Abstract
The Protein Circular Dichroism Data Bank (PCDDB) has been in operation for more than 5 years as a public repository for archiving circular dichroism spectroscopic data and associated bioinformatics and experimental metadata. Since its inception, many improvements and new developments have been made in data display, searching algorithms, data formats, data content, auxillary information, and validation techniques, as well as, of course, an increase in the number of holdings. It provides a site (http://pcddb.cryst.bbk.ac.uk) for authors to deposit experimental data as well as detailed information on methods and calculations associated with published work. It also includes links for each entry to bioinformatics databases. The data are freely available to accessors either as single files or as complete data bank downloads. The PCDDB has found broad usage by the structural biology, bioinformatics, analytical and pharmaceutical communities, and has formed the basis for new software and methods developments.
INTRODUCTION
The Protein Circular Dichroism Data Bank (PCDDB) (1) is an open access data bank for the deposition and dissemination of circular dichroism (CD) spectra and synchrotron radiation circular dichroism (SRCD) spectra and metadata. The data bank was created in 2009 (first as an accession-only data bank, and later as a deposition data bank), and has been in continuous use since then. Accessors of the data do not have to register to access or download files, although depositors must register (as in other deposition data banks) to ensure good practice and traceability of the data. Any user can make an account for the purposes of logging repeat searches, saving subsets of files, or receiving notification of content enhancements.
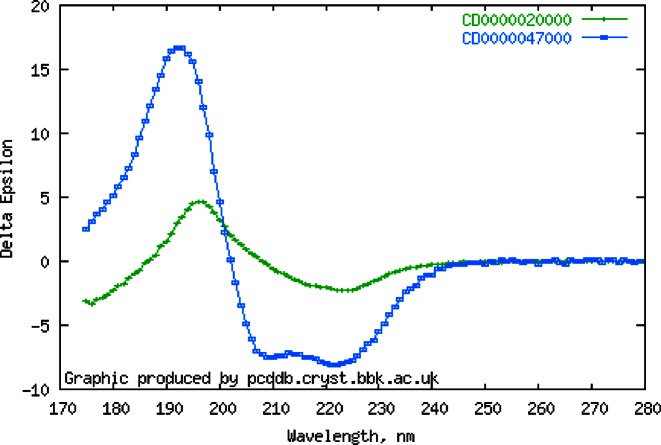
The PCDDB entries include spectral data (including both raw and final processed CD spectra, and associated HT/HV curves (which are, effectively, pseudo-absorbance spectra)), detailed information on the sample and experimental conditions used to obtain the spectra, and links to related bioinformatics databases, including the Protein Data Bank (PDB) (2), the UniProt sequence database (3), the CATH protein structure classification system (4), and the Enzyme Classification (EC) database (5), where available. Plots of the spectra (Figure 1) are available for display online. Entries may also include annotations in the ‘keywords’ which can provide useful identifiers; for example, ‘SMP180’ which indicates the spectrum is a component of the membrane protein reference dataset (6) used in empirical secondary structure analyses. The accession ID system allows for grouping of related spectra, such as thermal melt series of spectra undertaken for stability studies. Entries include information on the associated publication describing the work (citation to the original work is required of any user of the data). Each entry includes a validation section, as a guide to the data quality for both the depositor and user. The validation takes the form of a series of individual tests on the data and metadata, as well as providing an overall validation level. Individual entries can be downloaded in several ASCII formats, or the entire contents of the database can be downloaded as a single compressed archive file. The entries now include collections of proteins that cover both a wide range of secondary structures and folds (7), as well as more specialised types (such as membrane proteins (6) and beta-sheet rich proteins (8)), in addition to individual spectra of folded and unfolded proteins.
Figure 1.
Plots of overlaid selected circular dichroism spectra from the PCDDB, including (upper right hand corner) their PCDDB ID codes.
A number of individual publications (Biochemistry, Biophysical Journal) and publishers (Nature Publishing Group, and PLOS journals), research councils and funding agencies (i.e., http://www.bbsrc.ac.uk/funding/apply/application-guidance/justification-resources/resources/) suggest that PCDDB depositions be made in association with their publications or funding. The PCDDB is an approved component member of biosharing.org (https://biosharing.org/biodbcore-000613). The records within the data bank include a high proportion of SRCD spectra, which has been the direct consequence of SRCD beamline scientists (and synchrotrons) encouraging their users to deposit data (9). It is noteworthy that the PCDDB remains the only publically-available database for protein circular dichroism spectra.
NEW FEATURES
The features of the PCDDB were described in the initial report of its release (1). Since that time, a number of new or modified features, procedures, and associated materials have been developed based on user and developer suggestions, in order to make the resource more complete, user-friendly, and informative. These new features are described below.
Entry naming formats
The naming convention for PCDDB entries has been updated (on the basis of a recommendation from the International Scientific Advisory Board): Entries now begin with the initial letters CD (i.e. CDxxxxxxxxxx), to signify being associated with this data bank. The letters are followed by 10 digits which can be thought of as, sequentially, a seven-digit main code, a one-digit revision number and a two-digit series number. Grouped spectra therefore (e.g. a thermal melt series), may all have the same seven-digit main code and one-digit revision number, but sequential two-digit series numbers, so they are recognisable as part of related experiments. Old codes are aliased to these new ones in the searching facility.
New database fields
Additional database fields have been added: the sample supplier (person or company), mutation details (compared to wildtype), continuous or stepped scan (a relevant experimental detail), and final spectrum calibrated (to determine what processing has been applied to the spectra). These fields were added to improve data traceability and enable reproducibility. As both conventional CD and SRCD spectra may be deposited, additional fields specific for SRCD data collection have also been added.
Deposition improvements
Depositors are now sent notifications of release, and periodic reminders of entries that are still in pre-release. If depositions are released prior to publication, updates can be made to the entry to note the citing publication by contacting the PCDDB via email (PCDDB@mail.cryst.bbk.ac.uk). To improve the ease (and time required) for depositing multiple similar files, depositors may now use previous entries as templates (to be modified rather than re-created). To aid new depositors, tutorials about deposition, both online – (http://www.youtube.com/user/ThePcddb) and in print (10), have been created.
Validation
Each entry includes an online validation report, and following experience gained in analysing depositions, the validation procedures have been improved and extended. A validation report is provided to the depositor prior to the release of the entry so that they may assess the quality and completeness of their deposition, and make any changes/additions they deem necessary prior to release. Validation reports include flag (minor issues) or fail (major issues) tags for individual items and overall, and a completeness label. The PCDDB also provides the facility for publication reviewers (with permission) to anonymously access unreleased entries as a guide when considering a paper for publication. Alternatively, depositors can provide the validation report in .pdf format that was created as part of the deposition process to journals as additional data for reviewing purposes.
Enhanced searching
The database metadata can be searched using a number of parameters [Table 1 lists the current/new search terms and parameters] including protein names, source organism, a range of experimental parameters, bioinformatics information (including PDBIDs, Uniprot codes, EC number, and CATH class), with the option of excluding/including entries that have ‘failed’ or ‘flagged’ validation status. It can now also be searched for proteins with specific ranges of secondary structure types (as an example: >50% helix). Keywords can be searched for protein type (e.g., membrane protein), and whether the entry is one included in a CD analysis reference database (such as SP175 (7) or SMP180 (6)). Additionally, a novel development for searching the spectral data using a spectrum as the input has been added; this enables identification of related spectra (something which may have value in studies seeking to identify structurally-related proteins). Performing spectral matching relevant to circular dichroism is not necessarily as straight forward as ranking the potential matches by minimal RMSD differences, so several search methods are enabled, following the concepts previously described for the DichroMatch web server (11). Finally, the database may now be queried by deposition date (before, after, or between specific dates), which can be helpful in enabling accessors to identify new entries.
Table 1. Popular search parameters: (all text fields support * wildcards and most values include options for greater than, less than, or between).
| Identity: |
| PCDDBID |
| Deposition date (YYYYMMDD) |
| Protein name |
| Sample characteristics: |
| Source organism |
| Expression system |
| Molecular weight (Da) |
| Spectral/experimental characteristics: |
| Minimum wavelength (nm) |
| Protein concentration (min or max), mg/ml |
| Instrument/synchrotron |
| Temperature, °C |
| Protein purity, % |
| Sample secondary structures: |
| Alpha helix (%) |
| 3–10 helix (%) |
| Pi helix (%) |
| Beta strand (%) |
| Beta bridge (%) |
| Hydrogen-bonded bend (%) |
| Hydrogen-bonded turn (%) |
| Irregular (%) |
| Bioinformatics information: |
| Keyword/phrase |
| PDB ID |
| UniProt ID |
| Enzyme Classification (EC) number |
| CATH classification |
| Deposition information: |
| Depositor/principal investigator name |
| Depositor address |
| Utility: |
| Show all entries |
| Show all entries with a PDB record |
Enhanced operations on search results and enhanced plots
Search results can now be selected for inclusion in subsets via checkboxes. These subsets can be downloaded, and if the user is logged in (an option for accessors), they can be saved as lists for future use. Whilst online plots are not intended to be of publication quality since the spectral files can be downloaded by users for production purposes, it was recognised that the ability to facilely compare spectra of several proteins online was important. Hence, plotting spectra of multiple entries in the subset list on a single figure is now possible (Figure 1).
Associated tools/software
The main page of the site includes links to associated analytical tools such as the DichroWeb (12,13), 2Struc (14) and DichroMatch (11) analysis websites. There is also a link to the ValiDichro standalone data testing website (http://valispec.cryst.bbk.ac.uk/circularDichroism/ValiDichro/upload.html) (15), which enables users to perform validation analyses equivalent to the data checking software utilised in the curation of PCDDB entries (but in this case, not requiring deposition), as an aid to experimental good practice.
Associated YouTube informational videos on the ‘PCCDB Channel’
Instructional videos have been created to provide additional related information for users of the PCCDB. The channel may be accessed from the PCDDB home page or from the YouTube icon on the footer of all pages. The videos now include the following topics: An Introduction to the PCDDB, and information on how to make depositions (https://www.youtube.com/watch?v=NTblyIhwjog), information on analysing CD spectra using the DichroWeb server (https://www.youtube.com/watch?v=QZat_Wr2NGM), how to calibrate CD spectra (https://www.youtube.com/watch?v=ovY6yVxw-tI), how to calibrate CD instruments (https://www.youtube.com/watch?v=PEIDelWvSsg), how to calibrate the pathlengths of CD sample cells (https://www.youtube.com/watch?v=PEIDelWvSsg), and how to clean and load CD cells (https://www.youtube.com/watch?v=OhD50eiLzWI). Users are invited to submit suggestions for additional informational videos (to PCDDB@mail.cryst.bbk.ac.uk).
Information pages
A glossary of terms commonly used in the field has been added and may be accessed via the homepage. Software updates are noted on the ‘Version History’ page (http://pcddb.cryst.bbk.ac.uk/verhist.php), that is part of the ‘about’ section, which may be of particular use to software developers that access this database programmatically.
Spectrum of the Month feature
To recognise the contributions of our depositors, a selected ‘Featured Spectrum of the Month’ is displayed on the front page, with links to its entry. Featured spectra include both new and existing entries, along with graphic illustrations of their crystal structures (when available) from associated PDB codes. The selected spectra are chosen based on their having interesting spectral features that may be novel or representative of new classes of entries, or when a protein is highly accessed due to popular interest in its structure.
Bulletins
Below the ‘Featured Spectrum’ notices of relevant meetings and workshops are posted (suggestions from external users may be offered by contacting the PCDDB developer). Software and database holding updates are listed on the left-hand ‘Information’ panel.
USES
As a resource for structural biology, biochemistry and bioinformatics, the PCDDB has been accessed both by users obtaining individual spectra for specific applications and also as complete (or selected component) downloads. A number of the types of applications for the PCDDB data that were envisioned in the initial report describing the development of the PCDDB (1) have now been realised.
Individual files have been used for biochemical/structural biology studies of specific and related proteins, including comparisons with spectra of other proteins of known structure (16,17), comparisons of environmental effects on protein structure, where the structure of a given protein is known only in a single environment (crystal) (18), and using spectra and calculated secondary structure (and also thermal melt profiles) to identify the structure and stability of a protein as an aid to crystallisation studies (19).
As complete or selected subsets of downloads, they have been used for new method developments (both bioinformatics and physical methods), including: developing and/or testing new algorithms for secondary structure determination (8,20–22), using the ratio of the 222 and 200 nm peaks as a measure of identifying folded vs. unfolded proteins (23), proposing a new method for determining membrane protein helical content from the spectral slope between 230–240 nm (24), and testing of calculations of vibronic structure contributions to far UV CD spectra (25). Based on the large number of downloads that have been undertaken thus far, it is expected that many additional uses will be identified by users in the future.
FUTURE DEVELOPMENTS
Our aim is to expand the PCDDB holdings beyond traditional solution circular dichroism spectroscopy on proteins, by including entries in the rapidly growing spectral areas of oriented CD (oCD) and oriented SRCD (oSRCD) (26). The data bank will also be expanded to include other sample types, including nucleic acids (both RNAs and DNAs) and peptides. These will require the addition of new fields in the spectral parameters, different fields in the sample characteristics and bioinformatics links, and different validation procedures (likely including machine learning to identify parameter outliers). As a result we will be adding new experts in these areas to our advisory boards.
CONCLUSIONS
The PCDDB is an ongoing resource for the deposition and dissemination of CD spectral data and associated metadata of proteins. Its entries are linked to a range of other bioinformatics resources such as the Protein Data Bank, the CATH protein classification database, the EC database, and sequence databases. New developments are constantly being added (often in response to users’ requests made via the contact email PCDDB@mail.cryst.bbk.ac.uk). It has had >1 million files downloaded, and has been accessed by >10,000 unique users in the ∼5 years since its inception, and has been used for individual structural biology/biochemistry studies as well as for new bioinformatics methods developments.
Acknowledgments
The PCDDB would like to acknowledge the advice and support of members of its International Advisory Boards (http://pcddb.cryst.bbk.ac.uk/about.php).
FUNDING
U.K. Biotechnology and Biological Research Council [Bioinformatics and Biological Resources Fund grants J019135 and F010346 to B.A.W.; J019194 and F010362 to R.W.J., a Tools and Resources Development Fund grant I023887 to R.W.J.]; International Union of Pure and Applied Chemistry [2010-033-2-024 to B.A.W.]. Funding for open access charge: Biotechnology and Biological Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Whitmore L., Woollett B., Miles A.J., Klose D.P., Janes R.W., Wallace B.A. PCDDB: The protein circular dichroism data bank, a repository for circular dichroism spectral and metadata. Nucleic Acids Res. 2011;39:D480–D486. doi: 10.1093/nar/gkq1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H., Henrick K., Nakamura H., Markley J.L. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The UniProt, Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sillitoe I., Lewis T.E., Cuff A., Das S., Ashford P., Dawson N.L., Furnham N., Laskowski R.A., Lee D., Lees J.G., Lehtinen S., Studer R.A., Thornton J., Orengo C.A. CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015;43:D376–D381. doi: 10.1093/nar/gku947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann A., Darsow M., Degtyarenko K., Fleischmann W., Boyce S., Axelsen K.B., Bairoch A., Schomburg D., Tipton K.F., Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdul-Gader A., Miles A.J., Wallace B.A. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinformatics. 2011;27:1630–1636. doi: 10.1093/bioinformatics/btr234. [DOI] [PubMed] [Google Scholar]
- 7.Lees J.G., Miles A.J., Wien F., Wallace B.A. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 8.Micsonai A., Wien F., Kernya L., Lee Y.H., Goto Y., Réfrégiers M., Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace B.A., Bu¨rck J. Third international synchrotron radiation circular dichroism spectroscopy meeting. Synch. Rad. News. 2015;28:58–59. [Google Scholar]
- 10.Janes R.W., Miles A.J., Woollett B., Whitmore L., Klose D., Wallace B.A. Circular dichroism spectral data and metadata in the protein circular dichroism data bank (PCDDB): a tutorial guide to accession and deposition. Chirality. 2012;24:751–763. doi: 10.1002/chir.22050. [DOI] [PubMed] [Google Scholar]
- 11.Klose D.P., Wallace B.A., Janes R.W. DichroMatch: A website for similarity searching of circular dichroism spectra. Nucleic Acids Res. 2012;40:W547–W552. doi: 10.1093/nar/gks449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 13.Whitmore L., Wallace B.A. DICHROWEB, An online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klose D.P., Wallace B.A., Janes R.W. 2Struc:The secondary structure server. Bioinformatics. 2010;26:2624–2625. doi: 10.1093/bioinformatics/btq480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woollett B., Whitmore L., Janes R.W., Wallace B.A. ValiDichro: a website for validating and quality control of protein circular dichroism spectra. Nucleic Acids Res. 2013;41:W417–W421. doi: 10.1093/nar/gkt287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavassoly O., Lee J.S. Methamphetamine binds to α-synuclein and causes a conformational change which can be detected by nanopore analysis. FEBS Lett. 2012;586:3222–3228. doi: 10.1016/j.febslet.2012.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Muta H., Lee Y.H., Kardos J., Lin Y., Yagi H., Goto Y. Supersaturation-limited amyloid fibrillation of insulin revealed by ultrasonication. J. Biol. Chem. 2014;289:18228–18238. doi: 10.1074/jbc.M114.566950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thyparambil A.A., Wei Y., Latour R.A. Experimental characterization of adsorbed protein orientation, conformation, and bioactivity. Biointerphases. 2015;10:019002. doi: 10.1116/1.4906485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deller M.C., Kong L., Rupp B. Protein stability: a crystallographer's perspective. Acta Cryst. 2016;F72:72–95. doi: 10.1107/S2053230X15024619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis-Jeune C., Andrade-Navarro M.A., Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins: Struct. Funct. Bioinf. 2012;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 21.Wiedemann C., Bellstedt P., Görlach M. CAPITO - a web server based analysis and plotting tool for circular dichroism data. Bioinformatics. 2013;29:1750–1757. doi: 10.1093/bioinformatics/btt278. [DOI] [PubMed] [Google Scholar]
- 22.Uporov I.V., Forlemu N.Y., Nori R., Aleksandrov T., Sango B.A., Mbote Y.E.B., Pothuganti S., Thomasson K.A. Introducing DInaMo: a package for calculating protein circular dichroism using classical electromagnetic theory. Int. J. Mol. Sci. 2015;16:21237–21276. doi: 10.3390/ijms160921237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Motlagh H.N., Chakuroff C., Thompson E.B., Hilser V.J. Thermodynamic intrinsically disordered N-terminal domain of human glucocorticoid receptor. J. Biol. Chem. 2012;287:26777–26787. doi: 10.1074/jbc.M112.355651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y., Thyparambil A.A., Latour R.A. Protein helical structure determination using CD spectroscopy for solutions with strong background absorbance from 190 to 230 nm. Biochim. Biophys. Acta. 2014;1844:2331–2337. doi: 10.1016/j.bbapap.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z., Robinson D., Hirst J.D. Vibronic structure in the far-UV electronic circular dichroism spectra of proteins. Faraday Disc. 2015;177:329–344. doi: 10.1039/c4fd00163j. [DOI] [PubMed] [Google Scholar]
- 26.Bu¨rck J., Wadhwani P., Fangha¨nel S., Ulrich A.S. Oriented circular dichroism: a method to characterize membrane-active peptides in oriented lipid bilayers. Acc. Chem. Res. 2016;49:184–192. doi: 10.1021/acs.accounts.5b00346. [DOI] [PubMed] [Google Scholar]