Abstract
Whole-exome and whole-genome sequencing have facilitated the large-scale discovery of de novo variants in human disease. To date, most de novo discovery through next-generation sequencing focused on congenital heart disease and neurodevelopmental disorders (NDDs). Currently, de novo variants are one of the most significant risk factors for NDDs with a substantial overlap of genes involved in more than one NDD. To facilitate better usage of published data, provide standardization of annotation, and improve accessibility, we created denovo-db (http://denovo-db.gs.washington.edu), a database for human de novo variants. As of July 2016, denovo-db contained 40 different studies and 32,991 de novo variants from 23,098 trios. Database features include basic variant information (chromosome location, change, type); detailed annotation at the transcript and protein levels; severity scores; frequency; validation status; and, most importantly, the phenotype of the individual with the variant. We included a feature on our browsable website to download any query result, including a downloadable file of the full database with additional variant details. denovo-db provides necessary information for researchers to compare their data to other individuals with the same phenotype and also to controls allowing for a better understanding of the biology of de novo variants and their contribution to disease.
INTRODUCTION
Each person contains novel variants not present in either of their parents and these variants are termed de novo. Most of the ∼70 (1) de novo single-nucleotide variants and small insertions/deletions (indels) found in an individual genome have no obvious phenotypic impact, but there are cases where de novo variants have been found to contribute to disease. Well-described examples include achondroplasia where mutations occur in FGFR3 (2) and Rett syndrome where in most cases the variants arise de novo in MECP2 (3). With the advancement of next-generation sequencing into the study of the whole complement of human genes via whole-exome or whole-genome sequencing, researchers are getting a clearer picture as to the contribution of these variants to ‘complex’ diseases such as autism (4–15) and schizophrenia (16–18). For example, in autism, de novo single-nucleotide variants and indels contribute to ∼7% of the attributable fraction (6) and as much as 21% of simplex cases of the disease (5). Considerable overlap has also been noted between genes with de novo variants contributing to several neurodevelopmental disorders (NDDs) (19).
While the primary focus in the literature for disease-causing de novo mutations has been on NDDs and congenital heart disease, other phenotypes have also been assessed with smaller sample sizes. With application to these and other disorders and diseases on a large-scale, even more findings are sure to arise. denovo-db was designed with the objective of consolidating all published de novo germline variants, regardless of phenotype, and systematically annotating with standardized analytical pipelines. This provides the research community with a one-stop location for assessing the significance of particular genes or mutations as they relate to their phenotype of interest. The researcher could then ask questions relevant to a disease, such as whether the number of de novo variants seen in a gene is statistically significant using tools such as denovolyzeR (20), or one could ask whether the variants seen in a gene, across many individuals, are more clustered in disease than would be expected based on control data using tools such as CLUMP (21). A researcher could also ask questions unrelated to disease with patterns of de novo variants gathered across many individuals potentially providing novel insight into the biology of new mutation in the human genome. denovo-db, thus, provides a resource for specific and general analyses regarding de novo mutations.
Data collection
We searched the literature for published studies where human de novo variants had been identified by next-generation sequencing technology (4–7,9,10,13–18,22–49). These studies were then carefully curated to gather essential information on each de novo variant, including sample identifier (if possible), chromosome, chromosome position, reference allele, alternate allele, and orthogonal validation status. A validation status of ‘yes’ indicates that the variant has been validated as de novo in the child and absent in the parents. The sample identifiers used in denovo-db originate directly from the published literature, and if there is not one available, a simple nomenclature is assigned: LastAuthorNameSampleX where X is a number. If source coordinates were not mapped to GRCh37, the coordinates were lifted over for consistent annotation among all studies. The data from each paper was then aggregated into a study table with a yaml (http://www.yaml.org/) configuration file corresponding to information in the file required for our pipeline. If any data was not available or was unclear, we queried the authors for additional information. Care was taken to avoid duplication of samples within the database. One example is individuals from the Simons Simplex Collection (SSC). To date, sequencing information has been published from ∼2500 SSC families and the data aggregated into denovo-db from eight studies (5,6,8–12,14). For this collection alone, there have been thousands of duplicates. In cases where this duplication occurs, orthogonal validation status takes precedence.
Combination and annotation of data
Each study table was converted to a gcf file, a variant call format (VCF) file file with one sample per line, and then all gcf files were combined to make a master VCF file of all the studies. This data was then run through the SnpEff (50) program to add annotation information. Post-annotation, variants were removed that did not validate (based on the orthogonal validation) or were found to be inherited and therefore not de novo. All variants were subsequently re-annotated using SeattleSeq (51) so that we could get annotation for all available RefSeq transcripts. Whenever a variant did not have annotation by SeattleSeq, we converted the SnpEff annotation to another label as described in Supplementary Table S1. Finally, the annotated data was loaded into a PostgreSQL relational database via five tables.
Website
denovo-db is available at http://denovo-db.gs.washington.edu and requires no usernames or passwords. It is available to the public for querying and downloading data. We have tested it in Mozilla Firefox, Google Chrome and Apple Safari browsers. The download version of the full denovo-db dataset is available as a tab-delimited file on the ‘Download’ page of the website. It contains annotation to all transcripts, based on SeattleSeq, as well as additional columns related to scoring of variants. Each update of denovo-db is released with a version on it and old versions are maintained and archived by the Eichler laboratory using the git version control system. We will update the database and website four to six times per year depending on the number of new papers in the literature with de novo variant data. We have set up a mailing list at denovo-db@uw.edu for users with additional questions. Researchers can also use the mailing list to send us information on other published studies to include in the database. Upon receiving this information we will run the data through our pipeline and integrate into denovo-db.
RESULTS
denovo-db statistics
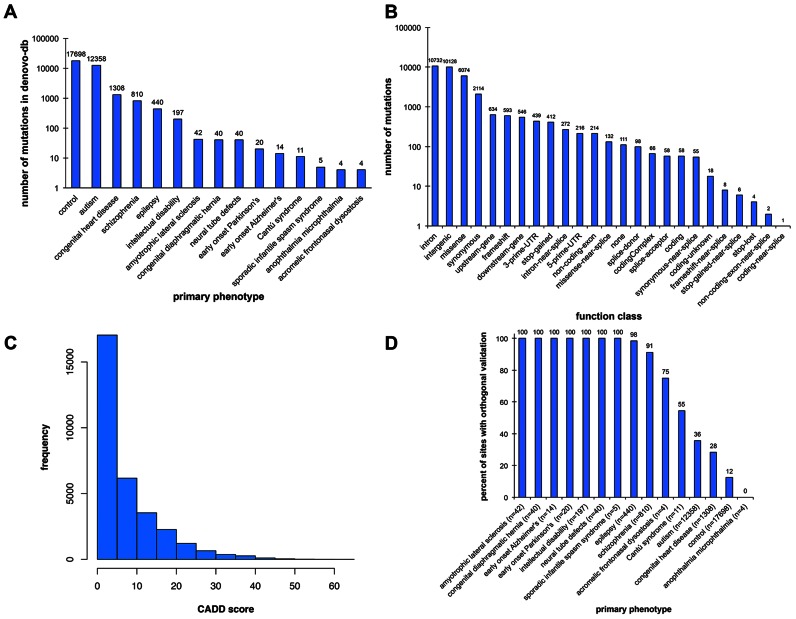
As of July 2016, denovo-db consisted of 32,991 variants (n = 8,541 orthogonally validated) collected from 40 studies and affecting 31,996 unique sites in the genome (Figure 1A). The majority of variants come from controls (n = 17,698), individuals with NDDs including autism (n = 12,358), schizophrenia (n = 810), epilepsy (n = 440), intellectual disability (n = 197) and congenital heart disease (n = 1,308). A number of other smaller studies have contributed variants found in people with amyotrophic lateral sclerosis (n = 42), congenital diaphragmatic hernia (n = 40), neural tube defects (n = 40), early onset Parkinson's (n = 20), early onset Alzheimer's (n = 14), Cantú syndrome (n = 11), sporadic infantile spasm syndrome (n = 5), anophthalmia microphthalmia (n = 4) and acromelic frontonasal dysostosis (n = 4).
Figure 1.
Statistics of denovo-db. (A) Shown are the number of mutations present in the database by the primary phenotype of the individual. (B) Number of mutations split out by the SeattleSeq function class of the variant. (C) CADD score distribution of all variants. (D) Percent of sites validated by primary phenotype.
From the 40 studies there are 16,605 individuals affected with a disorder or disease (n = 14 affected phenotypes represented) and 6,493 unaffected individuals. Thirty of the studies are from whole-exome sequencing, eight from whole-genome sequencing, and two from targeted resequencing. Annotation of variants corresponds to 10,170 genes and 20,108 transcripts. In total, there are 25 functional categories representing 1,161 likely gene-disrupting (LGD) events (412 stop-gained, 593 frame-shift, 58 splice-acceptor and 98 splice-donor) and 6,074 missense events (Figure 1B). A metric often used to assess variant severity is the Combined Annotation Dependent Depletion (CADD) (52) score. We have also included this in the database (Figure 1C) and there are notably 394 missense events with a CADD score >30.
Finally, we have included information on orthogonal validation, which is very important since true de novo variants are sometime difficult to detect due to undercalling in parents. By searching the literature and/or contacting authors, we identified a total of 8,541 validated variants. Some studies, particularly those that are smaller, tend to validate all variants (Figure 1D) while larger studies tend to validate only a subset and from these extrapolate a false positive rate of discovery.
Novelty of denovo-db
We know of two other databases that are similar to denovo-db. Both of these databases focus on NDDs, in contrast to our database that collects information on de novo variants regardless of phenotype. The first database is NPdenovo (53) that collects only de novo variants related to NDDs. The link listed in the paper does not seem to work anymore (http://122.228.158.106/NPdenovo/) but this appears to be the new link http://www.wzgenomics.cn/NPdenovo/. denovo-db does not limit collection of de novo data to neuropsychiatric disorders like NPdenovo. The second is the Developmental Brain Disorder (DBD) Gene Database (54) (http://geisingeradmi.org/care-innovation/studies/dbd-genes/), which collects information on variants in developmental brain disorders. In particular, it keeps only LGD events such as splice-donor, splice-acceptor, stop-gain, and frame-shift mutations. It is a very useful website in that it calculates the relevance of each gene for NDDs. Our database differs by collecting variation on de novo variants regardless of phenotype and functional class. denovo-db is meant to be a compendium of all de novo variants and does not make any assumptions on the researchers’ usage of the data.
denovo-db website
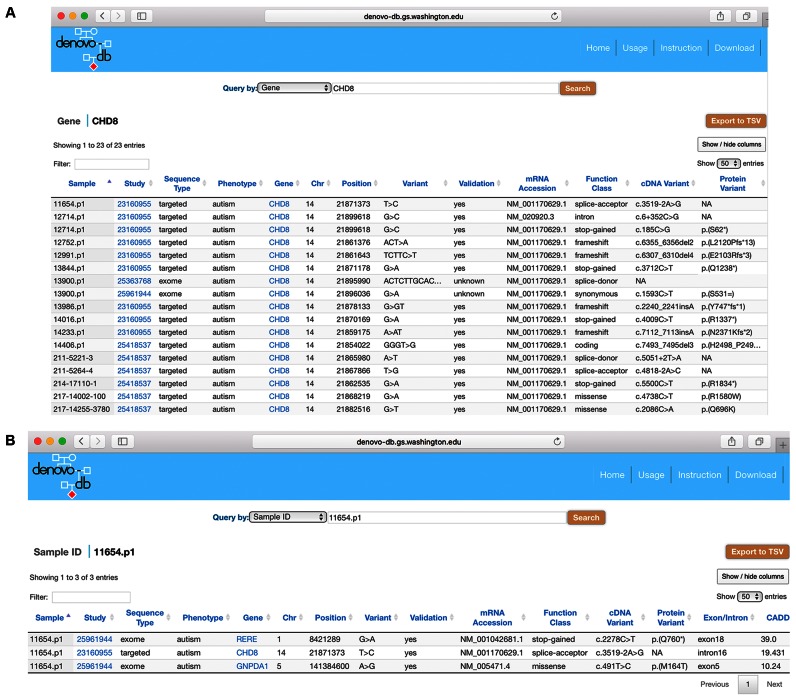
The denovo-db website consists of a number of options for querying the data. One way is to search by gene and this can be done by typing the gene name (e.g. CHD8) (Figure 2A), typing the beginning of the gene name and an asterisk (*) to identify all variants in genes beginning with that text (e.g. CHD*), and via a comma-separated list that can be pasted into the search (e.g. CHD8, MECP2, PAX4). The next way to search is by chromosome position; for this we have built in another option, including typing the base position (e.g. chr14:21871373) or by typing a genomic range (e.g. chr14:21806838-21946382). An alternative is to search by typing the name of a phenotype: the website has a built-in function to match the user's input text to the existing phenotypes in denovo-db and it provides a dropdown of available options. For example, by entering de, you get the following phenotypes to choose from: developmental_disorder and neural_tube_defects. Querying by function is also available and works like the phenotype search. For example, typing sp, you get the following function class options: splice-acceptor and splice-donor. The ability to search by CADD score greater than or equal to a designated value is also available. Another important search option is to enter a sample name (Figure 2B) and by doing so the website will return a list of all variants for that individual. Finally, the database can also be queried by study name.
Figure 2.
Browser shots of denovo-db. (A) Result of a gene search for CHD8. (B) Result of a sample search for 11654.p1.
There are other features available for browsing queried results on denovo-db. First, you can filter variants. This can be done by typing any term in the ‘Filter’ field on the top left side above the table and the variant table will display entries matching your term. For example, enter missense and only missense variants within the current queried result will be displayed in the table. Second, you can sort columns in ascending or descending fashion by clicking the arrows in the column headers. Third, you can select columns using the ‘Show/hide columns’ button on the top right side above the table. Fourth, you can select the table size per page by using the ‘Show entries’ pull-down menu on the top right side above the table. Finally, you can export data. The full queried data set can be exported to a tab-separated-value (TSV) file through the ‘Export to TSV’ button on the top right side above the table. The output TSV file may contain more entries than what is displayed online without filtering and may contain all annotated entries due to alternative transcripts. It also contains more attributes than what are displayed online. Of note, the results tables incorporate hyperlinks to PubMed study IDs, genecards for the gene name, and the dbSNP (55) variants when available.
DISCUSSION
We present denovo-db as a resource for human de novo variants found in the literature. Our new database provides a comprehensive collection and assessment of these variants with a standardized format for annotation. Getting data from the literature into this uniform annotation is a key benefit of our database as the original publications represent a number of formats. In some cases, the variants are presented in a table with the information readily usable but in many other cases, it is in a different format. Examples of these formats include a written description of the variant within the text of the paper, tables encoded into PDF documents that are not always exportable to Excel and require hand curation, variants listed only using their HGVS mRNA annotation, and variants containing the wrong reference base. In addition, some published variants are mapped to older versions of the human reference. We manage these formats through careful assessment of the publications and contact with authors as necessary.
Inclusion of orthogonal validation status is another unique aspect of denovo-db. While many studies reported the validated sites, in some cases the authors listed a number for validated events in the paper but then did not report the actual validation status of the actual sites. All of the authors that we emailed readily provided us with the validated events, if available, for their study and we were able to integrate this information into our database. As seen in Figure 1D, the percent of events validated varies greatly by phenotype. Variants from controls represent the largest representation of variants, but the majority of these events (88%) have not been tested for their validation status. This is very important for researchers to consider when using denovo-db.
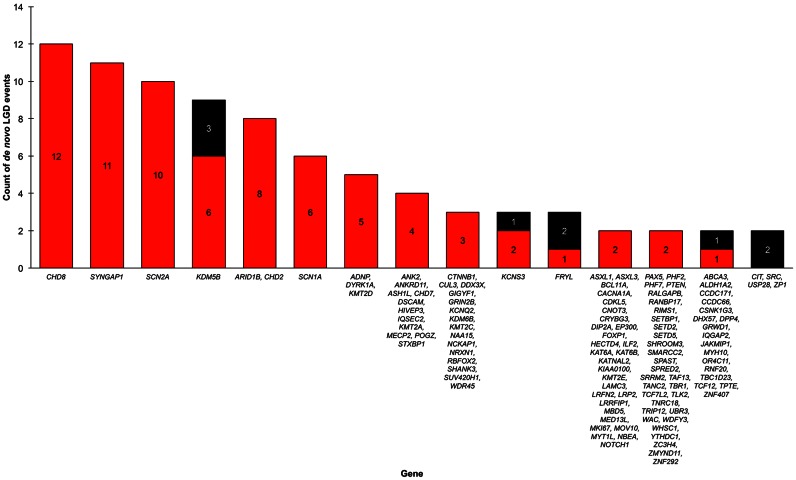
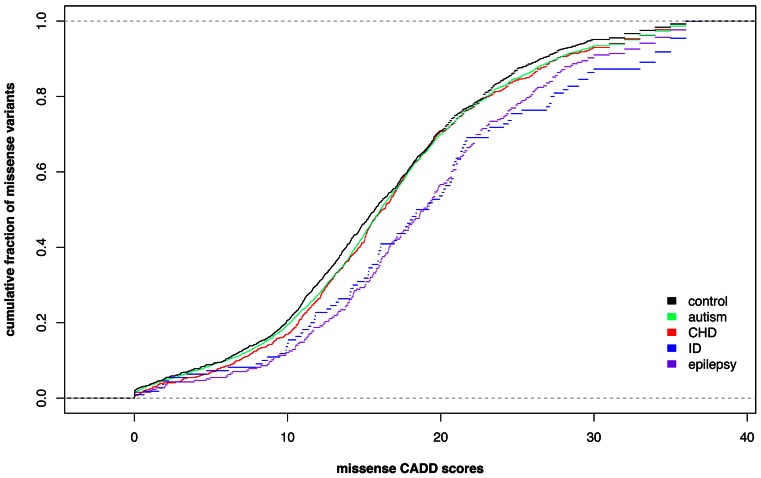
denovo-db is the first public database, to our knowledge, focusing on de novo variants irrespective of phenotype. It includes many features of the variants, including their basic annotation, and more advanced information including severity scores and orthogonal validation status. One way to analyze data from our database is to look at the number of LGD events by case or control status. We assessed those genes with two or more LGD events in denovo-db (Figure 3) and identified genes with only LGD events in cases, only LGD events in controls as well as genes with LGD events in both cases and controls. Genes with LGD events in controls may not be as interesting to researchers studying specific biology; this is a key reason why including various phenotypes is important. To assess missense mutations, we can examine the CADD score of missense variants by phenotype, particularly in controls, autism, congenital heart defects, intellectual disability, and epilepsy. By looking at empirical cumulative distribution function of these events (Figure 4), we see that in intellectual disability and epilepsy there are higher missense CADD scores than in the other phenotypes. These are just two ways to examine denovo-db and we look forward to seeing how other researchers are able to use this data to explore new biology.
Figure 3.
Likely gene-disrupting (LGD) events by cases (in red) and controls (in black). Shown are the counts of LGD events by cases (all phenotypes) and controls with the genes listed for each category. Note there are two bars for the genes with two counts in cases and zero in controls to allow for the full gene list to fit on the plot.
Figure 4.
Missense CADD scores in denovo-db. Empirical cumulative distribution functions of missense CADD scores in the following phenotypes: controls, autism, congenital heart defect (CHD), intellectual disability (ID), and epilepsy individuals.
Acknowledgments
We thank T. Brown for assistance in editing this manuscript and members of the Eichler lab for their feedback on database features.
Footnotes
Present address: Tychele N. Turner, University of Washington School of Medicine, Foege S413A, 3720 15th Ave NE, Box 355065, Seattle, WA 98195-5065, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Simons Foundation [SFARI 303241 to E.E.E.]; National Institute of Mental Health [R01MH101221 to E.E.E.]; National Human Genome Research Institute and the National Heart, Lung and Blood Institute [2UM1HG006493 to D.A.N.]; National Human Genome Research Institute [postdoctoral training grant 2T32HG000035 to T.N.T. and H.A.F.S.]. T.N.T. is an Autism Science Foundation postdoctoral fellow and E.E.E. is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: National Institute of Mental Health [R01MH101221].
Conflict of interest statement. E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
REFERENCES
- 1.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A., et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellus G.A., Hefferon T.W., Ortiz de Luna R.I., Hecht J.T., Horton W.A., Machado M., Kaitila I., McIntosh I., Francomano C.A. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am. J. Hum. Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 3.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 4.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I., O'Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E., et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X., et al. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaelson J.J., Shi Y., Gujral M., Zheng H., Malhotra D., Jin X., Jian M., Liu G., Greer D., Bhandari A., et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C., et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A., et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K., et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D., et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L., et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavassoli T., Kolevzon A., Wang A.T., Curchack-Lichtin J., Halpern D., Schwartz L., Soffes S., Bush L., Grodberg D., Cai G., et al. De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med. Genet. 2014;15:35. doi: 10.1186/1471-2350-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner T.N., Hormozdiari F., Duyzend M.H., McClymont S.A., Hook P.W., Iossifov I., Raja A., Baker C., Hoekzema K., Stessman H.A., et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuen R.K., Thiruvahindrapuram B., Merico D., Walker S., Tammimies K., Hoang N., Chrysler C., Nalpathamkalam T., Pellecchia G., Liu Y., et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 16.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M., et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gulsuner S., Walsh T., Watts A.C., Lee M.K., Thornton A.M., Casadei S., Rippey C., Shahin H., Nimgaonkar V.L., Go R.C., et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy S.E., Gillis J., Kramer M., Lihm J., Yoon S., Berstein Y., Mistry M., Pavlidis P., Solomon R., Ghiban E., et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatr. 2014;19:652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stessman H.A., Turner T.N., Eichler E.E. Molecular subtyping and improved treatment of neurodevelopmental disease. Genome Med. 2016;8:22. doi: 10.1186/s13073-016-0278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ware J.S., Samocha K.E., Homsy J., Daly M.J. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr. Prot. Hum. Genet. 2015;87 doi: 10.1002/0471142905.hg0725s87. doi:10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner T.N., Douville C., Kim D., Stenson P.D., Cooper D.N., Chakravarti A., Karchin R. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet. 2015;24:5995–6002. doi: 10.1093/hmg/ddv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veeramah K.R., O'Brien J.E., Meisler M.H., Cheng X., Dib-Hajj S.D., Waxman S.G., Talwar D., Girirajan S., Eichler E.E., Restifo L.L., et al. De novo pathogenic SCN8A mutation identified by whole-genome sequencing of a family quartet affected by infantile epileptic encephalopathy and SUDEP. Am. J. Hum. Genet. 2012;90:502–510. doi: 10.1016/j.ajhg.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H., Lin M.C., Kornblum H.I., Papazian D.M., Nelson S.F. Exome sequencing identifies de novo gain of function missense mutation in KCND2 in identical twins with autism and seizures that slows potassium channel inactivation. Hum. Mol. Genet. 2014;23:3481–3489. doi: 10.1093/hmg/ddu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto R., Nakazawa T., Tsurusaki Y., Yasuda Y., Nagayasu K., Matsumura K., Kawashima H., Yamamori H., Fujimoto M., Ohi K., et al. Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J. Hum. Genet. 2016;61:199–206. doi: 10.1038/jhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno-Ramos O.A., Olivares A.M., Haider N.B., de Autismo L.C., Lattig M.C. Whole-exome sequencing in a South American cohort links ALDH1A3, FOXN1 and retinoic acid regulation pathways to autism spectrum disorders. PLoS One. 2015;10:e0135927. doi: 10.1371/journal.pone.0135927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kranz T.M., Harroch S., Manor O., Lichtenberg P., Friedlander Y., Seandel M., Harkavy-Friedman J., Walsh-Messinger J., Dolgalev I., Heguy A., et al. De novo mutations from sporadic schizophrenia cases highlight important signaling genes in an independent sample. Schizophr. Res. 2015;166:119–124. doi: 10.1016/j.schres.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barcia G., Fleming M.R., Deligniere A., Gazula V.R., Brown M.R., Langouet M., Chen H., Kronengold J., Abhyankar A., Cilio R., et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat. Genet. 2012;44:1255–1259. doi: 10.1038/ng.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veeramah K.R., Johnstone L., Karafet T.M., Wolf D., Sprissler R., Salogiannis J., Barth-Maron A., Greenberg M.E., Stuhlmann T., Weinert S., et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia. 2013;54:1270–1281. doi: 10.1111/epi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helbig K.L., Farwell Hagman K.D., Shinde D.N., Mroske C., Powis Z., Li S., Tang S., Helbig I. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet. Med. 2016;18:898–905. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]
- 31.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C., et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 32.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T., Albrecht B., Bartholdi D., Beygo J., Di Donato N., et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J.D., Romano-Adesman A., Bjornson R.D., Breitbart R.E., Brown K.K., et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J., et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinberg K.M., Yu B., Koboldt D.C., Mardis E.R., Pamphlett R. Exome sequencing of case-unaffected-parents trios reveals recessive and de novo genetic variants in sporadic ALS. Sci. Rep. 2015;5:9124. doi: 10.1038/srep09124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chesi A., Staahl B.T., Jovicic A., Couthouis J., Fasolino M., Raphael A.R., Yamazaki T., Elias L., Polak M., Kelly C., et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dimassi S., Labalme A., Ville D., Calender A., Mignot C., Boutry-Kryza N., de Bellescize J., Rivier-Ringenbach C., Bourel-Ponchel E., Cheillan D., et al. Whole-exome sequencing improves the diagnosis yield in sporadic infantile spasm syndrome. Clin. Genet. 2016;89:198–204. doi: 10.1111/cge.12636. [DOI] [PubMed] [Google Scholar]
- 38.Lemay P., Guyot M.C., Tremblay E., Dionne-Laporte A., Spiegelman D., Henrion E., Diallo O., De Marco P., Merello E., Massicotte C., et al. Loss-of-function de novo mutations play an important role in severe human neural tube defects. J. Med. Genet. 2015;52:493–497. doi: 10.1136/jmedgenet-2015-103027. [DOI] [PubMed] [Google Scholar]
- 39.Rovelet-Lecrux A., Charbonnier C., Wallon D., Nicolas G., Seaman M.N., Pottier C., Breusegem S.Y., Mathur P.P., Jenardhanan P., Le Guennec K., et al. De novo deleterious genetic variations target a biological network centered on Abeta peptide in early-onset Alzheimer disease. Mol. Psychiatr. 2015;20:1046–1056. doi: 10.1038/mp.2015.100. [DOI] [PubMed] [Google Scholar]
- 40.Kun-Rodrigues C., Ganos C., Guerreiro R., Schneider S.A., Schulte C., Lesage S., Darwent L., Holmans P., Singleton A., Bhatia K., et al. A systematic screening to identify de novo mutations causing sporadic early-onset Parkinson's disease. Hum. Mol. Genet. 2015;24:6711–6720. doi: 10.1093/hmg/ddv376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu L., Sawle A.D., Wynn J., Aspelund G., Stolar C.J., Arkovitz M.S., Potoka D., Azarow K.S., Mychaliska G.B., Shen Y., et al. Increased burden of de novo predicted deleterious variants in complex congenital diaphragmatic hernia. Hum. Mol. Genet. 2015;24:4764–4773. doi: 10.1093/hmg/ddv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slavotinek A.M., Garcia S.T., Chandratillake G., Bardakjian T., Ullah E., Wu D., Umeda K., Lao R., Tang P.L., Wan E., et al. Exome sequencing in 32 patients with anophthalmia/microphthalmia and developmental eye defects. Clin. Genet. 2015;88:468–473. doi: 10.1111/cge.12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith J.D., Hing A.V., Clarke C.M., Johnson N.M., Perez F.A., Park S.S., Horst J.A., Mecham B., Maves L., Nickerson D.A., et al. Exome sequencing identifies a recurrent de novo ZSWIM6 mutation associated with acromelic frontonasal dysostosis. Am. J. Hum. Genet. 2014;95:235–240. doi: 10.1016/j.ajhg.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Bon B.W., Gilissen C., Grange D.K., Hennekam R.C., Kayserili H., Engels H., Reutter H., Ostergaard J.R., Morava E., Tsiakas K., et al. Cantu syndrome is caused by mutations in ABCC9. Am. J. Hum. Genet. 2012;90:1094–1101. doi: 10.1016/j.ajhg.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y.H., Yuen R.K., Jin X., Wang M., Chen N., Wu X., Ju J., Mei J., Shi Y., He M., et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francioli L.C., Menelaou A., Pulit S.L., van Dijk F., Palamara P.F., Elbers C.C., Nerrincx P.B.T, Ye K., Guryev V., Kloosterman W.P., et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 47.Besenbacher S., Liu S., Izarzugaza J.M., Grove J., Belling K., Bork-Jensen J., Huang S., Als T.D., Li S., Yadav R., et al. Novel variation and de novo mutation rates in population-wide de novo assembled Danish trios. Nat. Commun. 2015;6:5969. doi: 10.1038/ncomms6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrad D.F., Keebler J.E., DePristo M.A., Lindsay S.J., Zhang Y., Casals F., Idaghdour Y., Hartl C.L., Torroja C., Garimella K.V., et al. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramu A., Noordam M.J., Schwartz R.S., Wuster A., Hurles M.E., Cartwright R.A., Conrad D.F. DeNovoGear: de novo indel and point mutation discovery and phasing. Nat. Methods. 2013;10:985–987. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J., Cai T., Jiang Y., Chen H., He X., Chen C., Li X., Shao Q., Ran X., Li Z., et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatr. 2016;21:290–297. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Mantilla A.J., Moreno-De-Luca A., Ledbetter D.H., Martin C.L. A Cross-Disorder Method to Identify Novel Candidate Genes for Developmental Brain Disorders. JAMA Psychiatr. 2016;73:275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherry S.T., Ward M., Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]