Abstract
Soybean (Glycine max) is a legume crop with substantial economic value, providing a source of oil and protein for humans and livestock. More than 50% of edible oils consumed globally are derived from this crop. Soybean plants are also important for soil fertility, as they fix atmospheric nitrogen by symbiosis with microorganisms. The latest soybean genome annotation (version 2.0) lists 56 044 coding genes, yet their functional contributions to crop traits remain mostly unknown. Co-functional networks have proven useful for identifying genes that are involved in a particular pathway or phenotype with various network algorithms. Here, we present SoyNet (available at www.inetbio.org/soynet), a database of co-functional networks for G. max and a companion web server for network-based functional predictions. SoyNet maps 1 940 284 co-functional links between 40 812 soybean genes (72.8% of the coding genome), which were inferred from 21 distinct types of genomics data including 734 microarrays and 290 RNA-seq samples from soybean. SoyNet provides a new route to functional investigation of the soybean genome, elucidating genes and pathways of agricultural importance.
INTRODUCTION
Soybean (Glycine max) is a legume, and one of the most commonly cultivated crops in the world. Soybean seeds are an important source for human food, cooking oil, and animal feed, because of their abundant protein and oil content. Soybean plants are also important for soil fertility, as they fix atmospheric nitrogen through symbiosis with microorganisms. The first soybean draft genome, G. max var. Williams 82, was reported in 2010 (1), and the latest version of the genome assembly, version 2.0 (Wm82.a2.v1) lists 56 044 genes; yet their functional contribution to crop traits remains mostly unknown. Dozens of soybean coding genes are currently annotated for Gene Ontology biological process (GOBP) terms with experimental evidence. The number of research articles for G. max has increased every year since 2009, which suggests that availability of the assembled crop genomes indeed facilitates research progress in crop science. Recently, genome-wide association studies (GWAS), quantitative trait loci (QTL) analysis, and other genomics studies have suggested many candidate chromosomal regions and loci associated with important soybean agricultural traits such as seed content and stress responses (2,3). However, these unbiased genotype-to-trait analyses suffer from limited statistical power and difficulty in mechanistic interpretation.
Network-based approaches have proven useful to complement such limitations by guilt-by-association and other network algorithms in the study of various organisms including model plant and crops (4). Here, we present SoyNet (http://www.inetbio.org/soynet), a database of soybean co-functional networks and a companion web tool for network-based functional predictions. SoyNet maps 1 940 284 co-functional links between 40 812 soybean genes (covering 72.8% of the coding genome), which were inferred from 21 distinct types of genomics data including 734 microarrays and 290 RNA-seq samples from soybean. SoyNet freely provides edge information for not only the integrated network, but also all individual component networks inferred from each data type, including many microarray and RNA-seq data sets. These will allow users to construct an alternative version of the integrated network using different data integration methods, and to conduct network analysis for individual component networks.
To increase the usability of SoyNet, we implemented three network-based methods of generating functional hypotheses: (i) find new members of a pathway, (ii) find context-associated genes and (iii) find functional modules. The SoyNet server can take user input of Arabidopsis genes based on TAIR10 (5), as well as soybean genes based on genome v1.1 and v2 annotations. Indeed, SoyNet is the first network database to date that facilitates web-based hypothesis generation for soybean genes. We demonstrated the superiority of SoyNet in pathway predictions and crop traits over other previously published networks of soybean genes, PlaNet (6) and STRING v10 (7), using benchmarking based on independent test data.
NETWORK CONSTRUCTION
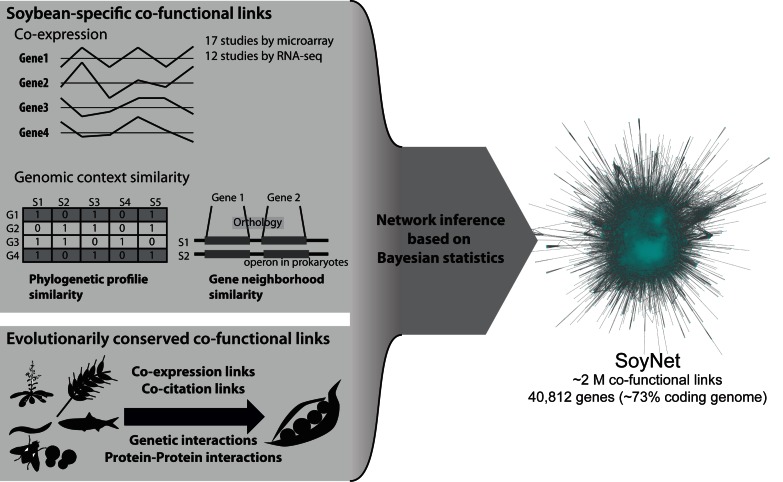
An overview of SoyNet construction is summarized in Figure 1. We considered soybean genes compiled from the latest genome assembly for G. max, version 2.0 (Wm82.a2.v1), which contains 56 044 protein-coding genes, distributed by Phytozome v10.0 (8). Soybean genes from the previous genome assembly version 1.1 were cross-mapped to those of version 2.0 using synonym information provided by Phytozome v10.0. To infer co-functional relationships by a supervised learning approach, we compiled gold-standard positive gene pairs, based on pathway databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) (9), SoyCyc (10) and MapMan (11), resulting in 726 709 gene pairs between 13 514 genes. A set of gold-standard negatives was then generated by pairing two genes that are annotated by pathway databases, albeit different pathway terms, resulting in 90 580 632 gene pairs. Using these gold-standard gene pairs, we inferred 21 networks that are based on 21 distinct data types (Table 1). The individual component networks are largely divided into three categories: (i) networks inferred from co-expression of G. max genes across samples from 17 microarray studies and 12 RNA-seq studies (Supplementary Table S1), comprising 734 array samples and 290 RNA-seq samples, available from Gene Expression Omnibus (GEO) (12) and NCBI Sequence Read Archive (SRA) (13). (ii) Networks inferred by genomic context similarity based on either gene neighborhood across bacterial genomes (14) or similarity of phylogenetic profiles (15), using 396 eukaryotic genomes and 1748 prokaryotic genomes. (iii) Networks inferred from evolutionarily conserved functional associations (associalogs) (16) in seven other species: Arabidopsis thaliana (17), Caenorhabditis elegans (18), Drosophila melanogaster (19), Danio rerio, Homo sapiens (20), Saccharomyces cerevisiae (21) and Oryza sativa (22). We transferred co-expression links, protein-protein interactions derived from the literature and high-throughput analysis, and genetic interactions, based on orthology relationships as measured by Inpranoid software version 4.1 (23). All individual networks were trained by log likelihood score (LLS) scheme and integrated by weighted sum methods (24). Integration of the 21 individual networks resulted in SoyNet, containing 1 940 284 links covering approximately 73% of the soybean coding genome. A more detailed description of network construction is available in Supplementary Online Methods.
Figure 1.
Overview of SoyNet construction. Co-functional networks of soybean genes were inferred from various soybean-specific genomics data and from evolutionarily conserved links in other species. The individual networks were then integrated into a single network, SoyNet, using Bayesian statistical framework.
Table 1. SoyNet and component networks inferred from 21 distinct data types.
| Network | Description | Genes | Links |
|---|---|---|---|
| SoyNet | Integrated network | 40 812 | 1 940 284 |
| GM-CX | By co-expression of Glycine max (soybean) genes | 38 300 | 539 521 |
| GM-GN | By gene neighborhood of two bacterial orthologs of Glycine max (soybean) genes in prokaryotic genomes | 6072 | 211 084 |
| GM-PG | By phylogenetic profile of Glycine max (soybean) gene similarity across species | 2665 | 30 695 |
| AT-CC | By co-citation of Arabidopsis thaliana orthologs in Pubmed articles | 11 482 | 256 676 |
| AT-CX | By co-expression of Arabidopsis thaliana orthologs | 10 494 | 125 109 |
| AT-HT | By high-throughput Arabidopsis thaliana orthologous PPI | 4966 | 22 140 |
| AT-LC | By literature curated Arabidopsis thaliana orthologous PPI | 4261 | 15 971 |
| CE-CC | By co-citation of Caenorhabditis elegans orthologs in Pubmed articles | 900 | 30 166 |
| CE-CX | By co-expression of Caenorhabditis elegans orthologs | 746 | 6694 |
| DM-CX | By co-expression of Drosophila melanogaster (Fly) orthologs | 4244 | 63 988 |
| DM-HT | By high-throughput Drosophila melanogaster (Fly) orthologous PPI | 6162 | 29 617 |
| DM-LC | By literature curated Drosophila melanogaster (Fly) orthologous PPI | 1379 | 10 494 |
| DR-CX | By co-expression of Danio rerio (Zebrafish) orthologs | 5934 | 281 220 |
| HS-HT | By high-throughput Homo sapiens (human) orthologous PPIs | 4695 | 85 126 |
| HS-LC | By literature curated Homo sapiens (human) orthologous PPIs | 7993 | 124 281 |
| OS-CX | By co-expression of Oryza sativa (rice) orthologs | 12 682 | 253 016 |
| SC-CC | By co-citation of Saccharomyces cerevisiae (yeast) orthologs in Pubmed articles | 7730 | 216 412 |
| SC-CX | By co-expression of Saccharomyces cerevisiae (yeast) orthologs | 6985 | 580 194 |
| SC-GT | By genetic interactions of Saccharomyces cerevisiae (yeast) orthologs | 5793 | 229 598 |
| SC-HT | By high-throughput Saccharomyces cerevisiae (yeast) orthologous PPIs | 6754 | 445 574 |
| SC-LC | By literature curated Saccharomyces cerevisiae (yeast) orthologous PPIs | 5760 | 103 626 |
NETWORK ASSESSMENT AND APPLICATION
Network assessment
To ensure that the co-functional links inferred by SoyNet are not simply based on memorizing gold-standard gene pairs or over-training, we needed to assess the network using test gene pairs that are independent from those used for the network training. We compiled gene pairs from GOBP annotations by the agriGO database (25) in May 2016, which was not used for training SoyNet. We noticed that several agriGO terms have so many member genes that potentially cause biased evaluation towards a few pathway terms (26). To avoid this pathway bias during network assessment, we excluded eight agriGO terms with more than 500 member genes in generating test gene pairs: ‘oxidation-reduction process’ (GO:0055114), ‘protein phosphorylation’ (GO:0006468), ‘regulation of transcription, DNA-templated’ (GO:0006355), ‘metabolic process’ (GO:0008152), ‘transmembrane transport’ (GO:0055085), ‘carbohydrate metabolic process’ (GO:0005975), ‘proteolysis’ (GO:0006508), and ‘translation’ (GO:0006412). Pairing two genes that are annotated by the same agiGO term resulted in a set of 745 683 gene pairs, of which only 82 919 gene pairs overlapped with the set of gene pairs used for network training (approximately 11% of 726 709 training gene pairs), confirming fair independence of the test gene pairs from those used for training SoyNet.
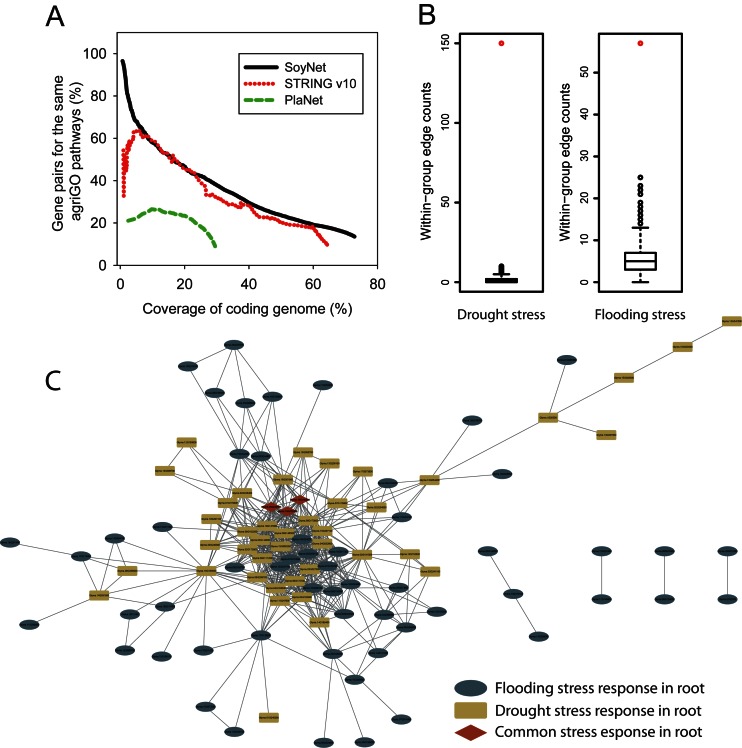
Two co-functional networks of soybean genes were previously published: PlaNet (6) and STRING v10 (7). Because the agriGO-based test gene pair set is also independent from the two other soybean gene networks, we compared SoyNet with those networks based on the same test gene pair set. The network assessment showed substantially higher performance for SoyNet compared to the other networks in retrieval rate of the test gene pairs, particularly for the top ranked gene pairs (Figure 2A).
Figure 2.
Assessment of SoyNet and other soybean functional networks. (A) Accuracies of gene pairs for the same agriGO pathways for the given genome coverage of each network are indicated for every bin of 1000 links. The resultant plot indicates that SoyNet outperforms STRING v10 and PlaNet in accuracy for most ranges of genome coverage. (B) Assuming genes connected in the network are functionally associated, SoyNet was assessed for functional modularity of proteins that are differentially expressed during specific abiotic stresses: drought and flooding. For both stress response proteomes, SoyNet shows significantly higher within-group edge counts than the distribution of those by 1000 random protein sets. (C) An abiotic stress response network of soybean genes based on SoyNet. Gene networks that respond to two different abiotic stresses, drought and flooding, have only three common genes, yet they are well-connected, suggesting that pathways for responding to different types of abiotic stresses are functionally interlaced.
In abiotic stress conditions, plants activate stress response pathways, often by enhancing protein biosynthesis. Then, differentially expressed proteins (DEPs) upon abiotic stress tend to be functionally associated with one another. Therefore, a significantly higher probability of functional association among DEPs for a specific stress condition would support the quality of the network. For example, the observed within-group edge count for stress-specific DEPs will be significantly higher than the count for random protein groups in a high quality co-functional network. For the analysis, we compiled two sets of soybean DEPs for two different abiotic stresses from a proteomics study: 48 proteins for drought response and 94 proteins for submergence response (27). We then tested the significance of the observed within-group edge counts for each stress-specific DEP group using 1000 random protein groups. We found that within-group edge count for both stress response proteomes are significantly higher than random chance (P < 0.001 for both DEPs by binomial distribution) (Figure 2B).
We also found that the two stress response proteomes have only few common response proteins in SoyNet, although they are highly interwoven to build an abiotic stress response network (Figure 2C). Notably, three response genes shared between drought and flooding stresses—Glyma.03G223000, Glyma.15G190500, and Glyma.19G220200—are located in the central region of the abiotic stress response network, suggesting their roles as common modulators in multiple stress responses. We found that Arabidopsis orthologs of the three genes are known to be involved in ethylene biosynthesis. Since ethylene is a core plant hormone involved in various stress responses including drought and submergence (28), the given network topology of the common stress response genes further supports the quality of SoyNet.
Network-based functional predictions by SoyNet
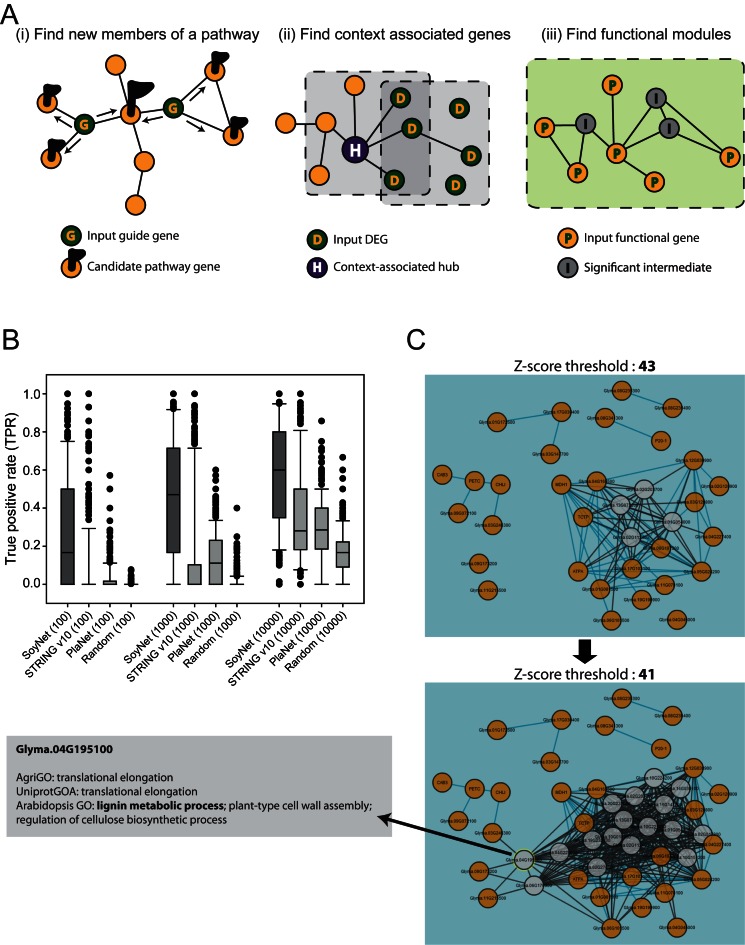
Integrative analysis of many co-functional links using various graph algorithms can effectively predict functions of individual genes (29). To maximize the utility of the co-functional links from SoyNet for the functional study of soybean genes and pathways involved in various crop traits, we implemented three complementary network-based algorithms for generating functional hypotheses: (i) find new members of a pathway, (ii) find context-associated genes and (iii) find functional modules (Figure 3A).
Figure 3.
Network-based methods for functional predictions implemented in the SoyNet server. (A) Overview of three network-based functional prediction methods. (B) Assessment of pathway predictions by ‘Find new members of a pathway’ with different soybean gene networks, SoyNet, STRING v10, and PlaNet. True positive rate (TPR) was measured for the top 100, 1000 and 10 000 retrieved genes for each of 338 agriGO pathways that have at least four member genes. Similar analyses were also conducted for random gene sets with the same number of member genes for each pathway. (C) Networks of 44 genes that respond to phosphorus deficiency and their intermediate nodes. A network obtained from a z-score threshold of 43 contains four intermediate nodes, whereas that by lower z-score threshold, 41, contains 13 more intermediate nodes. Clicking each gene or edge of the network shows additional information. For example, an intermediate node Glyma.04G195100 is annotated for lignin metabolic process.
Find new members of a pathway
Since two genes connected in co-functional networks have a high probability of being involved in the same pathways, new members of a pathway can be prioritized by closeness to the known pathway genes in the network. The same approach can predict new genes for a phenotype, because the majority of phenotypes are regulated by their associated pathways. In this network-based method, a functional search through the network is guided by the known genes for a target pathway or phenotype, called guide genes, which are usually provided by databases and the literature. A SoyNet network search can take guide genes based on not only soybean genes (both version 1.1 and 2) but also Arabidopsis genes by TAIR 10 (5) annotation, which are automatically converted into the soybean orthologs, using Inparanoid (23) software. The effectiveness of network-based functional prediction depends on the interconnectivity of pathway genes (30). Thus, once a user submits guide genes, the SoyNet server first measures the retrieval rate of the submitted guide genes by SoyNet connections among themselves, where a guide gene connected to the largest number of other guide genes is retrieved first. Overall performance of the network for retrieval of all guide genes is assessed by receiver operating characteristic (ROC) analysis, which can be summarized as an area under the ROC curve (AUC) score. For example, the AUC for 107 soybean genes for ‘fatty acid biosynthesis’ by agriGO (GO:0006633) is 0.743, which indicates that known genes for fatty acid biosynthesis are well connected to each other by SoyNet analysis, and other highly ranked genes are also likely to be involved in fatty acid biosynthesis. The SoyNet server then visualizes a network of guide genes and an extended network including their neighbors in SoyNet using Cytoscape Web (31). Lastly, novel candidate genes are prioritized based on sum of edge weights (log likelihood scores) to the guide genes are listed, along with additional information such as paralogs (32), functional annotations by agriGO (25), UniprotGOA (33) and GOBP of Arabidopsis orthologs (34).
We systematically assessed the prediction capability of SoyNet, PlaNet (6) and STRING v10 (7) for 338 agriGO pathways with at least four member genes. Because only top candidates are likely to be considered for the follow-up functional analysis, true positive rate (TPR) of top candidates is more significant. We therefore measured TPR for top 100, 1000 and 10 000 candidates by the method of ‘Find new members of a pathway’. We observed substantially higher TPR for the top 100 candidates from SoyNet than from other networks (Figure 3B). Notably, the TPR difference between SoyNet and other networks reduces as we consider a larger number of top candidates, indicating large improvement from previous soybean networks in prediction capability for more significant candidates.
Find context-associated genes
Plant transcriptome profiling for a particular biological context, such as abiotic stress, can reveal signature genes that are differentially expressed in that context. Network topology analysis with those signature genes can identify regulators for the cellular response. We hypothesize that if network neighbors of a certain hub gene are enriched among differentially expressed genes (DEGs) in a particular context, the hub gene is likely associated with the context. The identified hub gene could be associated with the context-specific response by regulating the DEGs directly or indirectly. We therefore implemented the ‘Find context associated genes’ method to identify candidate genes that regulate responses to the query biological context, which can be represented as DEGs. For this network-based method, only 15 444 hub genes with >50 direct neighbors in SoyNet are considered, and the significance of overlap between the neighbors of the hub and DEGs for the query context is measured by Fisher's exact test. For example, we submitted 94 DEGs observed after 6 h of incubating plant roots in iron deficient conditions (35). The SoyNet server returned a list of candidate hub genes that are associated with the root iron deficiency response along with additional information such as paralogs (32), transcription factor membership (36), functional annotations by agriGO (25), UniprotGOA (33) and GOBP of Arabidopsis orthologs (34). We found that 17 of the top 50 candidates were annotated by GOBP terms related to iron transport or iron homeostasis, and only one candidate gene (first rank) was a DEG itself, which indicates high complementarity between DEGs and the network-based predictions. To provide a closer look at the network-based candidates, the SoyNet server also visualizes the network of the DEGs and the candidate gene when the user clicks the candidate gene name in the table.
Find functional modules
Functionally coherent soybean genes are represented as a connected subnetwork or a module in SoyNet. However, the genetic part lists for pathways are often incomplete, and the missing member genes make the module fragmented into several disconnected graphs. Alternatively, the user may want to find a pathway by functional connections among signature genes such as DEGs derived from a relevant context, yet not all member genes of the pathway are DEGs in that context, resulting in disconnected graphs. Sometimes, including intermediate nodes between the disconnected graphs facilitates a more complete subnetwork for the pathway. Furthermore, these intermediate genes could be new candidates for the pathway. Therefore, we implemented a systematic way to choose intermediate nodes for the subnetwork based on a z-score threshold from the binomial proportion test as introduced previously (37), where a lower z-score permits more intermediate nodes. With real time network visualization for a user-selected z-score threshold, one may find optimal network modules by trials of various z-scores. However, due to the considerable time required for calculating coordinates of all network nodes, the SoyNet server allows up to only 50 intermediate nodes. For example, we submitted 44 genes that respond to phosphorus deficiency (38). When the z-score threshold was 43, we had four intermediate nodes (Figure 3C upper panel). By lowering the z-score down to 41, 13 more intermediate nodes were visible and the size of the largest connected graph increased (Figure 3C lower panel). Network viewer allows users to see additional information for a selected node of edge. We found that new intermediate nodes, Glyma.04G195100 and Glyma.06G170900 were annotated for lignin metabolic process, a cell wall biosynthesis process, which justifies the relevance of these intermediate genes to the module for phosphorus deficiency response.
CONCLUSIONS
In this study, we developed SoyNet, a database of co-functional networks of G. max genes, constructed by analyzing 21 distinct types of genomics big data and Bayesian integrations. The database contains not only evolutionarily conserved co-functional links transferred from other species but also many of those inferred from soybean-specific genomics data such as transcriptome profiles based on microarray and RNA-seq. The extensive network inference from heterogeneous data enabled us to obtain the most comprehensive view of the soybean pathway systems to date. Moreover, we confirmed the superiority of our novel database over other soybean functional network databases in network quality and pathway predictions. To facilitate network-based functional hypothesis generation in soybean, we implemented three complementary network-based algorithms in the database web server. To the best of our knowledge, SoyNet is the first genome-scale co-functional network database with companion web-based functional prediction tools. Users can test each network-based prediction method with available example input data in the web server. We believe that the substantially enhanced genome coverage, accuracy, and usability of SoyNet will facilitate systems biology approaches to study complex soybean traits. We also expect that similar genome-scale co-functional networks can be constructed for many other economic crops with the aid of the recent explosion of genomics data based on next-generation sequencing technology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [2012M3A9B4028641, 2012M3A9C7050151, 2015R1A2A1A15055859 to I.L.]. Funding for open access charge: National Research Foundation of Korea.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D.L., Song Q., Thelen J.J., Cheng J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhary J., Patil G.B., Sonah H., Deshmukh R.K., Vuong T.D., Valliyodan B., Nguyen H.T. Expanding Omics Resources for Improvement of Soybean Seed Composition Traits. Front. Plant Sci. 2015;6:1021. doi: 10.3389/fpls.2015.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshmukh R., Sonah H., Patil G., Chen W., Prince S., Mutava R., Vuong T., Valliyodan B., Nguyen H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014;5:244. doi: 10.3389/fpls.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee T., Kim H., Lee I. Network-assisted crop systems genetics: network inference and integrative analysis. Curr. Opin. Plant Biol. 2015;24:61–70. doi: 10.1016/j.pbi.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutwil M., Klie S., Tohge T., Giorgi F.M., Wilkins O., Campbell M.M., Fernie A.R., Usadel B., Nikoloski Z., Persson S. PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P.F., Dreher K., Karthikeyan A., Chi A., Pujar A., Caspi R., Karp P., Kirkup V., Latendresse M., Lee C., et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153:1479–1491. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodama Y., Shumway M., Leinonen R., International Nucleotide Sequence Database C. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin J., Lee T., Kim H., Lee I. Complementarity between distance- and probability-based methods of gene neighbourhood identification for pathway reconstruction. Mol. BioSyst. 2014;10:24–29. doi: 10.1039/c3mb70366e. [DOI] [PubMed] [Google Scholar]
- 15.Shin J., Lee I. Co-inheritance analysis within the domains of life substantially improves network inference by phylogenetic profiling. PLoS One. 2015;10:e0139006. doi: 10.1371/journal.pone.0139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim E., Kim H., Lee I. JiffyNet: a web-based instant protein network modeler for newly sequenced species. Nucleic Acids Res. 2013;41:W192–W197. doi: 10.1093/nar/gkt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee T., Yang S., Kim E., Ko Y., Hwang S., Shin J., Shim J.E., Shim H., Kim H., Kim C., et al. AraNet v2: an improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015;43:D996–D1002. doi: 10.1093/nar/gku1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho A., Shin J., Hwang S., Kim C., Shim H., Kim H., Kim H., Lee I. WormNet v3: a network-assisted hypothesis-generating server for Caenorhabditis elegans. Nucleic Acids Res. 2014;42:W76–W82. doi: 10.1093/nar/gku367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin J., Yang S., Kim E., Kim C.Y., Shim H., Cho A., Kim H., Hwang S., Shim J.E., Lee I. FlyNet: a versatile network prioritization server for the Drosophila community. Nucleic Acids Res. 2015;43:W91–W97. doi: 10.1093/nar/gkv453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee I., Blom U.M., Wang P.I., Shim J.E., Marcotte E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H., Shin J., Kim E., Kim H., Hwang S., Shim J.E., Lee I. YeastNet v3: a public database of data-specific and integrated functional gene networks for Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D731–D736. doi: 10.1093/nar/gkt981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee T., Oh T., Yang S., Shin J., Hwang S., Kim C.Y., Kim H., Shim H., Shim J.E., Ronald P.C., et al. RiceNet v2: an improved network prioritization server for rice genes. Nucleic Acids Res. 2015;43:W122–W127. doi: 10.1093/nar/gkv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnhammer E.L.L., Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee I., Date S.V., Adai A.T., Marcotte E.M. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 25.Du Z., Zhou X., Ling Y., Zhang Z., Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee I., Li Z., Marcotte E.M. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS One. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh M., Komatsu S. Characterization of proteins in soybean roots under flooding and drought stresses. J. Proteomics. 2015;114:161–181. doi: 10.1016/j.jprot.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Morgan P.W., Drew M.C. Ethylene and plant responses to stress. Physiol. Plantarum. 1997;100:620–630. [Google Scholar]
- 29.Mostafavi S., Morris Q. Combining many interaction networks to predict gene function and analyze gene lists. Proteomics. 2012;12:1687–1696. doi: 10.1002/pmic.201100607. [DOI] [PubMed] [Google Scholar]
- 30.Shim J.E., Hwang S., Lee I. Pathway-Dependent Effectiveness of Network Algorithms for Gene Prioritization. PLoS One. 2015;10:e0130589. doi: 10.1371/journal.pone.0130589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee T.H., Tang H., Wang X., Paterson A.H. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41:D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J., O'Donovan C. The GOA database: gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:D1057–D1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blake J.A., Christie K.R., Dolan M.E., Drabkin H.J., Hill D.P., Ni L., Sitnikov D., Burgess S., Buza T., Gresham C., et al. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran Lauter A.N., Peiffer G.A., Yin T., Whitham S.A., Cook D., Shoemaker R.C., Graham M.A. Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genomics. 2014;15:702. doi: 10.1186/1471-2164-15-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., Libault M., Joshi T., Valliyodan B., Nguyen H.T., Xu D., Stacey G., Cheng J. SoyDB: a knowledge database of soybean transcription factors. BMC Plant Biology. 2010;10:14. doi: 10.1186/1471-2229-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger S.I., Posner J.M., Ma'ayan A. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sha A.H., Li M., Yang P.F. Identification of phosphorus deficiency responsive proteins in a high phosphorus acquisition soybean (Glycine max) cultivar through proteomic analysis. BBA-Proteins Proteom. 2016;1864:427–434. doi: 10.1016/j.bbapap.2016.02.001. [DOI] [PubMed] [Google Scholar]