggregated genotype results now have to turn to controlled-access repositories instead for the information, and it would be available only after formal data access requests had been submitted, reviewed and approved by Data Access Committees (DACs).
The Browser was developed in response to requests from the scientific community for a resource that would more easily enable view-only access of genotypes, aggregate variant data, and individual-level genomic sequence data not available in unrestricted-access (Appendix – Glossary), and without having to download individual dbGaP data sets. This new tool allows approved users to find and view specific regions of the human genome, including all allele frequencies and subsets of individual-level genotype and sequence data stored in dbGaP within that region, without having to download the data sets of interest and perform additional analyses. In addition, because users can view and better understand features of these data sets through the Browser tools, users can make better informed decisions about whether to request full access. The Browser will also benefit other users for whom genotype frequencies may be sufficient for their research needs. The Browser, like the PheGenI tool (4) and the GWAS catalogue (http://www.ebi.ac.uk/gwas/), allows users to view samples having alleles with phenotypic associations; however, it also allows users to view those findings directly and see the underlying individual-level data, without having to download or transfer the data set. The Browser allows access to these data while preserving the data confidentiality and research participant privacy principles set forth in the GDS Policy. Therefore, the dbGaP Data Browser serves as a middle ground, providing researchers with view-only access to a compilation of individual-level data from general research use (GRU) studies through a simplified controlled-access approval process. As of August 30, 2016, the dbGaP GRU collection contained over 60 research study data sets, and 32 882 samples with sequence tracks available for display in this new dbGaP view-only access option. The GRU collection represents 13% of the 1 240 829 subjects in dbGaP. In the future, new GRU data sets that are submitted to dbGaP will be automatically accessible through the dbGap Browser. The remaining 87% of subjects in dbGaP have research use limitations associated with them, making them inappropriate to include in the Browser where limits on research use are not imposed. Browser access to these individuals, however, are separately provided to approved users (https://www.ncbi.nlm.nih.gov/gap/ddb/faq/#APPLY) as part of their individual-level data set access.
IMPLEMENTATION
The NIH is pursuing a phased approach to development and deployment of the Browser. This step-wise release allows the NIH to consider and resolve any technical or policy issues identified along the way, while gaining experience to inform future Browser development and scale-up plans. Phase I, implemented in August 2014, allows existing dbGaP users to access the Browser and view data sets for which they have approved access. Here, the Browser provides investigators the ability to quickly visualize relevant features and pertinent results of the genomic regions and phenotypes they are evaluating in their data analyses. Phase II, implemented in September 2016, allows investigators to apply for Browser-only access to a collection of GRU data sets. Browser access can be requested at https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=list_viewonly&login=NFL. To request access to the Browser, an investigator must be a permanent employee of his or her institution at a level equivalent to a tenure-track professor or senior scientist and hold a principal investigator (PI) electronic Research Administration (eRA Commons) account (https://grants.nih.gov/grants/ElectronicReceipt/preparing_grants_commons_roles.htm). Laboratory staff (e.g. laboratory technicians) and trainees (e.g. graduate students, postdoctoral fellows) are not permitted to submit Browser requests at this time. However, approved investigators may grant the trainees and staff that they directly supervise permission to access the Browser and are responsible for ensuring that any individuals granted permission to access the Browser will adhere to the Browser Code of Conduct and abide by the expectations outlined in the Browser Use Agreement (Appendix – Glossary). As feedback from Phase I and II of Browser implementation is obtained, the NIH may consider extending access request privileges to other researchers, such as staff scientists and postdoctoral fellows.
Features
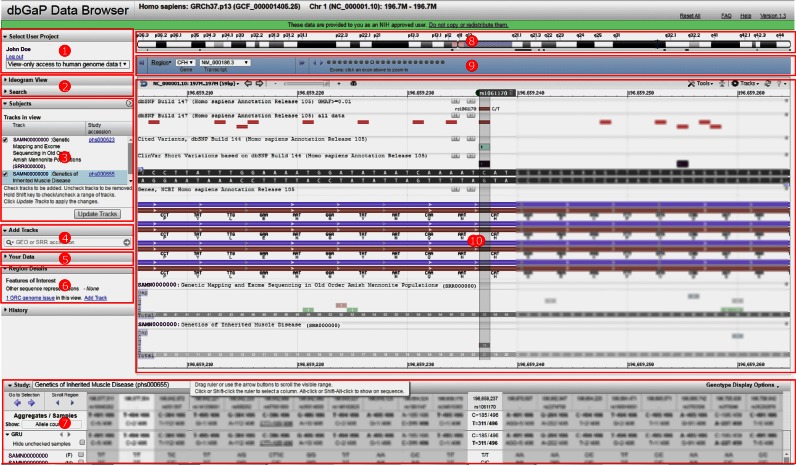
The Browser uses the standard National Center for Biotechnology Information (NCBI) graphical interface developed for data from the 1000 Genomes Project and dbGaP that combines sequence viewer track views with genotype tables and a novel sample/subject data selector that displays core phenotype data about the samples. The Browser webpage (Figure 1) consists of a series of page ‘widgets’ that display data from the dbGaP view-only data project, i.e. data from the dbGaP GRU collection. The widgets interact such that an action in one widget causes other widgets on the page to update. For instance, clicking on a chromosome in the ‘Ideogram Overview’ will update all other widgets on the page.
Figure 1.
The dbGaP Browser web page displays individual level sequence and genotype data in genome context. Component widgets are indicated by red boxes. Numbers are keyed to the feature descriptions. Some information in the figure has been intentionally blurred for publication.
Page widgets
Page widgets allow the user to maneuver around the page. The widgets listed below are keyed by number to Figure 1. For additional information on the widgets, see the online browser documentation (https://www.ncbi.nlm.nih.gov/gap/ddb/help/) and The NCBI YouTube channel (https://www.youtube.com/user/NCBINLM).
Select User Project [1] is used to define the dbGaP user project whose data will be shown in the browser. When accessing the browser via https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=list_viewonly&login=NFL, this defaults to ‘View-only access to human genome data’ for the Browser. If a user has permissions for other projects, they will be listed in this widget.
Ideogram View [2] provides genomic context for the displayed sequence and can be used for navigation. Clicking on a chromosome in the ideogram view will make it the selected chromosome and update the rest of the page to show data for this chromosome.
Search [2] will accept a location directive, such as chr1:1 500 000–2 000 000 or a search term (such as ‘PTEN’ or ‘rs13432’). The ‘Genes’ tab lists genes matching the search term. The ‘Other Features’ tab list transcripts, phenotypes and sequence tagged sites associated with the search term.
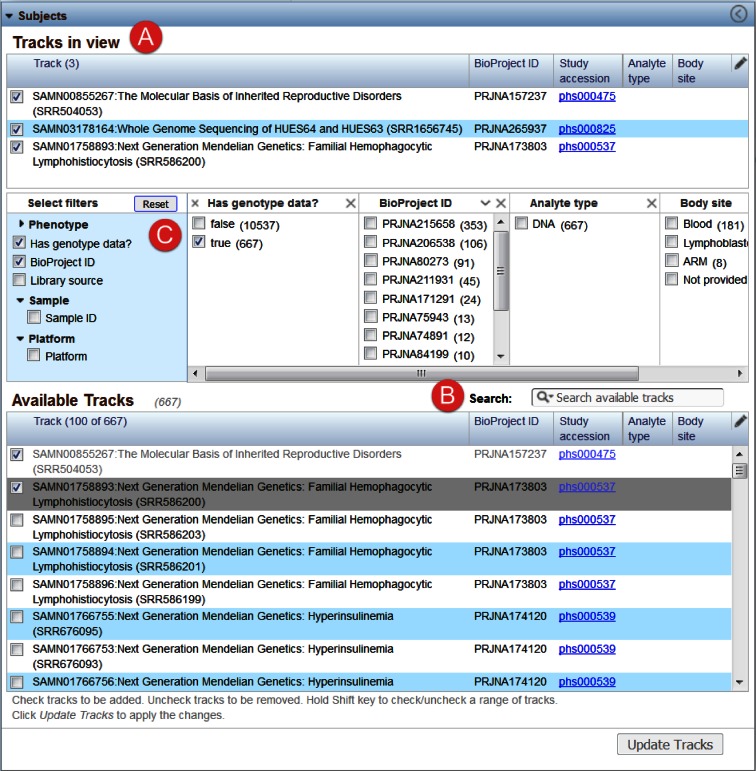
Subjects [3] allows the user to search for, add and remove alignment tracks to the sequence viewer and thereby view the read alignments generated for a particular sample. Alignment tracks currently displayed in the browser's graphical sequence viewer are listed in the ‘Tracks in View’ (A) section (Figure 2). Users can directly search the ‘Available Tracks’ table in the widget (B) or filter it or the results of a direct search by a variety of sample attributes (C). A description of the sample attributes available for filtering the available alignment tracks is included in Table 1. The primary source of data in the Browser is the ‘dbGaP Collection: Compilation of Individual-Level Genomic Data for General Research Use’ (Accession number – phs000688.v1.p1, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/collection.cgi?study_id=phs000688.v1.p1). The collection summary page includes a list of data sets, primary disease terms, main categories of phenotypes and number of participants. This study contains most individual-level genomic data sets currently in dbGaP that are designated as GRU and that therefore have no further use limitations beyond those outlined in the Browser Use Agreement.
Figure 2.
Subjects Widget, expanded view. The widget is used to add and remove alignment tracks from the sequence view. It is comprised of three parts: (A) Table of tracks currently displayed in the DDB sequence view; (B) Searchable table of all available tracks for the specified user project; and (C) filters used to restrict the tracks shown in B. Columns in A and B can be edited using the pencil icon in the upper right of each table. Selecting a filter from the expandable lists in the first column of C adds a column for the corresponding filter to this section that displays the filter values and counts. Selecting filter values in these additional columns updates the table in B.
Table 1. Participant sample attributes are derived from basic study phenotypes, sample properties, and experimental metadata. Attribute values are drawn from dbGaP, BioSample, and SRA as indicated.
| Data source | Attributes viewable | Example value(s) |
|---|---|---|
| From dbGaP | study accession | phs000500 |
| local sample ID | phs000500|xx-99999 | |
| consent group | GRU (General Research Use), UR (Unrestricted) | |
| study design | Longitudinal Cohort, Case-Control, Mendelian,... | |
| primary disease term for study | anemia sickle cell, muscular dystropohies, ... | |
| ancestry | African American, Hispanic, East or South Asian | |
| has genotype data | false, true | |
| From BioSample (public metadata) | BioProject ID | PRJNA75631 |
| sample ID | SAMN00000000 | |
| sex | female, male, not provided | |
| is subject affected | false, true, not provided | |
| is Tumor | false (germline sample), not provided | |
| study name | STAMPEED: Northern Finland Birth Cohort 1966 | |
| From Sequence Read Archive (experimental metadata) | analyte type | DNA, Genomic DNA, RNA, DNA:DNA Somatic... |
| library source | genomic, transcriptomic |
Add Tracks [4] allows users to directly add data tracks from other NCBI resources to the graphical sequence viewer display. The widget accepts GEO (5), SRA (6) and dbGaP accessions. In the case of controlled access SRA data, only tracks for SRA accessions associated with the selected user project (e.g. the GRU set) will be displayed. All tracks must be associated with the assembly displayed in the browser (e.g. currently GRCh37).
Your Data [5] opens a dialog box that allows users to upload their own data as a display track for comparison purposes. Supported file types are BED, GFF3, GTF, GVF, VCF, HGVS and ASN.1 (text and binary). If the user is logged in with a MyNCBI account, files uploaded via this widget will also be available in certain other NCBI resources such as Variation Viewer (https://www.ncbi.nlm.nih.gov/variation/view/) (7); Variation Reporter (https://www.ncbi.nlm.nih.gov/variation/tools/reporter); and the 1000 Genomes browser (http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/). User uploaded data will expire after 30 days.
Region Details [6] table provides the sequence identifiers of other assembly sequences (e.g. alternate loci and patch scaffolds) that provide other representations for this genomic region. Clicking on the identifiers in the table will display them in the Browser. The ‘Add Track’ link in this widget adds a track to the graphical view that shows the locations of regions in the assembly that are under review by the Genome Reference Consortium (https://genomereference.org), the group responsible for defining the human reference genome assembly.
Genotype table [7] provides both individual-level and aggregate GRU data set genotypes. The table rows list samples by BioSample id (https://www.ncbi.nlm.nih.gov/biosample/); hovering over the BioSample ids will reveal the sample name (e.g. NA12878) as reported in the VCF file. The checkbox next to each sample in this expanded view can be used to add alignment files for that sample to the Sequence Viewer display. The genotype table is only populated once a user has added an alignment track for a sample with genotype data to the graphical sequence display. Users can find such tracks through the ‘Has genotype data’ filter in the Subjects widget. The table displays genotypes for all samples in the same project as the displayed track. If displayed tracks come from different dbGaP projects, the project ids will be listed in the Genotype table menu, and can be selected to update the table display.
Chromosome Overview [8] provides context and navigation for the displayed chromosome (10). The blue overlay shown on the ideogram image covers the amount of the chromosome shown in the Sequence Viewer (described below). The blue overlay can be resized or moved to update the region displayed in the browser.
Gene and Exon Navigator [9] is located below the Chromosome Overview widget. When a region of sequence is displayed that includes one or more genes, the symbols for those genes are provided in the ‘Gene’ menu. Users can navigate quickly to a gene by clicking on its symbol and through exons of the selected gene by clicking the circles in this widget.
Sequence Viewer [10] provides graphical representation of features annotated on individual sequences. The ‘Tracks’ button in the upper right corner of the Sequence Viewer allows users to add additional NCBI tracks for sequence, genes, variation and other features to the display, in addition to the alignment tracks added from the Subjects or Genotypes table widgets. Other NCBI tools (such as BLAST) are also available within this widget. Additional information on using the Sequence Viewer (that is embedded on many NCBI pages) can be found at https://www.ncbi.nlm.nih.gov/tools/sviewer/. The NCBI YouTube channel (https://www.youtube.com/user/NCBINLM) also has videos that demonstrate Sequence Viewer features.
Process for requesting access to the browser
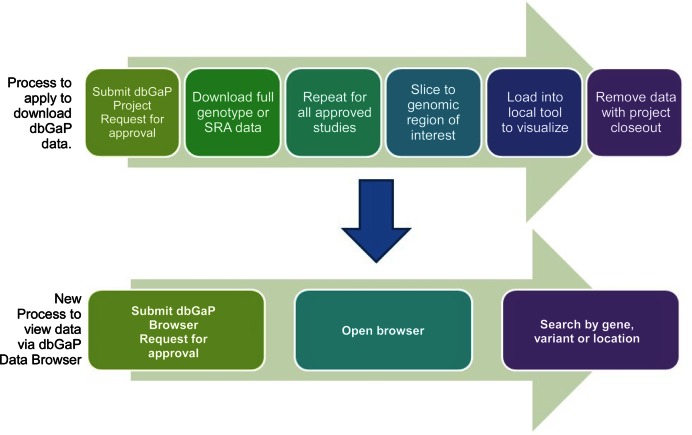
Investigators interested in obtaining access to the Browser must first sign-in to the dbGaP authorized-access portal using their eRA PI credentials to establish their account. Next, investigators should navigate to the ‘My View-only data’ tab on the dbGaP authorized access homepage and begin the Browser Request application process by clicking their institutional signing official (Appendix – Glossary), before the request is reviewed by the NIH Central Data Access Committee (CDAC) (https://gds.nih.gov/pdf/Central_DAC_Charge_and_Roster.pdf). This review process is more streamlined than the current dbGaP data access request process and investigators who are approved will be able to access the Browser and begin to visualize data. The current dbGaP (non-Browser) data access request process requires PIs to submit a dbGaP Project Request that includes a request for approval for each individual data set being requested, be approved for each, download or transfer them individually to their computational infrastructure or cloud, and load the data set into a separate tool for visualization (Figure 3). A typical request to dbGaP can take 1–2 weeks for approval once it is received in a Data Access Committee's queue. NIH will be periodically analyzing the data on approval wait times and potential reductions in data downloads as a part of ongoing efforts to streamline and simplify data access.
Figure 3.
The dbGaP Data Browser offers a streamlined request process to view dbGaP data. Users submit a dbGaP Data Browser Request using their existing eRA, institutional or NIH credentials. Once they are approved by their signing official and the NIH Central Data Access Committee (CDAC), they can open the Browser and immediately visualize data. In contrast, gaining download access to dbGaP data requires users to submit a dbGaP Project Request for approval for each individual data set, wait to be approved for each, download them individually to their local computer, and then load the data set into a local tool for visualization.
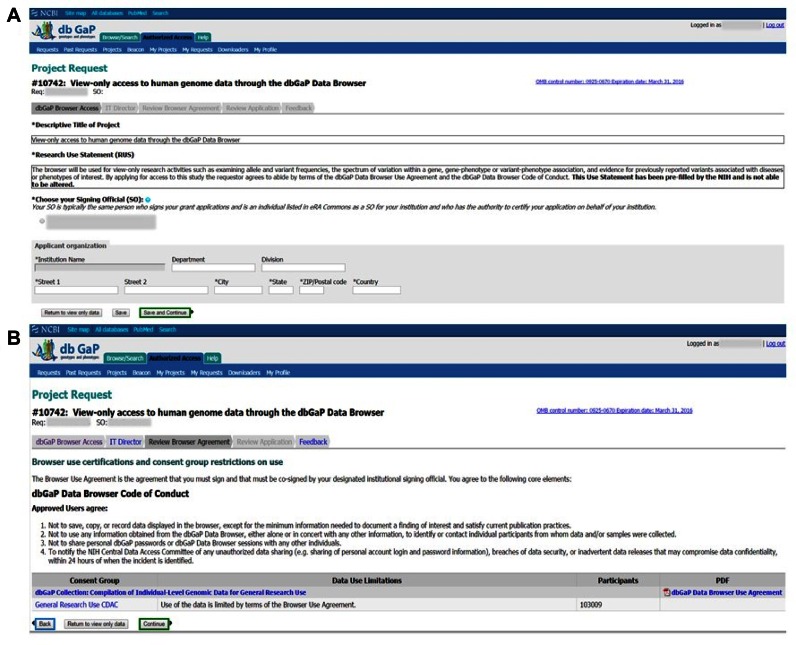
Once logged in to the Browser, a user can click the ‘Create New View-Only Data Browser project.’ Similar to requests to access and download data, investigators must provide their institutional information and identify their Institutional Signing Official (SO, see Appendix – Glossary) and an appropriate IT Director (Appendix – Glossary). However, unlike the regular dbGaP data access request, the project title and research use statement for requests to the Browser are prefilled. No further information is required. By submitting the Browser Request, the investigator agrees to abide by the Browser Code of Conduct (see Appendix – Glossary or https://dbgap.ncbi.nlm.nih.gov/aa/ddb_Code_of_Conduct.html) and Browser Use Agreement (see Appendix – Glossary or https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?bid=815018&view_blob=1) (Figure 4). The submitted application is then routed to the SO (Appendix – Glossary) for review, and once co-signed, the application is forwarded to the CDAC for a rapid, administrative review. The CDAC ensures that the levels of authority designated in the request are appropriate (i.e. investigator, SO and IT Director), but does not review a research use statement because it is pre-defined for all Browser users. Requesters of the Browser receive email notifications for each stage of the process, including CDAC approval. Once approved, investigators can access the Browser by logging into the dbGaP authorized-access portal and clicking the ‘Launch Browser’ button.
Figure 4.
(A) The Descriptive Title of Project and Research Use Statement are pre-filled in the browser request form. (B) Users must agree to the dbGaP Data Browser Code of Conduct and Browser Use Agreement in order to complete the browser request.
Consistent with NIH's policy to maintain transparency around the research use of its controlled-access data resources, once approved to access the Browser, the investigator's name, institution and date of access approval will be publicly posted on the dbGaP website. Access to the Browser is granted for one year, after which the user may renew access or simply let access permissions lapse, without penalty (Figure 5). In contrast with the process for users approved to download individual-level dbGaP data sets, Browser users are not required to submit a close-out report when access is no longer desired.
Figure 5.
To access the dbGaP Data Browser, users must request access annually. The Request form is a streamlined version of the normal dbGaP request form, with a pre-filled Research Use Statement. Each qualified Browser Request will be approved by the NIH CDAC.
DISCUSSION
The dbGaP Browser enables investigators to have rapid access to a collection of dbGaP GRU data, providing a rich resource of both individual-level and summary-level information for their exploration. The confidentiality of participant data available through the Browser is protected by a number of features and policies carefully developed to balance the interests of participants, whose data are entrusted to dbGaP, and the interests of investigators seeking access to these data. Features and policies include restricting the scope of available data to only those participants who gave consent for General Research Use (the most permissive permission category in dbGaP), limiting the associated phenotype data to basic demographic and disease status information, implementing a Browser Use Code of Conduct to establish clear expectations for use, disabling browser data download features, and displaying a watermark on the browser that images should not be captured or published. The Browser request process is simplified compared to the access request process for approval to download or transfer dbGaP data sets because the restricted format and view-only design of Browser access mediates the latent privacy risks associated with potential third-party access to insufficiently secured downloaded copies of full genome data sets from dbGaP.
NIH anticipates that removing the need to download data for preliminary research explorations for which Browser access is granted will avoid unnecessary data downloads, which, may in turn decrease overall vulnerabilities to participant data; fewer data downloads to individual computers may result in less exposure of the data to security risks. In addition, re-identification risks are decreased by limiting available subjects (Table 1), and restricting the size of the table of genomic variants to the monitor's screen resolution.
NIH intends to conduct periodic assessments on use of the Browser, gathering information from research publications that cite the Browser as well as user feedback. These assessments will be included as a part of NIH's regular oversight the GDS Policy. Assessments will allow NIH to consider how the Browser is being used and whether updated versions of the Browser need to be released, to meet the needs of the research community.
Genomic variant information is being used in clinical care, particularly to accurately diagnose and treat inherited disorders, as well as for pharmacogenomics and targeted drug therapy for somatic tumors (8). However, more evidence is needed to help understand the clinical significance of the thousands of known genes and variants associated with disease. Clinical annotation of sequence variants is being generated by the Clinical Genome Resource as well as by other clinical sequencing efforts funded by the NIH and deposited in dbGaP (8). However, it may not be appropriate or practical for the clinical community (e.g. directors of testing laboratories, medical geneticists, genetic counselors) to download these data sets individually to view for clinical purposes, and the appropriateness of research data use in non-research contexts should be considered. In the future, NIH will consider how the Browser may be useful to the clinical community and what modifications, if any, would be appropriate to facilitate their ability to access this resource.
In addition, NIH may consider expanding the criteria by which dbGaP study data may be accessed via the Browser beyond GRU. However, this will require a number of considerations, such as the scope of uses for which participants have consented in relation to the list of approved uses in the Browser Use Statement.
Finally, in the future, NIH may consider allowing postdoctoral fellows and other researchers to apply for Browser access under their own credentials. Currently, these researchers may view the data under the supervision of an approved senior investigator, but in consideration of the structure of the Browser and the policy limitations on Browser use, the rationale for requiring only supervised access may not apply.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health. We thank Fiona Cunningham and three anonymous reviewers for their comments.
APPENDIX – GLOSSARY
Unrestricted-access, also known as “open access", to dbGaP data is access available to anyone with no restrictions. (https://dbgap.ncbi.nlm.nih.gov/aa/dbgap_request_process.pdf)
Controlled-access. Controlled access to dbGaP data requires preauthorization of the Investigator and the Institution to assure responsible, secure use of the data. (https://dbgap.ncbi.nlm.nih.gov/aa/dbgap_request_process.pdf)
Individual-level data. For the purposes of dbGaP, these are data from a single individual that have been deidentified (i.e. no personal identifiers such as name, dates, social security numbers, phone numbers, etc.)
Aggregated data. For the purposes of dbGap, aggregateD genomic data sets have been approved for general research use and have no further limitations beyond the Data Use Certification. Collections linked here: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000501.v1.p1
Institutional signing official. A senior official at an Institution who is authorized to enter the Institution in to a legally binding contract and sign on behalf of an investigator who plans to submit data to NIH, e.g. Dean, Vice President for Research.
Information Technology (IT) Director. Generally, a senior IT official with the necessary expertise and authority to affirm the IT capacities at an academic institution, company or other research entity. The IT Director is expected to have the authority and capacity to ensure that the NIH Security Best Practices for Controlled-Access Data Subject to the NIH GDS Policy and the institution's IT security requirements and policies are followed by the Approved Users.
Browser Code of Conduct. dbGaP Data Browser Code of Conduct, Approved Users agree: (i) Not to save, copy or record data displayed in the browser, except for the minimum information needed to document a finding of interest and satisfy current publication practices. (ii) Not to use any information obtained from the dbGaP Data Browser, either alone or in concert with any other information, to identify or contact individual participants from whom data and/or samples were collected. (iii) Not to share personal dbGaP passwords or dbGaP Data Browser sessions with any other individuals. (iv) To notify the NIH Central Data Access Committee of any unauthorized data sharing (e.g. sharing of personal account login and password information), breaches of data security or inadvertent data releases that may compromise data confidentiality, within 24 h of when the incident is identified.
Browser Use Agreement. The browser will be used for view-only research activities such as examining allele and variant frequencies, the spectrum of variation within a gene, gene–phenotype or variant–phenotype association and evidence for previously reported variants associated with diseases or phenotypes of interest. By applying for access to this study the requestor agrees to abide by terms of the Browser Use Agreement and the dbGaP Data Browser Code of Conduct. This Use Statement has been pre-filled by the NIH and is not able to be altered.
FUNDING
Intramural Research Program of the National Library of Medicine at the National Institutes of Health (NIH) [in part]. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mailman M.D., Feolo M., Jin Y., Kimura M., Tryka K., Bagoutdinov R., Hao L., Kiang A., Paschall J., Phan L., et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tryka K.A., Hao L., Sturcke A., Jin Y., Wang Z.Y., Ziyabari L., Lee M., Popova N., Sharopova N., Kimura M., et al. NCBI's Database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Homer N., Szelinger S., Redman M., Duggan D., Tembe W., Muehling J., Pearson J.V., Stephan D.A., Nelson S.F., Craig D.W. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramos E.M., Hoffman D., Junkins H.A., Maglott D., Phan L., Sherry S.T., Feolo M., Hindorff L.A. Phenotype-genotype integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet. 2014;1:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database Collaboration The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., et al. ClinGen — The Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]