Abstract
Mutations at the ligand binding sites (LBSs) can influence protein structure stability, binding affinity with small molecules, and drug resistance in cancer patients. Our recent analysis revealed that ligand binding residues had a significantly higher mutation rate than other parts of the protein. Here, we built mutLBSgeneDB (mutated Ligand Binding Site gene DataBase) available at http://zhaobioinfo.org/mutLBSgeneDB. We collected and curated over 2300 genes (mutLBSgenes) having ∼12 000 somatic mutations at ∼10 000 LBSs across 16 cancer types and selected 744 drug targetable genes (targetable_mutLBSgenes) by incorporating kinases, transcription factors, pharmacological genes, and cancer driver genes. We analyzed LBS mutation information, differential gene expression network, drug response correlation with gene expression, and protein stability changes for all mutLBSgenes using integrated genetic, genomic, transcriptomic, proteomic, network and functional information. We calculated and compared the binding affinities of 20 carefully selected genes with their drugs in wild type and mutant forms. mutLBSgeneDB provides a user-friendly web interface for searching and browsing through seven categories of annotations: Gene summary, Mutated information, Protein structure related information, Differential gene expression and gene-gene network, Phenotype information, Pharmacological information, and Conservation information. mutLBSgeneDB provides a useful resource for functional genomics, protein structure, drug and disease research communities.
INTRODUCTION
Molecular recognition plays a fundamental role in all biological processes (1). Mutation-induced conformational change and induced fit with the ligand are the key factors of protein–ligand interactions in cancer cells (2,3). Point mutations at spatially distinct sites lead to conformational changes and exert hinge effects (4). Some point mutations at ligand binding sites may dramatically change the binding affinities of the ligands (5,6). Studies also reported that mutations at ligand binding sites could link to the resistance to small molecule drugs in patient care (7,8). Recently, we also found a significantly higher mutation rate at ligand binding residues than in other parts of the protein sequence across 16 cancer types (9). Therefore, comprehensive annotations of all ligand binding site mutations in pan-cancer will allow for investigators to better understand cancer mechanisms and identify targetable mutations at ligand binding sites.
Many researchers have identified mutation-induced molecular modifications in ligand-protein interactions. For example, mutations in epidermal growth factor receptor (EGFR) in glioblastoma increased ligand binding affinity for EGF (10). A point mutation in neuraminidase 1 gene (NEU proto-oncogene) conferred high ligand binding affinity (6). Moreover, a few studies reported the roles of ligand binding domain mutations. The association between the ligand binding sites and disease related mutations in the type I collagen was observed (11) and the ligand-binding-domain mutations of androgen receptor (AR) gene led to the disruption of interaction between N- and C-terminal domains (12). Recently, several studies showed that ligand binding site mutations could lead to drug resistance. For example, ligand-binding domain mutations in estrogen receptor 1 gene (ESR1) were found in hormone-resistant breast cancer (7). Two major ligand binding site mutations in ESR1 can confer partial resistance to the currently available endocrine treatments (13). Consequently, the cancer and drug research community has recognized the importance of ligand binding site mutations and called for systematic and comprehensive analyses of genes with ligand binding site mutations (14), which are still largely not done yet despite the exponential growth of cancer and other biomedical data recently.
This paper introduces mutLBSgeneDB (mutated Ligand Binding Site gene DataBase), the web interface, and its applications. As the first database encompassing all human ligand binding site mutations with bioinformatics analyses, it provides unique and useful information for functional genomics, protein structure, disease and drug research communities.
DATABASE OVERVIEW
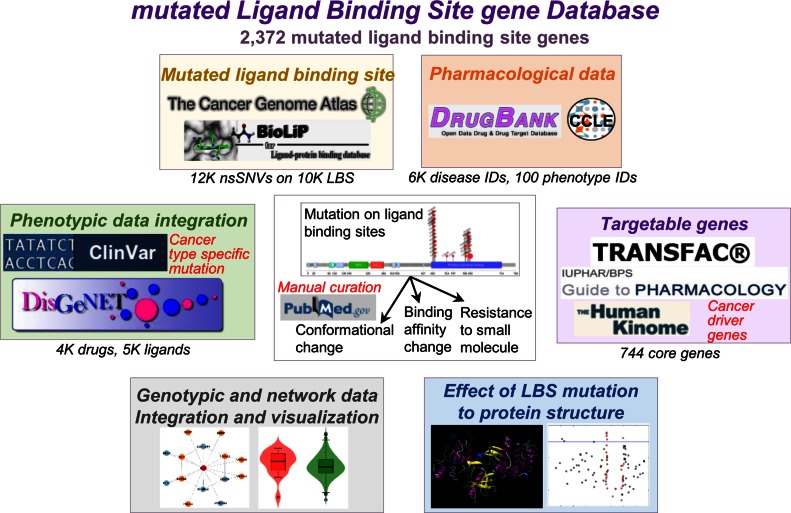
mutLBSgeneDB contains over 2300 genes with ligand binding site mutations that are annotated with seven categories (Figure 1). (i) Gene summary category provides basic gene information with diverse hyperlinks and the literature evidence in ligand binding site mutations for each gene. (ii) Ligand binding site mutation information category presents detailed information of somatic mutations that occur at the ligand binding sites only. The current version of mutLBSgeneDB includes 11 873 non-synonymous mutations at 10 108 ligand binding sites that were extracted from The Cancer Genome Atlas (TCGA) (15) and a semi-manually curated database for biologically relevant ligand-protein interactions (BioLiP) (16). (iii) Protein structure related information category shows relative stability of proteins encoded by all mutLBSgenes and ligand binding affinity changes with their drugs after the occurrence of mutation at the ligand binding site of carefully selected 20 genes. (iv) Differential gene expression and gene-gene network category shows expressional differences between mutated and non-mutated samples based on co-expressed protein interaction network (CePIN). (v) Phenotype information category includes disease related information on the genetic and mutation level. (vi) Pharmacology information category provides heat map using the top 20 correlated drugs between mutLBSgene expressions and 138 anti-cancer drug responses across 790 cell-lines from Cancer Cell Line Encyclopedia (CCLE) (17). mutLBSgeneDB shows the druggable features of mutLBSgenes covering a total of 1324 drugs from DrugBank. (vii) Conservation information category offers conserved sequences for each ligand binding site residue across eight species.
Figure 1.
Overview of mutLBSgeneDB. mutLBSgeneDB is composed of seven annotation categories that cover the genotypic, transcriptomic, proteomic, phenotypic, pharmacological data and network analyses of 2372 mutLBSgenes with manual curation of 744 targetable_mutLBSgenes.
Table 1 summarizes the statistics of 2372 genes having mutations at ligand binding sites (mutLBSgenes) and 744 drug targetable mutLBSgenes (targetable_mutLBSgenes) for each annotation category. mutLBSgeneDB can be used to explore and predict cancerous features and possible drug repurposing. All aforementioned entries and annotation data are available for browsing and searching on the mutLBSgenes web site (http://zhaobioinfo.org/mutLBSgeneDB).
Table 1. Annotation entry statistics for mutLBSgenes and targetable_mutLBSgenes.
| Data type | # Entries | # mutLBSgenesa | # targetable_mutLBSgenesb |
|---|---|---|---|
| Total 2372 (%) | Total 744 (%) | ||
| Targetable genes | # genes | ||
| Human Kinomec | 267 | 267 (11.3%) | 267 (35.9%) |
| TRANSFACd | 216 | 216 (9.1%) | 216 (29.0%) |
| IUPHARe | 579 | 579 (24.4%) | 579 (77.8%) |
| Cancer driver genesf | 179 | 179 (7.5%) | 179 (24.1%) |
| Ligand binding site | # LBS | ||
| BioLiPg | 10 108 | 2372 (100.0%) | 744 (100.0%) |
| Mutation | # nsSNV | ||
| TCGAh | 11 873 | 2372 (100.0%) | 744 (100.0%) |
| Expression | # genes | ||
| TCGAi | 20 502 | 2372 (100.0%) | 744 (100.0%) |
| Expression with drug treatment | # genes | ||
| CCLEj | 19 931 | 2372 (100.0%) | 744 (100.0%) |
| Molecule | # molecules | ||
| DrugBankk | 8206 drugs | 865 (36.5%) | 378 (50.8%) |
| UniProtl | 2374 proteins | 2372 (100.0%) | 743 (99.9%) |
| BioLiPm | 6108 ligands | 1780 (75.0%) | 572 (76.9%) |
| Phenotype | # phenotype | ||
| DisGeNetn | 6761 disease ID | 1449 (61.1%) | 662 (85.5%) |
| ClinVaro | 107 phenotype ID | 80 (3.4%) | 49 (6.6%) |
| Conservation | # LBS | ||
| MUSCLE resultsp | 27 269 | 2371 (100.0%) | 744 (100.0%) |
aNumber of genes having ligand binding site mutations (mutLBSgenes).
bTargetable mutLBSgenes.
cKinases from The Human Kinome database.
dTranscription factors from TRANSFAC database.
eDrug target genes from IUPHAR database.
fSignificantly mutated genes in 18 TCGA cancer types.
gLigand binding sites from Ligand-protein binding database (BioLiP).
hSomatic non-synonymous single nucleotide variants (nsSNVs) from TCGA in 16 cancer types.
iExpression values from TCGA.
jAnti-cancer drug treated cell-line's gene expression data.
kmutLBSgenes related drug IDs from DrugBank database.
lProtein accession data from UniProt database.
mLigand binding to mutLBSgenes.
nGene-level disease annotation from DisGeNet database.
oMutation-level pathogenic information from ClinVar.
pConservation information across 8 species from MUSCLE.
DATA INTEGRATION
mutLBSgenes and targetable_mutLBSgenes
A total of 145 531 ligand–protein binding interactions for 2874 proteins of UniProt (18) were downloaded from BioLiP (data version January 2016) (16). Somatic point mutation data was retrieved from TCGA (March 2016). Mutations that occur on the direct protein–ligand binding site residue or on its immediate two flanking residues at both upstream and downstream sides were considered to be ligand binding site mutations. There were 4660, 4472 and 2741 nsSNVs located at the direct protein–ligand binding site residues, the immediate flanking residues (±1 aa), and the immediate second flanking residues (±2 aa), respectively. After this data processing, 2372 genes with 11 873 non-synonymous mutations at 10 108 ligand binding sites were obtained. Furthermore, 744 drug targetable mutLBSgenes were identified by incorporating kinase genes from the Human Kinome (19), transcription factors from TRANSFAC (20), all drug target genes from the concise guide to pharmacology (IUPHAR, International Union of Basic and Clinical pharmacology) (21), and cancer driver genes from cancer type specific, significantly mutated genes that we collected and curated previously (22). As a result, the targetable_mutLBSgenes are composed of 220 human kinases, 216 human transcription factors, 579 IUPHAR target genes, and 101 cancer-type specific significantly mutated genes (Supplementary Table S1). Ten common genes among the five gene sets were CREBBP, EP300, ESR1, EZH2, FGFR1, HDAC3, PGR, RXRA, SMARCA4 and SMO.
Manual curation of PubMed articles
For the 744 targetable_mutLBSgenes, a literature query of PubMed was performed in June 2016 using the search expression that applied to each mutLBSgene (using BRAF as an example here: ‘((BRAF[Title/Abstract]) AND mutation[Title/Abstract]) AND ligand[Title/Abstract])’. The abstracts of over 1000 articles were manually reviewed. We found literature evidence (138 articles) for 98 genes (∼4.0%) that support the role of these ligand binding site mutations in cancer or drug response. For the 301 genes annotated as kinase or cancer driver genes in mutLBSgenes, we added 3D structure images by searching the Protein Data Bank (PDB) (23). For the most recurrent mutation in each targetable_mutLBSgene, we added related clinical information from genetically informed cancer medicine (My Cancer Genome) (24). Using this curation method, we created a classification system for the genes in the database to show reliability. Class A consists of genes with literature evidence and is part of the targetable_mutLBSgenes. Class B consists of only targetable_mutLBSgenes without additional evidence. The remaining genes belong to Class C.
Expression data preparation
Gene expression data was downloaded from TCGA (January 2015). Normalized gene expression data from RNASeqV2 was extracted using the R package TCGA-Assembler (25). In addition, microarray gene expression data from over 790 cancer cell lines was extracted from CCLE (October 2012).
Co-expressed protein interaction network (CePIN)
The protein interaction network (PIN) reported in our previous study included 113 473 unique protein-protein interactions connecting 13 579 protein-coding genes (26,27). It was used in conjunction with the Pearson Correlation Coefficient (PCC) calculated for each gene-gene pair to build a CePIN. Co-expressed network figures were drawn using the igraph package in R (28). For each gene, the top 20 neighbor genes with the highest PCC values were kept in the network to reflect the genetic signals.
Gene-drug and gene-ligand interaction networks
Drug–target interactions (DTIs) were extracted from DrugBank (29) and the duplicated DTI pairs were excluded. All drugs were grouped using Anatomical Therapeutic Chemical (ATC) classification system codes (30). Two-dimensional chemical structure images of all drugs were generated using the chemical toolbox, OpenBabel (v2.3.1) (31). Ligand–target interactions (LTIs) were extracted from BioLiP and the duplicated LTI pairs were excluded.
Calculating drug binding affinity for top 20 ranked genes and their drugs
We selected 20 genes ranked by the following information: recurrences in the samples, targeted by the drugs of ‘approved and investigational’ status from DrugBank, and number of mutated ligand binding sites. We further selected the most studied drugs (2 or 3) for each of these genes by searching DrugBank and PubMed and downloaded PDB structure file of drugs and proteins. Using these data sets, we searched the drug binding affinities for these 20 genes. We downloaded the crystal structures of genes and three-dimensional structures of drugs from PDB and a free database of commercially available compounds for virtual screening (ZINC) in mol2 format (32). Individual mol2 files were converted into pdbqt files using the python script prepare_ligand4.py available in the Autodock Tools package (33). Using Autodock package, we computed the free energy and studied the docking. Lastly we searched the optimal fit of each drug into targets. The details about this method were described in previous studies (34,35).
Correlation between drug response and gene expression using CCLE data
Drug response data in 714 cell lines on 142 drugs was extracted from Genomics of Drug Sensitivity in Cancer (http://www.cancerrxgene.org/) (36) (October 2012). Pearson Correlation Coefficient (PCC) between drug response and gene expression was calculated for each drug-gene pair.
Conservation information
All sequences used in comparative alignment and specific positions of amino acid were downloaded from the Conserved Domain Database (CDD) of NCBI. Comparison of homologous sequences was obtained by using the multiple sequence alignment tool with high accuracy and high throughput (MUSCLE) (37).
Database architecture
The mutLBSgeneDB system is based on a three-tier architecture: client, server, and database. It includes a user-friendly web interface, Perl's DBI module, and MySQL database. This database was developed on MySQL 3.23 with the MyISAM storage engine.
WEB INTERFACE AND APPLICATIONS
Ligand binding site mutation information category
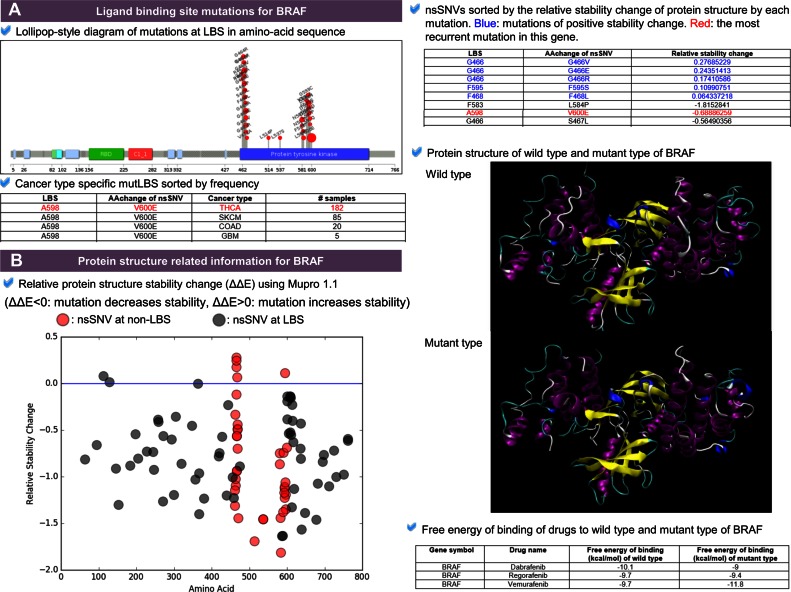
This category presents detailed information of non-synonymous mutations (i.e. nsSNVs) located at the ligand binding sites (Figure 2A) such as the lollipop-style plot showing the mutations that only occurred at the ligand binding sites, cancer type specific mutLBS table giving the sorted mutation frequency information across cancer types, and clinical information table showing the specific clinical information for the most frequently recurrent mutations. We obtained clinical information for 74 genes among 744 targetable_mutLBSgenes using My Cancer Genome. For example, the most frequently observed nsSNV of v-raf murine sarcoma viral oncogenes homolog B1 (BRAF) is the V600E driver mutation (BRAFv600E), activates the MAPK pathway in 50% of melanoma patients (38). This mutation is located near the ligand binding site (A598) (39). The cancer type specific mutLBS table shows the consistent results with the previous studies that the two most frequently mutated cancer types of BRAFv600E are thyroid carcinoma (THCA) and skin cutaneous melanoma (SKCM) (40–42). Another example of ESR1 shows the possible usage of cancer type specific mutLBS table for user to examine whether and how the mutLBS present in different cancer types (43) (Supplementary Figure S1).
Figure 2.
Ligand binding site mutation information category and protein structure related information category. (A) Ligand binding site mutation information category. The lollipop plot shows non-synonymous single nucleotide variants (nsSNVs) at the ligand binding sites only in amino acid sequence of protein B-Raf. Cancer type specific mutLBS table shows the frequency of each LBS mutation in each cancer type. (B) Protein structure related information category. The dot plot shows the relative protein stability change after occurrence of each mutation. Red dots represent ligand binding site mutations and grey dots for other nsSNVs. This category also shows the predicted protein structure of wild type and mutant protein with their drugs. For the top 20 selected genes, we also calculated the binding affinity.
To provide a weighted gene list, we sorted mutLBSgenes based on the number of mutated ligand binding site. Among the total 2372 mutLBSgenes, 1891 genes and 203 had more than two and ten ligand binding sites of mutation, respectively. Among these, the top 20 genes were ERGR, ABL, TP53, BCHE, CTNNB1, VHL, CSNK2A1, FOLH1, KRAS, THBS2, CD1D, F2, EP300, HGF, RUNX1T1, ABL1, AGO2, XDH, CD1B and CES1 (Supplementary Table S2). Gene set enrichment tests were performed for the 203 genes to infer the active pathways of mutLBSgenes (WebGestalt, adjusted P-value (i.e. q-value) <0.05, hypergeometric test followed by multiple test correction using Benjamini–Hochberg's method) (44). There were 40, 53 and 54 genes that were enriched in ‘negative regulation of cell death’ pathway, ‘response to endogenous stimulus’ pathway and ‘protein phosphorylation’ pathway with q-value 2.80 × 10−15, 3.58 × 10−16 and 6.35 × 10−15, respectively (Supplementary Table S3). From these pathways, we could infer that the top genes with the most ligand binding site mutations were significantly involved in tumorigenesis and phosphorylation.
Protein structure related information category
This category shows protein stability changes after occurrence of mutation at the ligand binding site for all proteins encoded by mutLBSgenes (Figure 2B). One study comparing protein mean square deviation (MSD) between wild type and mutant proteins of B-Raf V600E showed that even a single mutation of the protein could lead to much different molecular characteristics (45). To this end, we calculated the relative stability of protein structure after one ligand binding site mutation using MuPro1.1, a computational tool that predicts the protein stability changes for single-site mutation using support vector machines and neural networks (46). Our annotation results showed that five mutations (G466V, G466E, G466R, F468L and F595S) at three ligand binding sites of B-Raf may cause the change of protein structure toward a more stable form with a positive stability change value (Figure 2B).
Furthermore, to annotate mutation-induced modifications on protein-drug binding, we selected top 20 genes ranked by recurrences, targeted by the drugs of ‘approved and investigational’ status, and number of mutated ligand binding sites. These genes are BRAF, CDK2, CPS1, CYP11B2, CYP2B6, CYP2C19, CYP2C8, CYP3A4, EGFR, ERBB2, FGFR2, IDE, ITK, KEAP1, KIT, MET, RET, SULT1E1, VDR and XDH. Binding affinity (kcal/mol) between wild type and mutant proteins with their respective drugs (Supplementary Table S4) was calculated for each of these genes. For example, mutated protein encoded by BRAFV600E has a lower free energy of binding to Vemurafenib, a FDA-approved BRAF kinase inhibitor in the treatment of melanoma, compared to other drugs such as Dabrafenib and Regorafenib (Figure 2B).
Differential gene expression and gene–gene network category
This category provides differential gene expression and gene-gene network of mutated versus wild type samples to show expressional differences in each cancer type. First, a violin plot shows the differential gene expression between mutated and wild type samples for each mutLBSgene (Supplementary Figure S2A). Second, a co-expressed protein interaction network (CePIN) was created by calculating the Pearson Correlation Coefficient (PCC) using gene expression values from TCGA. Using this method, we found significantly different gene-gene networks of BRAF between mutated and non-mutated samples in colon adenocarcinoma (COAD). Gene set enrichment analysis using CePIN gene elements of mutated samples showed that two co-expressed genes (BRAF and PAK2) enriched in ‘positive kinase activity’ pathway with q-value 0.0116. In the wild type samples, 10 co-expressed genes were enriched in ‘regulation of defense response’ and ‘activation of immune response’ pathways with q-value 9.86 × 10−06 and 2.04 × 10−05, respectively (Supplementary Figure S2A, Supplementary Table S5). This result suggested that the occurrence of point mutation at the ligand binding sites of BRAF in COAD may not activate the immune response; instead, protein kinases were activated for the tumorigenesis.
Phenotype information category
This category includes two phenotype information tables. The first table shows the related disease information for each gene retrieved from a database of gene-disease associations (DisGeNet) (47). As shown in Supplementary Figure 2B, the most studied disease name for BRAF is ‘Melanoma’ and less frequently studied diseases include ‘Colon cancer’, ‘Thyroid cancer’, and ‘Cardiofaciocutaneous syndrome’. This is consistent with the previous finding that BRAF mutations are present in approximately 50% of melanoma, 60% of thyroid, and 10% of colorectal carcinomas and are less prevalent in other tumor types (48). On the other hand, the mutation-level pathogenic information table shows pathogenic mutation information from a public archive of relationships between sequence variation and human phenotype (ClinVar) (49).
Pharmacological information category
This category provides pharmacological information such as the correlation between drug response and gene expression and the network visualization of genes with their interacting small molecules (Supplementary Figure S3A). Each gene expression profile in cell lines treated with anti-cancer drugs summarizes the correlation between drug response and altered gene expression by integrating microarray gene expression data from the CCLE database. For example, the expression of BRAF was positively correlated with the treatment effect of drug NVP-TAE684. From the network and information table for the relating drugs and ligands of each mutLBSgene, user can retrieve more detailed information including drug structures. Overall, mutLBSgeneDB includes 1198 FDA-approved drugs targeting 961 mutLBSgenes (Supplementary Table S6).
Conservation information category
This category presents the homologous protein sequences of its flanking region for each ligand binding site obtained from MUSCLE to indicate if the ligand binding site is conserved among different species (Supplementary Figure S3B). For example, protein B-Raf has three mutated ligand binding sites: A481, A598, and C532, all of which are conserved in Homo sapiens (common name: human), Mus musculus (mouse), Gallus (chicken), Caenorhabditis elegans (roundworm), and Drosophila melanogaster (fly). In comparison, all ligand binding site mutations in human EGFR have shown conservation in mice, but not in chicken.
DISCUSSION AND FUTURE DIRECTION
This study introduces a unique resource, mutLBSgeneDB, for the systematic annotation of genes having ligand binding site mutations. To serve functional genomics, protein structure, and drug research communities and advance precision medicine research, we will continuously update mutLBSgeneDB in the following directions. (i) Update routinely by checking the new data of mutations and ligand binding sites from TCGA and BioLiP. (ii) Collect high-quality drug pharmacological data from high-throughput screening and drug resistance studies. (iii) Continue to collect articles on ligand binding site mutations. (iv) Add more protein-ligand 3D structures highlighting ligand binding site mutations with their drugs. (v) Collect and curate germline mutations at ligand binding sites and make the data interactive to somatic mutations. (vi) Perform additional integrative analysis by using other omics data such as methylation, microRNA, and proteomics data. mutLBSgeneDB will be useful for many investigators in functional genomics, protein structure, and drug and therapeutic research.
Acknowledgments
We thank Han Chen for improving the English of the manuscript and website content and his assistance in the manual curation of PubMed articles and PDB data. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health grants [R01LM011177 and R21CA196508]. Funding for open access charge: Dr. Doris L. Ross Professorship Funds to Dr. Zhao from the University of Texas Health Science Center at Houston.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogt A.D., Di Cera E. Conformational selection is a dominant mechanism of ligand binding. Biochemistry. 2013;52:5723–5729. doi: 10.1021/bi400929b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrol R., Trzaskowski B., Goddard W.A., 3rd, Nesterov A., Olave I., Irons C. Ligand- and mutation-induced conformational selection in the CCR5 chemokine G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 2014;111:13040–13045. doi: 10.1073/pnas.1413216111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morando M.A., Saladino G., D'Amelio N., Pucheta-Martinez E., Lovera S., Lelli M., Lopez-Mendez B., Marenchino M., Campos-Olivas R., Gervasio F.L. Conformational selection and induced fit mechanisms in the binding of an anticancer drug to the c-Src kinase. Sci. Rep. 2016;6:24439. doi: 10.1038/srep24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellingson S.R., Miao Y., Baudry J., Smith J.C. Multi-conformer ensemble docking to difficult protein targets. J. Phys. Chem. B. 2015;119:1026–1034. doi: 10.1021/jp506511p. [DOI] [PubMed] [Google Scholar]
- 5.Ogris W., Poltl A., Hauer B., Ernst M., Oberto A., Wulff P., Hoger H., Wisden W., Sieghart W. Affinity of various benzodiazepine site ligands in mice with a point mutation in the GABA(A) receptor gamma2 subunit. Biochem. Pharmacol. 2004;68:1621–1629. doi: 10.1016/j.bcp.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Levy R., Peles E., Goldman-Michael R., Yarden Y. An oncogenic point mutation confers high affinity ligand binding to the neu receptor. Implications for the generation of site heterogeneity. J. Biol. Chem. 1992;267:17304–17313. [PubMed] [Google Scholar]
- 7.Toy W., Shen Y., Won H., Green B., Sakr R.A., Will M., Li Z., Gala K., Fanning S., King T.A., et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013;45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marasca R., Zucchini P., Galimberti S., Leonardi G., Vaccari P., Donelli A., Luppi M., Petrini M., Torelli G. Missense mutations in the PML/RARalpha ligand binding domain in ATRA-resistant As(2)O(3) sensitive relapsed acute promyelocytic leukemia. Haematologica. 1999;84:963–968. [PubMed] [Google Scholar]
- 9.Zhao J., Cheng F., Wang Y., Arteaga C.L., Zhao Z. Systematic Prioritization of Druggable Mutations in approximately 5000 Genomes Across 16 Cancer Types Using a Structural Genomics-based Approach. Mol. Cell. Proteomics. 2016;15:642–656. doi: 10.1074/mcp.M115.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessman N.J., Bagchi A., Ferguson K.M., Lemmon M.A. Complex relationship between ligand binding and dimerization in the epidermal growth factor receptor. Cell Rep. 2014;9:1306–1317. doi: 10.1016/j.celrep.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Lullo G.A., Sweeney S.M., Korkko J., Ala-Kokko L., San Antonio J.D. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 12.Jaaskelainen J., Deeb A., Schwabe J.W., Mongan N.P., Martin H., Hughes I.A. Human androgen receptor gene ligand-binding-domain mutations leading to disrupted interaction between the N- and C-terminal domains. J. Mol. Endocrinol. 2006;36:361–368. doi: 10.1677/jme.1.01885. [DOI] [PubMed] [Google Scholar]
- 13.Fanning S.W., Mayne C.G., Dharmarajan V., Carlson K.E., Martin T.A., Novick S.J., Toy W., Green B., Panchamukhi S., Katzenellenbogen B.S. Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation. Elife. 2016;5:e12792. doi: 10.7554/eLife.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pires D.E., Blundell T.L., Ascher D.B. Platinum: a database of experimentally measured effects of mutations on structurally defined protein-ligand complexes. Nucleic Acids Res. 2015;43:D387–D391. doi: 10.1093/nar/gku966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research, N. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J., Roy A., Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013;41:D1096–D1103. doi: 10.1093/nar/gks966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 20.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K., et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Southan C., Sharman J.L., Benson H.E., Faccenda E., Pawson A.J., Alexander S.P., Buneman O.P., Davenport A.P., McGrath J.C., Peters J.A., et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res. 2016;44:D1054–D1068. doi: 10.1093/nar/gkv1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim P., Cheng F., Zhao J., Zhao Z. ccmGDB: a database for cancer cell metabolism genes. Nucleic Acids Res. 2016;44:D959–D968. doi: 10.1093/nar/gkv1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Allen E.M., Wagle N., Levy M.A. Clinical analysis and interpretation of cancer genome data. J. Clin. Oncol. 2013;31:1825–1833. doi: 10.1200/JCO.2013.48.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y., Qiu P., Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat. Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng F., Jia P., Wang Q., Lin C.C., Li W.H., Zhao Z. Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol. Biol. Evol. 2014;31:2156–2169. doi: 10.1093/molbev/msu167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng F., Liu C., Lin C.C., Zhao J., Jia P., Li W.H., Zhao Z. A Gene Gravity Model for the Evolution of Cancer Genomes: A Study of 3,000 Cancer Genomes across 9 Cancer Types. PLoS Comput. Biol. 2015;11:e1004497. doi: 10.1371/journal.pcbi.1004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Csardi G., Nepusz T. The igraph software package for complex network research. Inter. Comp. Syst. 2006;1695:1–9. [Google Scholar]
- 29.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sterling T., Irwin J.J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu P., Hontecillas R., Abedi V., Kale S., Leber A., Heltzel C., Langowski M., Godfrey V., Philipson C., Tubau-Juni N., et al. Modeling-Enabled Characterization of Novel NLRX1 Ligands. PLoS One. 2015;10:e0145420. doi: 10.1371/journal.pone.0145420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu P., Hontecillas R., Horne W.T., Carbo A., Viladomiu M., Pedragosa M., Bevan D.R., Lewis S.N., Bassaganya-Riera J. Computational modeling-based discovery of novel classes of anti-inflammatory drugs that target lanthionine synthetase C-like protein 2. PLoS One. 2012;7:e34643. doi: 10.1371/journal.pone.0034643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R., et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia J., Jia P., Hutchinson K.E., Dahlman K.B., Johnson D., Sosman J., Pao W., Zhao Z. A meta-analysis of somatic mutations from next generation sequencing of 241 melanomas: a road map for the study of genes with potential clinical relevance. Mol. Cancer Ther. 2014;13:1918–1928. doi: 10.1158/1535-7163.MCT-13-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ascierto P.A., Kirkwood J.M., Grob J.J., Simeone E., Grimaldi A.M., Maio M., Palmieri G., Testori A., Marincola F.M., Mozzillo N. The role of BRAF V600 mutation in melanoma. J. Transl. Med. 2012;10:85. doi: 10.1186/1479-5876-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fibbi B., Pinzani P., Salvianti F., Rossi M., Petrone L., De Feo M.L., Panconesi R., Vezzosi V., Bianchi S., Simontacchi G., et al. Synchronous occurrence of medullary and papillary carcinoma of the thyroid in a patient with cutaneous melanoma: determination of BRAFV600E in peripheral blood and tissues. Report of a case and review of the literature. Endocr. Pathol. 2014;25:324–331. doi: 10.1007/s12022-014-9303-1. [DOI] [PubMed] [Google Scholar]
- 41.Kim C.Y., Lee S.H., Oh C.W. Cutaneous malignant melanoma associated with papillary thyroid cancer. Ann. Dermatol. 2010;22:370–372. doi: 10.5021/ad.2010.22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oakley G.M., Curtin K., Layfield L., Jarboe E., Buchmann L.O., Hunt J.P. Increased melanoma risk in individuals with papillary thyroid carcinoma. JAMA Otolaryngol. Head Neck Surg. 2014;140:423–427. doi: 10.1001/jamaoto.2014.78. [DOI] [PubMed] [Google Scholar]
- 43.Alluri P.G., Speers C., Chinnaiyan A.M. Estrogen receptor mutations and their role in breast cancer progression. Breast Cancer Res. 2014;16:494. doi: 10.1186/s13058-014-0494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirov S., Ji R., Wang J., Zhang B. Functional annotation of differentially regulated gene set using WebGestalt: a gene set predictive of response to ipilimumab in tumor biopsies. Methods Mol. Biol. 2014;1101:31–42. doi: 10.1007/978-1-62703-721-1_3. [DOI] [PubMed] [Google Scholar]
- 45.Tang H.C., Chen Y.C. Insight into molecular dynamics simulation of BRAF(V600E) and potent novel inhibitors for malignant melanoma. Int. J. Nanomed. 2015;10:3131–3146. doi: 10.2147/IJN.S80150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 47.Pinero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holderfield M., Deuker M.M., McCormick F., McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]