Abstract
Plant genera with both diploid and polyploid species are a common evolutionary occurrence. Polyploids, especially allopolyploids such as cotton and wheat, are a great model system for heterosis research. Here, we have integrated genome sequences and transcriptome data of Gossypium species to construct co-expression networks and identified functional modules from different cotton species, including 1155 and 1884 modules in G. arboreum and G. hirsutum, respectively. We overlayed the gene expression results onto the co-expression network. We further provided network comparison analysis for orthologous genes across the diploid and allotetraploid Gossypium. We also constructed miRNA-target networks and predicted PPI networks for both cotton species. Furthermore, we integrated in-house ChIP-seq data of histone modification (H3K4me3) together with cis-element analysis and gene sets enrichment analysis tools for studying possible gene regulatory mechanism in Gossypium species. Finally, we have constructed an online ccNET database (http://structuralbiology.cau.edu.cn/gossypium) for comparative gene functional analyses at a multi-dimensional network and epigenomic level across diploid and polyploid Gossypium species. The ccNET database will be beneficial for community to yield novel insights into gene/module functions during cotton development and stress response, and might be useful for studying conservation and diversity in other polyploid plants, such as T. aestivum and Brassica napus.
INTRODUCTION
Plant genera with both diploid and polyploid species are a common evolutionary occurrence. Polyploidy, especially allopolyploidy, having a greater evolutionary divergence, is a great model system for heterosis research in crop plant domestication, agricultural improvement and evolution (1,2). Because of the large and complex genome assembly, the functional analyses of polyploid species have been great challenges. At present, transcriptomics supports heterosis research at the gene expression profiling level, such as the comparison of homolog expression changes between allopolyploid and diploid species in Triticum-Aegilops (3), and making global or modularized comparison between species. With large amounts of transcriptome data being released, network-based co-expression analyses have been used for gene function predictions on whole genome levels (4). For example, several co-expression network databases and web servers provide comparative analyses and evolutionary investigations to help identify context-associated hubs to prioritize the candidate genes related to vital biological processes (5,6).
As an important crop with economic value, cotton is associated with the agriculture and textile industries. CottonGen, a very good reference database for cotton genomics and breeding studies, has gathered assemblies and annotations of several species, including the diploid cotton Gossypium raimondii (D genome) (7,8), the diploid cotton Gossypium arboreum (A genome) (9) and their allotetraploid cotton Gossypium hirsutum (AD genome) (10,11). However, more refined gene functional annotations, for aspects such as regulation or roles involved in metabolism, disease resistance and stress responses, are limited and the mechanisms behind the evolutionary alteration of characteristics from the ancestral diploid cotton to allotetraploid cotton are not clear. Fortunately, high-throughput transcriptome data in cotton have accumulated, including samples of tissues and selective water stresses in G. arboreum, and samples of development stage tissues and leaf responses to stresses in G. hirsutum. These RNA-seq samples from multiple growth stages and different stress treatments make it possible to detect cotton gene functions on a genome-wide basis. Instead of integrating large transcriptome data sets for global regulatory networks, we overlayed the expression results from multiple development stages and stress-treatment conditions onto our co-expression networks, to help pinpoint candidates for follow-up molecular studies. Thus, the large amount of transcriptome data makes cotton a suitable organism for use in a co-expression network with gene expression views in multi-dimensions (development and stress) and for a network evolutionary relationship analysis between diploid and polyploid species. Furthermore, combining comparative genomics with co-expression networks can help to more easily identify shared features among orthologous genes between species, including the sub-network size, component member functions and gene expression profiling differentials, which may help to support the analysis of gene functions, especially when comparing polyploid organisms with their ancestors and determining their evolutionary relationships.
To date, existing plant network databases, like ATTED-II, PlaNet, AraNet, RiceNet and BAR, have successfully explored or classified gene functions based on networks (5,6,12–14). However, none of them includes cotton data. Thus, we developed the ccNET database to provide an online database server for comparative gene function analyses in multi-dimensional co-expression networks across diploid and polyploid Gossypium species. The algorithm of co-expression network construction (PCC and MR) and the method of function prediction were used to improve the cotton gene annotation. As a result, ccNET facilitates network analysis and gene annotation by (i) presenting co-expression networks with gene expression views in multiple dimensions (tissue-preferential and stress-differential expression profiling), (ii) establishing a comparative analysis between diploid and allotetraploid cotton, such as sub-network features and histone modifications of genes, and (iii) using functional enrichment tools, such as functional co-expression modules and gene set analyses.
DATABASE ARCHITECTURE
Data resources
Multi-dimensional omics data, including genome, transcriptome, epigenome and functional annotation, of two cotton species were integrated for ccNET construction (Table 1). For the genomes, that for G. arboreum was based on the BGI-CGP (Beijing Genomics Institute) genome assembly and annotation; while that for G. hirsutum was based on the NAU-NBI (Nanjing Agricultural University, Novogene Bioinformatics Institute) genome assembly and annotation.
Table 1. Collection, prediction and analyses results in ccNET.
| Database content | G. arboreum | G. hirsutum | Reference |
|---|---|---|---|
| Co-expression nodes | 33 413 (80.8%) | 65 870 (93.4%) | - |
| Positive co-expression edges | 338 049 | 593 758 | - |
| Positive co-expression edges | 207 938 | 291 801 | - |
| Tissue-preferential network | seedling, seed, root, leaf, stem, fiber | root, stem, leaf, cotyledon, calycle, petal, torus, stamen, ovule, pistil, seed | (11,49–52) |
| Stress-response network | NaCl, PEG | Salt, PEG, Cold, Hot | (11,51) |
| Protein-protein interaction (nodes/edges) | 18 004/338 863 | 27 422/696 090 | (14,18–23) |
| Co-expression function modules (>3 nodes overlap) | 1155 (493) | 1884 (1080) | (32,33) |
| miRNA target modules | 213 | 135 | (34–37) |
| Orthologous pairs (genes in each species) | 528 794 (29 086) | 528 794 (42 109) | - |
| Comparison sub-networks (> 3 orthologous pairs) | 62 971 (4012) | 62 971 (4012) | - |
| GO annotation entries (genes) | 72 812 (22 938) | 119 867 (41 939) | (17) |
| KEGG pathways (genes) | 188 (6164) | 391 (36 934) | (17) |
| Transcription regulators (families) | 3305 (81) | 6422 (81) | (39) |
| Kinases (families) | 1598 (87) | 2999 (87) | (40) |
| Carbohydrate-active enzymes | 1604 (94) | 2719 (95) | (41) |
| Cis-elements (kinds of existed motifs) | 742 | 747 | (24–30) |
| H3K4me3 modification profiling | root | root | - |
For transcriptome data, 29 samples of G. arboreum expression profiling data, including tissue (seed, seedling, fiber, root, stem and leaf) and stress-treated samples (dehydration and salinity) were collected from NCBI and our previous works; 115 samples of G. hirsutum expression profiling data, including tissue (root, stem, leaf, cotyledon, calycle, pistil, stamen, petal, torus, ovule, fiber and seed) and stress-treated leaf samples (dehydration, salinity, heat and cold) were collected from NCBI, which covered most growth stages and multiple levels of cotton. Details of these RNA-seq data are listed in Supplementary Tables S1 and S2.
For epigenome data, we have successfully obtained H3K4me3 ChIP-seq sequencing results from root tissues in two cotton species, which provides data for epigenome comparisons.
For the functional annotation, parts of the Gene Ontology (GO)(15) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations (16), which were publicly-available, were used (17); over 18 000 protein–protein interaction of Arabidopsis were integrated from several databases (14,18–22) and literature (23); and 930 plant cis-regulatory elements (discovered from Arabidopsis, Oryza sativa, Glycine max, Triticum aestivum, etc.) with functional annotations have been integrated from several groups, including the Plant Cis-acting Regulatory DNA Elements (PLACE) database (24), the AthaMap webserver (25), the PlantCARE (26) database and previous reports (27–30).
Co-expression network analysis with gene expression view
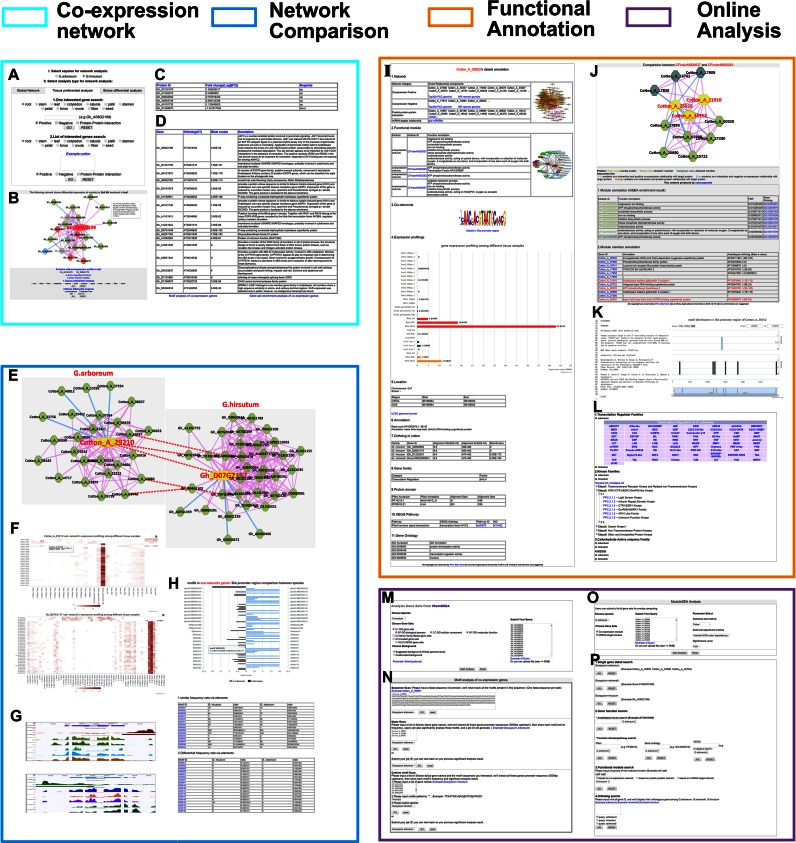
Based on the PCC and MR algorithm (5), the global co-expression network covered 80.8% (33 413/41 331) and 93.4% (65 870/70 478) of genes in G. arboreum and G. hirsutum, respectively (detailed method in Supplementary Material). Then, we overlayed the gene expression results onto the co-expression network. The tissue-preferentially expressed genes and stress-differentially expressed genes were classified for the gene expression view, which covered multi-dimensions (development and stress) in different Gossypium species. The classification rule was based on the gene expression value and fold change between treatment and wild-type samples. Finally, FPKM 0.24 and FPKM 0.17 were selected as cutoffs to identify whether the gene was expressed in G. arboreum and G. hirsutum, respectively (detailed method in Supplementary Material). Genes with log2 fold change (stress/control) ≤ −1 or ≥ 1 and a P-value ≤ 0.05 (t-test) were selected as stress-response genes. For G. arboreum, the co-expression network supports tissue-preferential analyses of six growth stages (seedling, root, stem, leaf, seed and fiber) and stress-differential analyses of two kinds of stress treatments (PEG and NaCl) among three tissues (root, stem and leaf). For G. hirsutum, 12 growth stages (root, stem, leaf, cotyledon, calycle, pistil, stamen, petal, torus, ovule, fiber and seed) and four kinds of stress treatments (PEG, NaCl, cold and hot) at four time points (1 h, 3 h, 6 h and 12 h) in leaves are supplied for the co-expression network analysis (Figure 1A). Instead of displaying the sub-network in which a single gene or multiple genes were involved, the changes in the sub-network during different developmental stages and stress treatments were supplied to mimic regulatory mechanisms under various conditions (Figure 1B). The user-friendly interface enables network analyses in which different conditions may be selected and additional information can be displayed, like whether a given gene is expressed, or its expression changes, along with its functional annotation. This information is listed in tables on the screen (Figure 1B–D). Members in the sub-network are ready for cis-element and gene sets’ enrichment analyses (GSEA) (Figure 1D).
Figure 1.
Description of database functions. (A) The gene network supports three kinds of co-expression or PPI network searches in two cotton species, global, gene tissue-preferential expression and stress-differential expression views. Single gene and gene list inputs are allowed. (B) An example of stress-differential analysis results in G. hirsutum. In a co-expression network with a gene expression view, a red node indicates up-regulated gene expression after the stress treatment; a blue node indicates down-regulated gene expression after the stress treatment; a green node indicates a gene without significant differences in expression level; a grey node indicates an no expression under the conditions; a pink colored edge links two genes with a positive co-expression relationship; a blue colored edge links two genes with a negative co-expression relationship; a grey colored edge links genes without expression values. The biggest node represents the gene submitted; the red-colored node indicates that the gene was up-regulated in the leaves 6 h after a salt treatment; the blue-colored node indicates that the gene was down-regulated in the leaves 6 h after a salt treatment; the grey-colored node indicates that the gene was not expressed in leaves. The locus ID is listed. Details of co-expression gene pairs and gene annotations are linked to the tables in (C) and (D). Moreover, all of the genes in the network could be subjected to cis-element and gene sets’ enrichment analyses (GSEA) analyses directly. (E) In this network comparison result, the two yellow-colored nodes indicated the compared orthologous pair and red-dotted lines connect the orthologous pairs in the two sub-networks. (F) The expression heatmaps of co-expression sub-network members from the orthologous pairs above. (G) Histone modifications and gene expression profiling of the orthologous pairs displayed in the UCSC genome browser. (H) The bidirectional bar chart represents the differential occurrence of motifs between the gene members in the two sub-networks: blue indicates a high occurrence in G. arboreum and black indicates a high occurrence in G. hirsutum. Details of the cis-elements’ occurrence are listed at the bottom. (I) Details of a gene annotation. (J) The two functional modules have overlapping nodes that are highlighted in yellow. The three dark green nodes, together with the three yellow nodes, constitute module CFinderM000263 and the other light green nodes, together with three yellow nodes, constitute module CFinder000037. Annotations are listed for download. (K) The description of the cis-elements on the left includes the name, description (DE), keywords (KW), reference title (RT) and sequence (SQ). On the right, highchart.js displays the motif distribution in the promoter region. (L) Gene families, KEGG pathways and functional modules in the two cotton species can be browsed. Tools, like GSEA analysis (M), cis-element analysis (N), functional module enrichment (O), quick search (P) and BLAST alignment, are supported in ccNET.
Comparison between diploid and allotetraploid cotton
In total, 528 794 ortholog pairs in 16 431 homologous groups, including 29 086 (70% coverage) and 42 109 (60% coverage) genes in G. arboreum and G. hirsutum, respectively, have been established through a bidirectional BLAST algorithm-based alignment and strict E-value cutoff (1E-55) (31). Four aspects of the co-expression networks of homologous genes between diploid and allotetraploid cotton can be compared: (i) all of the orthologous gene pairs are linked and highlighted in red color to exhibit the regulatory network's conservation and diversification during cotton evolution (Figure 1E, Supplementary Figure S9); (ii) gene expression profiles of the homologous sub-networks are compared, exhibiting the conservation and diversity of expression and the abiotic stresses in tissues during cotton evolution (Figure 1F); (iii) cis-elements in the promoters of genes in the sub-networks are compared, exhibiting regulatory elements and sequence conservation and diversity during cotton evolution (Figure 1H); and (iv) distribution of H3K4me3 histone modifications in the genes of the homologous sub-networks are displayed and compared, revealing regulatory mechanisms on the epigenomic level (Figure 1G).
Functional modules and gene annotation
Functional module identification and annotation
The Clique Percolation Method (32) was used to identify clusters or modules that contained more densely connected nodes to each other than to nodes outside the group in cotton co-expression networks. Parameters were established based on more modules, more gene coverage and more overlap. Here, we selected k = 6 communities, indicating that each node contains co-expression interactions with at least six nodes in a module (detailed method in Supplementary Material). The functions of the modules were predicted through integrating gene set annotations, like GO, gene families (transcription regulators, kinases and carbohydrate-active enzymes) and KEGG pathways, and non-significant entries were filtered by Fisher's tests and multiple testing correction method ‘Yekutieli’ (FDR), as referred to in the PlantGSEA toolkit (33). There are 1155 modules containing 6 to 99 genes in G. arboreum and 1884 modules containing 6 to 357 genes in G. hirsutum, which cover functions like metabolism, pathogen and stress responses, hormone, development, transcriptional regulation, etc. Connections between functional modules may represent crosslinks among different pathways in vivo. Thus, modules with three nodes connected to other modules were selected and, as a result, 493 functional modules in G. arboreum and 1080 functional modules in G. hirsutum were revealed as having connections with other modules (Figure 1J).
In addition, we clustered microRNA targets as another kind of module to expand the microRNA and gene functional annotations. Cotton miRNAs were integrated from public databases, like miRBase (34) and research articles (35–37), and the modules consist of the miRNAs’ target genes and their related co-expressed genes. We identified 213 and 135 miRNA target modules in G. arboreum and G. hirsutum, respectively.
Gene annotation
ccNET's gene annotation integrates all known and predicted information (Figure 1I). First, co-expression genes, PPI members and related miRNAs are listed. The annotations of these network members and miRNA target modules are presented on a separate web page. Second, functional modules with brief description are included and linked to module annotation pages. When a cross-linked module exists, there will be a detail page displaying elements, annotations and overlaps for the two modules (Figure 1J). Third, cis-elements located in the 3 kb promoter region, their frequencies of occurrence, location sites and reference annotations are shown on a separate page (Figure 1K). Fourth, gene expression profiles are exhibited as bar charts and tissues are distinguished by color for clarity. Fifth, gene location sites and structures (exon and intron regions) in the chromosome are summarized. The University of California at Santa Cruz (UCSC) (38) genome browser provides visualizations of gene structures, locations, expression profiles and histone modifications as well (Figure 1G). Last, functional annotations have been integrated and predicted, including orthologs in other cotton species, orthologous annotations of the model plant Arabidopsis, gene family classifications, protein domains and alignment sites, KEGG pathways and GO entries. In addition, all of the proteins in gene families and enzymes of KEGG pathways have been listed on the annotation summary page (Figure 1L). Here, gene families, including transcription factor and kinase family classifications, were predicted by the standalone iTAK program (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi), which was based on the rules of the PlnTFDB (39) and PlantsP databases (40), while carbohydrate-active enzymes were predicted using orthologs in Arabidopsis and confirmed by the presence of Pfam domains (41). The GO of G. arboreum was generated using BGI-CGP annotations, Blast2GO software (42), Pfam ID to GO ID translation, and searches for orthologs using the BLAST algorithm, while the GO of G. hirsutum was obtained from the NBI annotation of CottonGen. In addition to the gene annotations of G. hirsutum and G. arboreum, gene details of Gossypium raimondii are linked to GraP, a platform for the functional genomics analysis of G. raimondii (31).
Functional analysis and tools
Functional enrichment analysis of a gene list
Three categories of multiple gene functional annotations are presented in ccNET, including gene set enrichment, functional module enrichment and cis-element enrichment. The gene set enrichment analysis was based on PlantGSEA (33) data processing (Figure 1M). Here, 72 812 GO annotation entries with 22 938 genes, 188 KEGG pathways with 6164 genes, 81 transcription regulator families with 3305 genes, 87 kinase families with 1598 genes and 94 carbohydrate-active enzyme families with 1604 genes, were collected as gene sets in G. arboreum; 119 867 GO annotation entries with 41 939 genes, 391 KEGG pathways with 36 934 genes, 81 transcription regulator families with 6422 genes, 87 kinase families with 2999 genes and 95 carbohydrate-active enzyme families with 2719 genes, were collected as gene sets in G. hirsutum. The annotation entries with FDRs < 0.05 would be enriched and displayed.
The functional module enrichment analysis was based on previously annotated functional modules (Figure 1O), such as the 1155 co-expression modules and 213 miRNA-target modules in G. arboreum, and the 1884 co-expression modules and 135 miRNA-target modules in G. hirsutum. Fisher's exact, hypergeometric and Chi-square tests were used to calculate P-values, and multiple testing correction methods, such as Yekutieli, were used to adjust P-values to prevent possible false-positives. The modules with FDRs < 0.05 may be regarded as significantly enriched and the links can be clicked to browse details, including the genes in the module, gene annotations, module annotations, crosslinked modules, gene expression profiling heatmaps and overlapping genes in the query list and module (Figures 1J and 2B and 2C).
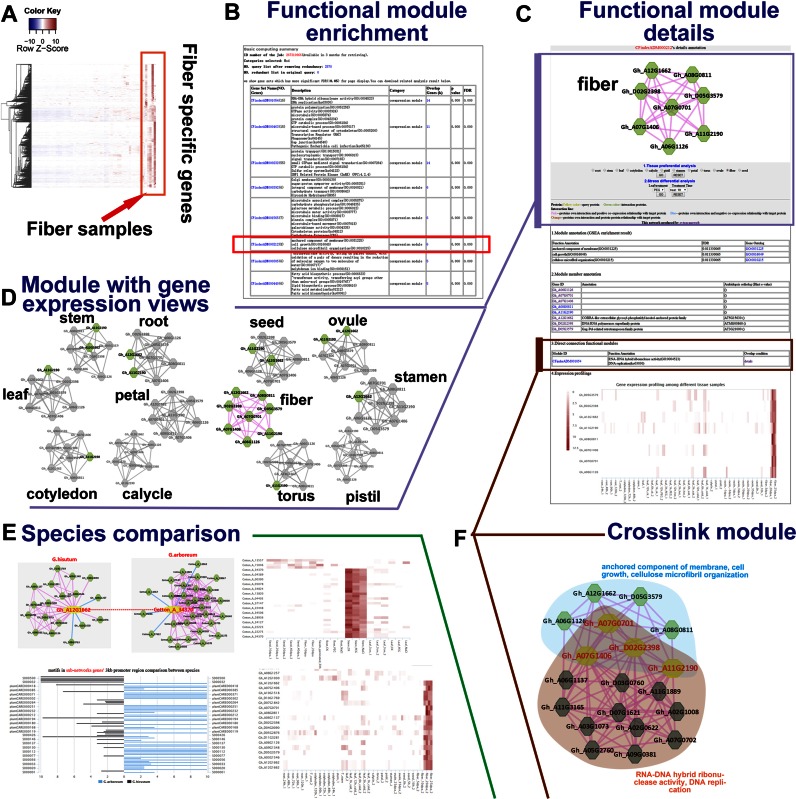
Figure 2.
Novel fiber-related functional module prediction. (A) The heatmap of 2570 fiber-specific expressed genes in G. hirsutum. (B) Functional module enrichment results of the 2750 fiber-specific expressed genes showing that CFinderADM000212 is significantly enriched and this co-expression module is annotated with cellulose microfibril organization. (C) Detailed annotation of the cellulose microfibril organization functional module. (D) Gene expression views of the functional module. Grey- and green-colored nodes represent un-expressed and expressed gene, respectively, in the tissues. The module is complete only in fiber tissues. (E) Comparison between Gh_A12G1662 (one gene of CFinderADM000212) and its orthologous gene Cotton_A_34370 in G. arboreum, including a MR network comparison (top left), expression profiling comparison (right) and cis-element comparison (bottom left). (F) A cross-linked functional module, which overlaps four genes with the CFinderADM000212 module.
The cis-element enrichment analysis was developed to identify the binding sites of regulators in a set of gene promoters and predict the gene set's function (Figure 1N). In total, 930 plant cis-regulatory elements (discovered in Arabidopsis, O. sativa, G. max, T. aestivum, etc.) with functional annotations have been integrated from several groups, including the PLACE database (24), AthaMap webserver (25), PlantCARE database (26) and text-mining results. The significance test is a statistical algorithm based on Z scores and P-value filtering (43,44) that can identify significance elements involved in a gene set. The cotton gene promoter region was taken to be 3000 bp and the Z-score can be calculated based on the frequency of motif occurrence. In the end, motifs with P-values < 0.05 may be considered significantly enriched in the inquiry gene set as compared to gene promoters on the whole genome level of cotton.
Tools supported in ccNET
A quick search, UCSC Genome Browser and a database manual have been developed for ease of use. In the quick search tool, the searches include single gene details of three cotton species, gene functions, model plant orthologs, protein domains and pathways and annotations of functional modules; while the ortholog search among the three cotton species allows a list of genes to be inputted (Figure 1P). In the UCSC Genome Browser, the gene structure, location, expression profile and histone modification are clearly displayed. Gene locus IDs, miRNA IDs and chromosome positions can be searched.
Database function summary
Our ccNET database, which can be accessed at http://structuralbiology.cau.edu.cn/gossypium, contains (i) a co-expression network analysis tool, featuring overlays of tissue-preferential and stress-differential expression levels of one gene or a list of genes, and network comparisons between diploid and allotetraploid cotton, such as module sizes and components, orthologous pairs, gene expression profiles, histone modifications and cis-elements of sub-networks; (ii) a functional enrichment analysis tool, including functional modules for co-expression, miRNA-target and text-mining and functional gene sets, such as GO sets, KEGG pathway sets, gene family sets and cis-element sets; and (iii) other user-friendly features for functional annotation integration, such as UCSC Genome Browser, quick search, BLAST, gene annotation summaries and a database manual (Supplementary Figure S1).
FUNCTIONAL APPLICATIONS
Rather than obtaining collected information of the two cotton species, including function domains, KEGG pathways, gene families, GO, annotation of orthologous genes, cis-regulatory elements, miRNAs, co-expression network and function modules (Figure 1), ccNET database can support gene function prediction mainly based on analysis tools like co-expression network comparison, motif and gene sets analysis. Users can construct functional modules by integrating reported annotation and information in ccNET, such as ‘Fiber elongation regulatory network construction and comparison’ and ‘Water-stress functional module construction and comparison’ (detailed in Supplementary Material). Here, we displayed an example of predicting novel fiber-related gene function by module enrichment online.
Key genes or functional modules associated with fiber synthesis are of interest to researchers, as cotton quality is directly influenced by fiber synthesis parameters. Here, we collected eight expression profiling samples from the fiber tissues of G. hirsutum during the growth stage, from 5 to 25 days post anthesis, and selected 2570 fiber-specific expressed genes using a Z-score test with a P-value < 0.05 (Figure 2A, Supplementary Table S3). The functional module enrichment tool was used to annotate the fiber-specific genes, and several modules related to fiber development were significantly enriched. For example, a NAC transcription factor (45), lipid biosynthesis (46) and sugar and carbohydrate-active enzymes (47) have been reported to play roles in modulating cotton fiber development (Figure 2B). Notably, there was a significant functional module (CFinderADM000212), consisting of eight nodes, that was annotated with ‘cellulose microfibril organization’. In the detailed annotation page of the CFinderADM000212 module, only three members had orthologous genes and a predicted annotation in Arabidopsis, but the expression profile heatmap showed high gene expression values in fiber tissues, meaning this co-expression based functional module might be involved in fiber development in cotton (Figure 2C). In addition, the overlays with tissue-preferential gene expression information in the co-expression network showed that the module was complete only in fiber tissue rather than in other tissues, with few or none of the genes in the module having expression values in the remaining 11 tissues (Figure 2D). This fiber-related module also contained a crosslinked module that had four genes in common with CFinderADM000212 and showed functions in ‘RNA-DNA hybrid ribonuclease activity’ and ‘DNA replication’ pathways, meaning they could be involved in the regulation between different modules or processes (Figure 2F). To compare regulatory functional modules among different species (diploid G. arboreum and allotetraploid G. hirsutum), the orthologous search tool was used to identify orthologous genes of the G. hirsutum fiber-related module (CFinderADM000212) in G. arboreum. However, only one gene, Gh_A12G1662, had an orthologous gene, Cotton_A_34370, in G. arboreum. According to the network comparison tool, no other orthologous genes existed in the two species’ co-expression sub-networks, and gene expression profile heatmaps displayed great differences. The genes in the G. hirsutum sub-network were highly expressed in fiber tissues, while genes in the G. arboreum sub-network were highly expressed in stem tissues. Additionally, the frequencies of cis-elements in the 3-kb promoters of each network gene were different. For example, in G. arboreum there were several light-related motifs, like MCACGTGGC (G box, S000041), and stem internode response motifs, like TAGTGGAT (NRRBNEXTA, S000242), which may modulate stem development. In G. hirsutum, cis-elements, like the sugar repression-related motif TACGTA (A-box, S000130) and the gibberellin response motif GATGAYRTGG(OPAQUE2ZMB32, S000077) (48), occurred more often and we hypothesize these modulate cotton fiber development.
Thus, the orthologous sub-network comparison illustrated possible differences in regulatory mechanisms between diploid and allotetraploid cotton species. There may be more fiber-related genes and regulatory modules involved in G. hirsutum, and gene functions may have changed over the course of cotton evolution, leading to a higher fiber quality in G. hirsutum than in G. arboreum.
DISCUSSION AND FUTURE DIRECTIONS
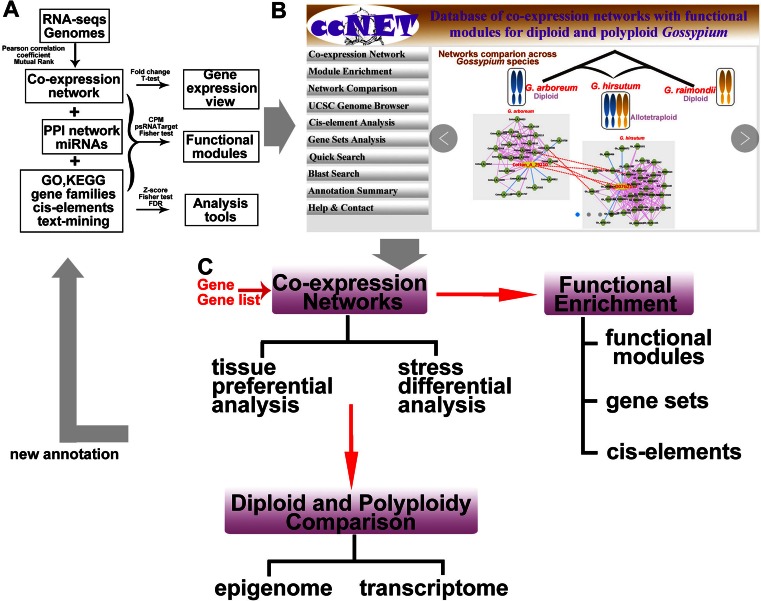
The ccNET database aims to provide an online database server for comparative gene functional analyses at a multi-dimensional network and epigenomic level across diploid and polyploid Gossypium species. After integrating genomic and transcriptomic data from public platforms (such as CottonGen and NCBI), and incorporating methods and algorithms commonly used in other network databases, ccNET permits the exploration of co-expression networks with gene expression views in multiple dimensions (development and stress) in cotton. We have prepared comprehensive functional annotations to predict gene functions. For instance, gene families, including transcriptional regulators, kinases, P450, catalytic and carbohydrate-binding modules (or functional domains) of enzymes; GO, functional domains (Pfam), KEGG enzymes and gene annotations from ortholog/homolog; about 1000 plant cis-elements with functional annotations have been collected. In addition, 1155 and 1884 co-expression modules, 213 and 135 miRNA target modules were identified in G. arboreum and G. hirsutum, respectively, which cover multiple functions like metabolism, pathogen and stress responses, hormone regulation, development and transcriptional regulation. Furthermore, in-house epigenomic data were integrated to study the conservation and variation in co-expression networks across the diploid and allotetraploid cotton species. All of the functional annotation categories are stored in ccNET as background information, in order to improve gene annotation in cotton (Figure 3A). To manage and update data effectively, ccNET has been built as a platform for cotton gene functional identification and analysis (Figure 3B). Three main sections, the co-expression network, diploid and polyploid comparisons and function analysis tools, have been designed (Figure 3C).
Figure 3.
Cotton co-expression network (ccNET) analysis server construction. (A) Data, methods and strategy for the network construction are listed. Transcriptomics data, including RNA-seq and genome sequences, were used to construct the co-expression network. Several types of networks, including co-expression, miRNA target and protein–protein and annotations, including GO, KEGG and cis-elements, were used for functional module prediction and enrichment analysis tool development. (B) The main page of the co-expression network analysis server. (C) Three main kinds of functions in the server, gene expression view, functional enrichment analysis tools and network comparison between diploid and polyploid species. The three tools have relationships with each other and are supported by a multiple gene analysis. Furthermore, functions predicted by the server will be added to the annotation background and expand the annotation ratios of cotton genes.
In the co-expression network, tissue-preferential and stress-differential gene expression views clearly indicate regulation during growth stages and under stress conditions on a transcriptional level.
For species comparisons, the co-expression network, expression profiles, cis-elements and histone modifications have been compared on genomic, transcriptomic and epigenomic levels, which can be used to comprehensively uncover conservation and variation during cotton evolution. For example, the regulatory module of HOX3 for fiber development and the stress-response modules (ABA signaling pathway) for water stress were identified by combining co-expression network comparisons with the expression view, cis-element search, text-mining and H3K4me3 modification analyses.
There are still limitations and possible improvements to ccNET. For instance, more RNA-seq samples for other growth stages, tissues and stress treatments with longer time-courses could be integrated into the dynamic network analysis on the transcriptomic level. Other cotton species or landraces, like Gossypium barbadense, and cultivars could be introduced into functional networks and module analyses, thereby linking networks with variation and evolution more closely. Multiple DNA or histone modification data, which typically show active or repressive correlations with gene expression, could be gathered to shed light on the complex associations between gene expression and chromatin structure. These future additions will contribute to gene function mining and breeding in cotton.
We hope our ccNET database will be beneficial to the community and help to yield novel insights into gene/module functions during cotton development and stress response. Furthermore, the network analysis strategy with conditional dissection, functional module classification and comparisons might be useful for studying conservation and diversity in other polyploid plants, such as T. aestivum and Brassica napus.
Acknowledgments
The authors thank Qunlian Zhang for technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [31571360, 31371291, U1303282 and 31171276]. Funding for open access charge: Ministry of Science and Technology of China [31571360, 31371291, U1303282 and 31171276].
Conflict of interest statement. None declared.
REFERENCES
- 1.Washburn J.D., Birchler J.A. Polyploids as a ‘model system’ for the study of heterosis. Plant Reprod. 2014;27:1–5. doi: 10.1007/s00497-013-0237-4. [DOI] [PubMed] [Google Scholar]
- 2.Renny-Byfield S., Wendel J.F. Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- 3.Wang X., Zhang H., Li Y., Zhang Z., Li L., Liu B. Transcriptome asymmetry in synthetic and natural allotetraploid wheats, revealed by RNA-sequencing. New Phytol. 2015;209:1264–1277. doi: 10.1111/nph.13678. [DOI] [PubMed] [Google Scholar]
- 4.Ma C., Xin M., Feldmann K.A., Wang X. Machine learning-based differential network analysis: A study of stress-responsive transcriptomes in Arabidopsis. Plant Cell. 2014;26:520–537. doi: 10.1105/tpc.113.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoki Y., Okamura Y., Tadaka S., Kinoshita K., Obayashi T. ATTED-II in 2016: A plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol. 2016;57:e5. doi: 10.1093/pcp/pcv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutwil M., Klie S., Tohge T., Giorgi F.M., Wilkins O., Campbell M.M., Fernie A.R., Usadel B., Nikoloski Z., Persson S. PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S., et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 8.Paterson A.H., Wendel J.F., Gundlach H., Guo H., Jenkins J., Jin D., Llewellyn D., Showmaker K.C., Shu S., Udall J., et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492:423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 9.Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., Li Q., Ma Z., Lu C., Zou C., et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014;46:567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 10.Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J., Ma Z., Shang H., Ma X., Wu J., et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 11.Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., Zhang J., Saski C.A., Scheffler B.E., Stelly D.M., et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 12.Lee T., Yang S., Kim E., Ko Y., Hwang S., Shin J., Shim J.E., Shim H., Kim H., Kim C., et al. AraNet v2: an improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015;43:D996–D1002. doi: 10.1093/nar/gku1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T., Oh T., Yang S., Shin J., Hwang S., Kim C.Y., Kim H., Shim H., Shim J.E., Ronald P.C., et al. RiceNet v2: An improved network prioritization server for rice genes. Nucleic Acids Res. 2015;43:W122–W127. doi: 10.1093/nar/gkv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel R.V., Nahal H.K., Breit R., Provart N.J. BAR expressolog identification: expression profile similarity ranking of homologous genes in plant species. Plant J. 2012;71:1038–1050. doi: 10.1111/j.1365-313X.2012.05055.x. [DOI] [PubMed] [Google Scholar]
- 15.The Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J., Jung S., Cheng C.H., Ficklin S.P., Lee T., Zheng P., Jones D., Percy R.G., Main D. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014;42:D1229–D1236. doi: 10.1093/nar/gkt1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A.P., Santonico E., et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xenarios I., Salwinski L., Duan X.J., Higney P., Kim S.M., Eisenberg D. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiser L., Berardini T.Z., Li D., Muller R., Strait E.M., Li Q., Mezheritsky Y., Vetushko A., Huala E. Sustainable funding for biocuration: The Arabidopsis Information Resource (TAIR) as a case study of a subscription-based funding model. Database (Oxford) 2016;2016:baw018. doi: 10.1093/database/baw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lumba S., Toh S., Handfield L.F., Swan M., Liu R., Youn J.Y., Cutler S.R., Subramaniam R., Provart N., Moses A., et al. A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev. Cell. 2014;29:360–372. doi: 10.1016/j.devcel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hehl R., Bulow L. AthaMap web tools for the analysis of transcriptional and posttranscriptional regulation of gene expression in Arabidopsis thaliana. Methods Mol. Biol. 2014;1158:139–156. doi: 10.1007/978-1-4939-0700-7_9. [DOI] [PubMed] [Google Scholar]
- 26.Rombauts S., Dehais P., Van Montagu M., Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27:295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao X., Franks R.G., Levin J.Z., Liu Z. Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell. 2004;16:1478–1489. doi: 10.1105/tpc.021147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W., Provart N.J., Glazebrook J., Katagiri F., Chang H.S., Eulgem T., Mauch F., Luan S., Zou G., Whitham S.A., et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teakle G.R., Manfield I.W., Graham J.F., Gilmartin P.M. Arabidopsis thaliana GATA factors: Organisation, expression and DNA-binding characteristics. Plant Mol. Biol. 2002;50:43–57. doi: 10.1023/a:1016062325584. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Zhang T., Wu Y., Jiang J. Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell. 2012;24:2719–2731. doi: 10.1105/tpc.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Guo J., You Q., Yi X., Ling Y., Xu W., Hua J., Su Z. GraP: Platform for functional genomics analysis of Gossypium raimondii. Database (Oxford) 2015;2015:bav047. doi: 10.1093/database/bav047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palla G., Derenyi I., Farkas I., Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 33.Yi X., Du Z., Su Z. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41:W98–W103. doi: 10.1093/nar/gkt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A., Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie F., Wang Q., Sun R., Zhang B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015;66:789–804. doi: 10.1093/jxb/eru437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong L., Kakrana A., Arikit S., Meyers B.C., Wendel J.F. Composition and expression of conserved microRNA genes in diploid cotton (Gossypium) species. Genome Biol. Evol. 2013;5:2449–2459. doi: 10.1093/gbe/evt196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan M.B., Zhao Y.T., Meng Z.H., Wang X.J., Yang W.C. Conserved miRNA analysis in Gossypium hirsutum through small RNA sequencing. Genomics. 2009;94:263–268. doi: 10.1016/j.ygeno.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Rodriguez P., Riano-Pachon D.M., Correa L.G., Rensing S.A., Kersten B., Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–D827. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tchieu J.H., Fana F., Fink J.L., Harper J., Nair T.M., Niedner R.H., Smith D.W., Steube K., Tam T.M., Veretnik S., et al. The PlantsP and PlantsT Functional Genomics Databases. Nucleic Acids Res. 2003;31:342–344. doi: 10.1093/nar/gkg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conesa A., Gotz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu J., Zhang Z., Wei J., Ling Y., Xu W., Su Z. SFGD: A comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genomics. 2014;15:271. doi: 10.1186/1471-2164-15-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You Q., Zhang L., Yi X., Zhang Z., Xu W., Su Z. SIFGD: Setaria italica Functional Genomics Database. Mol. Plant. 2015;8:967–970. doi: 10.1016/j.molp.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Huang D., Wang S., Zhang B., Shang-Guan K., Shi Y., Zhang D., Liu X., Wu K., Xu Z., Fu X., et al. A Gibberellin-Mediated DELLA-NAC Signaling Cascade Regulates Cellulose Synthesis in Rice. Plant Cell. 2015;27:1681–1696. doi: 10.1105/tpc.15.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu G.J., Xiao G.H., Liu N.J., Liu D., Chen P.S., Qin Y.M., Zhu Y.X. Targeted lipidomics studies reveal that linolenic acid promote cotton fiber elongation by activating phosphatidylinositol and phosphatidylinositol monophosphate biosynthesis. Mol. Plant. 2015;8:911–921. doi: 10.1016/j.molp.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Li L., Huang J., Qin L., Huang Y., Zeng W., Rao Y., Li J., Li X., Xu W. Two cotton fiber-associated glycosyltransferases, GhGT43A1 and GhGT43C1, function in hemicellulose glucuronoxylan biosynthesis during plant development. Physiologia Plantarum. 2014;152:367–379. doi: 10.1111/ppl.12190. [DOI] [PubMed] [Google Scholar]
- 48.Bai W.Q., Xiao Y.H., Zhao J., Song S.Q., Hu L., Zeng J.Y., Li X.B., Hou L., Luo M., Li D.M., et al. Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PloS One. 2014;9:e96537. doi: 10.1371/journal.pone.0096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao T., Zhao L., Lv Y., Chen J., Hu Y., Zhang T., Zhou B. Transcriptome sequencing and differential gene expression analysis of delayed gland morphogenesis in Gossypium australe during seed germination. PloS One. 2013;8:e75323. doi: 10.1371/journal.pone.0075323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo M.J., Szadkowski E., Wendel J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity. 2013;110:171–180. doi: 10.1038/hdy.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X., Yao D., Wang Q., Xu W., Wei Q., Wang C., Liu C., Zhang C., Yan H., Ling Y., et al. mRNA-seq analysis of the Gossypium arboreum transcriptome reveals tissue selective signaling in response to water stress during seedling stage. PloS One. 2013;8:e54762. doi: 10.1371/journal.pone.0054762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renny-Byfield S., Gallagher J.P., Grover C.E., Szadkowski E., Page J.T., Udall J.A., Wang X., Paterson A.H., Wendel J.F. Ancient gene duplicates in Gossypium (cotton) exhibit near-complete expression divergence. Genome Biol. Evol. 2014;6:559–571. doi: 10.1093/gbe/evu037. [DOI] [PMC free article] [PubMed] [Google Scholar]