Abstract
RNA editing is a widespread post-transcriptional mechanism that can make a single base change on specific nucleotide sequence in an RNA transcript. RNA editing events can result in missense codon changes and modulation of alternative splicing in mRNA, and modification of regulatory RNAs and their binding sites in noncoding RNAs. Recent computational studies accurately detected more than 2 million A-to-I RNA editing sites from next-generation sequencing (NGS). However, the vast majority of these RNA editing sites have unknown functions and are in noncoding regions of the genome. To provide a useful resource for the functional effects of RNA editing in long noncoding RNAs (lncRNAs), we systematically analyzed the A-to-I editing sites in lncRNAs across human, rhesus, mouse, and fly, and observed an appreciable number of RNA editing sites which can significantly impact the secondary structures of lncRNAs and lncRNA–miRNA interactions. All the data were compiled into LNCediting, a user-friendly database (http://bioinfo.life.hust.edu.cn/LNCediting/). LNCediting provides customized tools to predict functional effects of novel editing sites in lncRNAs. We hope that it will become an important resource for exploring functions of RNA editing sites in lncRNAs.
INTRODUCTION
Recent studies have revealed that most of the human genome nucleotides are capable of being transcribed, producing numerals of noncoding RNAs (ncRNAs) (1). Among these, RNA molecules with >200 nucleotides in length are refered as long noncoding RNAs (lncRNAs), which are typically transcribed by RNA polymerase II and are often multiexonic and polyadenylated (2). With the development of high-throughput sequencing as well as the computational prediction algorithms, an enormous amount of lncRNAs is being identified. Therefore, several databases have been developed to annotate lncRNAs, such as NONCODE (3), LNCipedia (4) and GENCODE (5). The most comprehensive lncRNA database, LNCat, combined 24 lncRNA annotation resources and annotated >300 000 lncRNA transcripts in over 50 tissues and cell lines in the human genome (6). LncRNAs play key roles in various biological processes, such as imprinting control, cell differentiation, immune response and chromatin modification (7,8). Dysregulation of lncRNAs is relevant to tumorigenesis, neurological disorders, cardiovascular disorders, developmental disorders, and other diseases (9). Variants in lncRNA sequences, such as single nucleotide polymorphisms (SNPs), are associated with a wide range of human diseases (10).
Similar to SNPs, RNA editing, which can also make a single base change on specific nucleotide sequence, is a post-transcriptional modification of RNA sequences. Among different types of RNA editing, adenosine to inosine (A-to-I) RNA editing is the most common one in mammals (11). Genome-wide identification of RNA editing from high thoughput transcriptome sequencing showed that up to 85% of the high-confidence RNA variants are A-to-I editing (12,13). The events of A-to-I editing are catalyzed by the adenosine deaminase acting on RNA (ADAR) family of enzymes, that convert A to I, which in turn is interpreted as guanosine (G) by both the translation and splicing machineries (14). In recent years, A-to-I editing sites have been identified not only in protein-coding genes, but also in ncRNAs, such as microRNAs, small interfering RNAs, transfer RNAs and lncRNAs (15–17). Editing events increase the diversity of the transcriptome and may result in several consequences, such as affecting splicing, creating or disrupting microRNA/mRNA binding sites, and affecting the biogenesis, stability, and target recognition of ncRNAs (18,19). The aberrations of the A-to-I editing are associated with various human diseases, including neurodegenerative, metabolic and carcinogenic diseases (20–22). For example, Maas et al. analyzed the editing level of malignant gliomas tissue samples and revealed that the GluA2 Q/R site was significantly under-edited compared to control samples (23). Chen et al. discovered that RNA editing of AZIN1 (encoding antizyme inhibitor 1) was increased in hepatocellular carcinoma (24).
Due to the functional importance of RNA editing sites, several databases presented collections of RNA editing sites, which greatly accelerated the research in the field. In 2007, dbRES collected 5437 experimentally validated editing sites of 251 transcripts across 96 organisms (25). In 2010, DARNED collected 42 063 editing sites in human genome based on the EST data (26). With the development of next-generation sequencing (NGS) technology, the number of RNA editing sites increased exponentially (27–29). In 2013, RADAR collected more than 2 million RNA editing sites, creating the most comprehensive collection of RNA editing sites so far (30). However, there is no functional annotation for RNA editing sites in the aforementioned databases. Furthermore, the research focus on RNA editing has moved from the identification of novel sites to the characterization of the functional impacts (31). Recently, we characterized the global A-to-I RNA editing profiles of 6236 patient samples of 17 cancer types from The Cancer Genome Atlas (32). An appreciable number of clinically relevant editing events was identified in noncoding regions. These results highlighted the necessity to study the functional roles of editing sites because of mechanistic, prognostic and therapeutic implications, potentially in lncRNAs (33).
In this study, we constructed the LNCediting database, aiming to provide comprehensive information about A-to-I editing sites in lncRNAs across human, rhesus, mouse, and fly, and to explore their potential functions. We characterized all editing sites in lncRNAs and predicted their effects on lncRNA secondary structures and lncRNA–miRNA interactions. The LNCediting database is freely accessible at http://bioinfo.life.hust.edu.cn/LNCediting/.
DATA COLLECTION AND DATABASE CONTENT
RNA editing sites in lncRNA transcripts
The annotation of more than 300 000 human lncRNAs (hg19) was obtained from LNCat (6), the most comprehensive annotation resources for human lncRNAs. The lncRNA annotation of mouse (mm10) was downloaded from NONCODE (version 2016) (3) and GENCODE (M10) (34), while the lncRNA annotations of rhesus (mac3) and fly (dm6) were downloaded from NONCODE (version 2016).
The editing sites of human, mouse, and fly were obtained from RADAR (30), while the editing sites of the rhesus were obtained from Chen et al. (35). By using the LiftOver tool from the UCSC Genome Browser (36), we converted the coordinates of RNA editing sites to be consistent with lncRNA annotation. We obtained 2 576 459, 8822, 26 861 and 5025 editing sites in human (hg19), mouse (mm10), rhesus (mac3) and fly (dm6), respectively.
We then mapped the coordinates of the editing sites to lncRNA, and identified 191 991, 1922, 165 and 1829 editing sites in human, mouse, rhesus, and fly lncRNAs, respectively (Table 1). In humans, the number of the editing sites in lncRNAs ranges from 0 to 1248, with a mean of 16 and a median of five editing sites per lncRNA.
Table 1. The statistics in LNCediting database.
| Species | Species full name (genome build) | Editing sites | LncRNAs | Editing sites in lncRNAs | Sites affect structuresa | Sites affect miRNA bindingb |
|---|---|---|---|---|---|---|
| Human | Homo sapiens (hg19) | 2 576 459 | 301 896 | 191 991 | 123 950 | 109 788/114 814 |
| Mouse | Mus musculus (mm10) | 8822 | 59 574 | 1922 | 1585 | 819/956 |
| rhesus | Macaca mulatta (mac3) | 26 861 | 15 450 | 165 | 148 | 52/50 |
| Fly | Drosophila melanogaster (dm6) | 5025 | 68 706 | 1829 | 1454 | 157/160 |
aNumber of editing sites affect lncRNA secondary structures.
bNumber of editing sites affect miRNA–lncRNA interaction (loss/gain).
Impacts of RNA editing sites on lncRNA structures
The secondary structure of lncRNAs dictates their functions for sensory, guiding, scaffolding and allosteric capacities (37), indicating that a change in the secondary structure may influence their stability and function. Therefore, we assessed the impacts of editing sites on lncRNA secondary structures. Whole genome sequences of human (hg19), mouse (mm10), rhesus (mac3) and fly (dm6) were downloaded from the UCSC genome browser, and unedited-type (UT) sequences of lncRNAs were extracted based on lncRNA annotation. For each editing site, we changed the corresponding base of editing site from A to G to obtain the edited-type (ET) lncRNA transcript. We then applied RNAfold program (38) to predict secondary structures, and calculated the minimal free energy (MFE, ΔG) of UT and ET lncRNA transcript. Energy change of RNA structures (ΔΔG) was calculated by the MFE differences using ΔΔG = ΔGET − ΔGUT, where ΔGET and ΔGUT are the MFEs of ET and UT lncRNA transcript, respectively. We only analyzed editing sites in lncRNAs with length <10kb limited by RNAfold. In human, we predicted the MFEs of 156 502 editing sites in 26 554 lncRNAs. Among them, we identified 123 950 (79.2%) editing sites with impacts on the MFE of lncRNA secondary structures, and 51 489 (32.9%) editing sites altered the MFE for more than 2 kcal/mol, which can significantly change the local structure of lncRNAs.
Previous studies indicated that A-to-I editing sites tend to be clustered together in human transcriptome (39,40). Therefore, we further predicted the impact of multiple editing sites on the structure of this lncRNA by changing multiple editing sites simultaneously. The absolute values of ΔΔG can be up to 364.3 kcal/mol, with a mean of 14.7 kcal/mol and a median of 5.9 kcal/mol per lncRNA. Linear regression analysis suggested that ΔΔG is highly correlated with the number of editing sites (0.96 kcal/mol per editing site, Pearson correlation r = 0.92, P< 0.001).
Impacts of RNA editing sites on miRNA–lncRNA interactions
lncRNAs can be directly regulated by miRNAs that starBase collected >10 000 miRNA–ncRNA interactions from high-throughput studies (41). These lncRNAs can function as miRNA decoy, thereby indirectly regulate the expression of certain protein-coding genes by competing shared miRNAs (42). For example, miRNA-192 and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma (43). Several cases have been reported that in the miRNA–lncRNA binding region, a single base change, such as SNPs and somatic mutations, can influence the miRNA–lncRNA interactions (44,45). Similar to SNPs, we hypothesized that editing sites also have the potential to influence the miRNA–lncRNA interactions. We downloaded the miRNA annotations of the four species from miRBase (v21) (46). For each editing site, we extracted editing site and the flanking 25 bps from lncRNA transcript (in total 51 bp) as unedited-type (UT) sequence, and changed the editing site from A to G as edited-type (ET) sequence. We then used TargetScan and miRanda to predict the miRNA target sites on UT and ET sequences by a similar method described in our previous studies (45,47). We generated four target datasets, which are TU (TargetScan predicted targets on UT sequence), MU (miRanda predicted targets on UT sequence), TE (TargetScan predicted targets on ET sequence), and ME (miRanda predicted targets on ET sequence). The miRNA–lncRNA interactions that exist in both TU and MU, but in neither TE nor ME, were defined as interaction losses (loss = (TU and MU) − (TE or ME)). On the contrary, the miRNA–lncRNA interactions that exist in both TE and ME, but in neither TU nor MU, were defined as interaction gains (gain = (TE and ME) − (TU or MU)). We found a large number of editing sites with the potential to disturb original miRNA target sites and/or to create new potential miRNA target sites. In human, we identified 114 814 and 109 788 editing sites which can cause 742 855 and 731 035 human miRNA–lncRNA interactions gain and loss of functions, respectively (Table 1). Similarly, we identified >1000 editing sites that can change the miRNA–lncRNA interactions in other three species (Table 1).
The expression of miRNAs has been widely profiled by The Cancer Genome Atlas (TCGA) (48) and other studies (49). Previously, we studied the miRNA expression profiles based on 9566 and 410 human small RNA sequencing data from TCGA and NCBI SRA, respectively (50). Our analysis revealed that the pool of miRNAs was dominated by a small proportion of abundantly expressed miRNAs and ∼70% of known miRNAs were expressed at low level (RPM < 1) or only expressed in specific tissues. Therefore, we provided the miRNA expression profiles across different tissues from TCGA for better selection of miRNA–lncRNA interactions.
Customized tools
The numbers of annotated lncRNAs and RNA editing sites increased sharply every year. To help users to predict the function of novel editing sites in lncRNAs, we provided two customized tools: (i) predict the impact of editing site on the structure of the lncRNA and (ii) predict the impact of editing site on the miRNA–lncRNA interaction. For each tool, users can submit an lncRNA sequence and the relative position of the editing site. The first tool will display the secondary structure of the UT/ET lncRNA and their folding MFE. The second tool will invoke the TargetScan and miRanda to predict the miRNA targets on the UT/ET lncRNA, then show the lists of ‘Unedited-type lncRNA binding miRNAs’, ‘Edited-type lncRNA binding miRNAs’, ‘The lost miRNAs of unedited-type lncRNA’, and ‘The gain miRNAs of edited-type lncRNA’. All the results can be easily downloaded from the result page.
DATABASE ORGANIZATION AND WEB INTERFACE
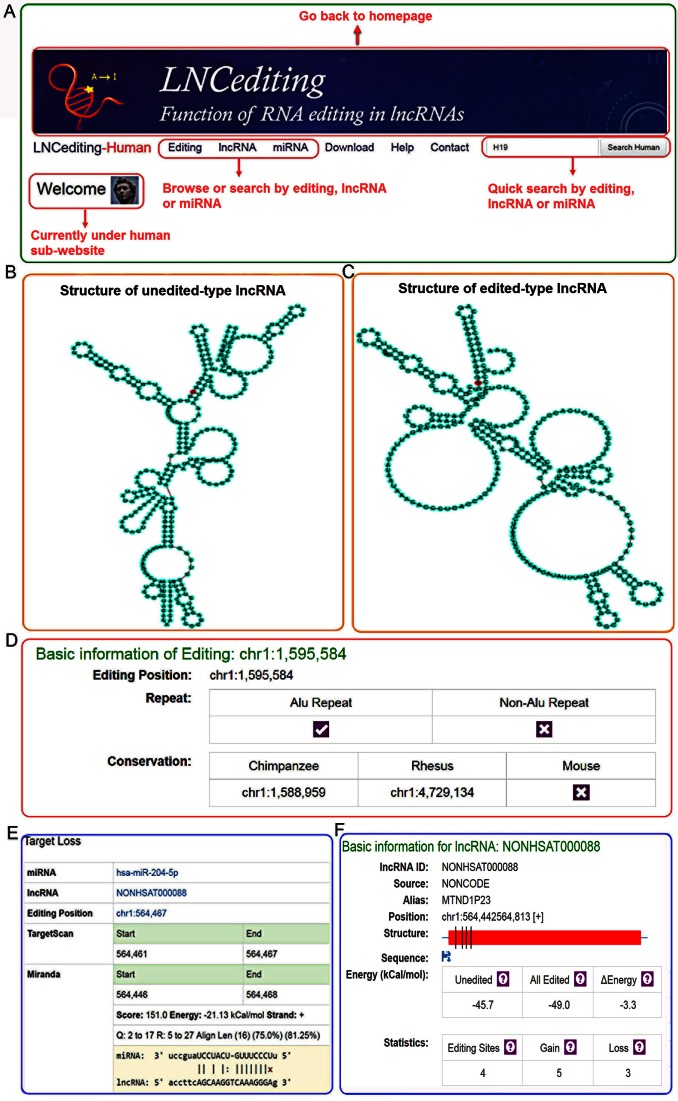
We predicted the functions of editing sites in lncRNAs across four species and all the data were organized into a set of relational MySQL tables. Django (v1.9.7), an open-source web framework based on the WSGI and Apache (v2.4.18), was used to construct the LNCediting database. To make the data acquisition more convenient, we organized our database into four sub-databases by species, namely LNCediting-human, LNCediting-rhesus, LNCediting-mouse, and LNCediting-fly. Users can go to the sub-databases by clicking the photo of each species. Users can come back to the homepage and choose other sub-databases by clicking the header logo (Figure 1A).
Figure 1.
Overview of the LNCediting database. (A) The homepage of the LNCediting database. (B) Secondary structure of unedited-type lncRNA. (C) Secondary structure of edited-type lncRNA. (D) The basic information for RNA editing sites in editing section. (E) An example of miRNA target loss by an editing sites in lncRNA. (F) The basic information of lncRNA provided in the lncRNA section.
In each sub-database, the species-specific customized tools are provided in the homepage. The structures of UT and ET lncRNA are provided (Figure 1B and C), as well as the interactions between miRNA and UT/ ET lncRNA. All data for browse or search are displayed mainly through three sections, which are RNA editing section, lncRNA section, and miRNA section. (i) In the RNA editing section, users can view the basic information for RNA editing site, such as genomic location, conservation, and repetitive information (Figure 1D). Users can also view the impact of RNA editing site on lncRNA secondary structure and the miRNA–lncRNA interaction (Figure 1E). (ii) In the lncRNA section, users can view the basic information for lncRNA (Figure 1F), and the impacts of all RNA editing sites on lncRNA secondary structures and the miRNA–lncRNA interactions. (iii) In the miRNA section, users can choose specific miRNAs to view the miRNA–lncRNA interactions altered by RNA editing sites. Furthermore, the human miRNAs are divided into four categories based on their expression levels, which are highly expressed miRNA group (RPM ≥ 1000), moderately expressed miRNA group (10 ≤ RPM < 1000), lowly expressed miRNA group (1 ≤ RPM < 10), and unexpressed miRNA group (RPM < 1). This feature enables users to choose specific miRNAs based on their expression levels.
At the top right of each page, a quick search box is designed for searching by the position of editing site, lncRNA ID (by either symbol or ID annotated by NONCODE, GENCODE and LNCipedia), miRNA ID, and any defined chromosome regions.
SUMMARY AND FUTURE DIRECTIONS
With the development of high-throughput technologies, the number of RNA editing sites and lncRNAs increased significantly in the past several years. It is worthwhile to explore the function of editing sites, especially in noncoding regions. In this study, we identified the editing sites in lncRNAs across four species and predicted their potential functions through their impacts on secondary structures and miRNA–lncRNA interactions. We observed an appreciable number of editing sites which can influence the lncRNA secondary structures and miRNA–lncRNA interactions. LNCediting presented an important resource for exploring functions of RNA editing sites in lncRNAs. We expect that the number of editing sites will accumulate rapidly in human, mouse, and many other species. We will update LNCediting regularly to include more species, novel editing sites, novel lncRNAs, and other editing types such as C-to-U.
Acknowledgments
The authors gratefully acknowledge FATMA MÜGE ÖZGÜÇ for the editorial assistance.
FUNDING
Cancer Prevention & Research Institute of Texas [RR150085 to L.H.]; National Natural Science Foundation of China [31471247, 81402744]; Funding for open access charge: Cancer Prevention & Research Institute of Texas [RR150085].
Conflict of interest statement. None declared.
REFERENCES
- 1.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q., et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J., Bai J., Zhang X., Lv Y., Gong Y., Liu L., Zhao H., Yu F., Ping Y., Zhang G. A comprehensive overview of lncRNA annotation resources. Brief Bioinform. 2016:bbw015. doi: 10.1093/bib/bbw015. [DOI] [PubMed] [Google Scholar]
- 7.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 8.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 10.Ning S., Zhao Z., Ye J., Wang P., Zhi H., Li R., Wang T., Li X. LincSNP: a database of linking disease-associated SNPs to human large intergenic non-coding RNAs. BMC Bioinformatics. 2014;15:152. doi: 10.1186/1471-2105-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park E., Williams B., Wold B.J., Mortazavi A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012;22:1626–1633. doi: 10.1101/gr.134957.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahn J.H., Lee J.H., Li G., Greer C., Peng G., Xiao X. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rueter S.M., Dawson T.R., Emeson R.B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 15.Picardi E., D'Erchia A.M., Gallo A., Montalvo A., Pesole G. Uncovering RNA Editing Sites in Long Non-Coding RNAs. Front Bioeng. Biotechnol. 2014;2:64. doi: 10.3389/fbioe.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grewe F., Herres S., Viehover P., Polsakiewicz M., Weisshaar B., Knoop V. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011;39:2890–2902. doi: 10.1093/nar/gkq1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakurai M., Ueda H., Yano T., Okada S., Terajima H., Mitsuyama T., Toyoda A., Fujiyama A., Kawabata H., Suzuki T. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 2014;24:522–534. doi: 10.1101/gr.162537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazloomian A., Meyer I.M. Genome-wide identification and characterization of tissue-specific RNA editing events in D. melanogaster and their potential role in regulating alternative splicing. RNA Biol. 2015;12:1391–1401. doi: 10.1080/15476286.2015.1107703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galeano F., Tomaselli S., Locatelli F., Gallo A. A-to-I RNA editing: the ‘ADAR’ side of human cancer. Semin. Cell Dev. Biol. 2012;23:244–250. doi: 10.1016/j.semcdb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Alon S., Mor E., Vigneault F., Church G.M., Locatelli F., Galeano F., Gallo A., Shomron N., Eisenberg E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012;22:1533–1540. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slotkin W., Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maas S., Patt S., Schrey M., Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L., Li Y., Lin C.H., Chan T.H., Chow R.K., Song Y., Liu M., Yuan Y.F., Fu L., Kong K.L., et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He T., Du P., Li Y. dbRES: a web-oriented database for annotated RNA editing sites. Nucleic Acids Res. 2007;35:D141–144. doi: 10.1093/nar/gkl815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiran A., Baranov P.V. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 27.Li J.B., Levanon E.Y., Yoon J.K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G.M. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 28.Ramaswami G., Lin W., Piskol R., Tan M.H., Davis C., Li J.B. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramaswami G., Zhang R., Piskol R., Keegan L.P., Deng P., O'Connell M.A., Li J.B. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramaswami G., Li J.B. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shipman L. Cancer genomics: The relevance of extensive editing in tumour transcriptomes. Nat. Rev. Cancer. 2015;15:698. doi: 10.1038/nrc4044. [DOI] [PubMed] [Google Scholar]
- 32.Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M., Eterovic A.K., Yuan Y., et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han L., Liang H. RNA editing in cancer: mechanistic, prognostic, and therapeutic implications. Mol. Cell. Oncol. 2016;3:e1117702. doi: 10.1080/23723556.2015.1117702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright J.C., Mudge J., Weisser H., Barzine M.P., Gonzalez J.M., Brazma A., Choudhary J.S., Harrow J. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat. Commun. 2016;7:11778. doi: 10.1038/ncomms11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J.Y., Peng Z., Zhang R., Yang X.Z., Tan B.C., Fang H., Liu C.J., Shi M., Ye Z.Q., Zhang Y.E., et al. RNA editome in rhesus macaque shaped by purifying selection. PLoS Genet. 2014;10:e1004274. doi: 10.1371/journal.pgen.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novikova I.V., Hennelly S.P., Sanbonmatsu K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Ge Y., Yan X., Jin Y., Yang X., Yu X., Zhou L., Han S., Yuan Q., Yang M. MiRNA-192 [corrected] and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharya A., Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA-ceRNA interactions. Nucleic Acids Res. 2016;44:D1005–D1010. doi: 10.1093/nar/gkv1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong J., Liu W., Zhang J., Miao X., Guo A.Y. lncRNASNP: a database of SNPs in lncRNAs and their potential functions in human and mouse. Nucleic Acids Res. 2015;43:D181–D186. doi: 10.1093/nar/gku1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong J., Tong Y., Zhang H.M., Wang K., Hu T., Shan G., Sun J., Guo A.Y. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum. Mutat. 2012;33:254–263. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- 48.Jacobsen A., Silber J., Harinath G., Huse J.T., Schultz N., Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat. Struct. Mol. Biol. 2013;20:1325–1332. doi: 10.1038/nsmb.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 50.Gong J., Wu Y., Zhang X., Liao Y., Sibanda V.L., Liu W., Guo A.Y. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. RNA Biol. 2014;11:1375–1385. doi: 10.1080/15476286.2014.996465. [DOI] [PMC free article] [PubMed] [Google Scholar]