Abstract
HieranoiDB (http://hieranoiDB.sbc.su.se) is a freely available on-line database for hierarchical groups of orthologs inferred by the Hieranoid algorithm. It infers orthologs at each node in a species guide tree with the InParanoid algorithm as it progresses from the leaves to the root. Here we present a database HieranoiDB with a web interface that makes it easy to search and visualize the output of Hieranoid, and to download it in various formats. Searching can be performed using protein description, identifier or sequence. In this first version, orthologs are available for the 66 Quest for Orthologs reference proteomes. The ortholog trees are shown graphically and interactively with marked speciation and duplication nodes that show the inferred evolutionary scenario, and allow for correct extraction of predicted orthologs from the Hieranoid trees.
INTRODUCTION
Orthology inference plays an important role in predicting unknown functions of newly sequenced genes (1) and in establishing evolutionary relations between organisms (2). The well-known definition of orthology introduced by Fitch (3) states that genes originating from the single gene in their last common ancestor are true orthologs. This distinguishes them from paralogs arising from duplications within one species. Paralogs are often further divided into either inparalogs or outparalogs (4). Inparalogs are defined as duplication products after a speciation event, and can be co-orthologs of a gene in another species. On the another hand, outparalogs, which duplicated before the speciation, can not be orthologs.
Many methods for finding orthologs like OMA (5), OrthoInspector (6), RoundUp (7), EggNog (8), KEGG-OC (9), OrthoDB (10) or InParanoid (11) use a variant of the two-way best matching approach between sequences in two proteomes, followed by a clustering step. One such method is InParanoid (12), which finds seed orthologs and clusters inparalogs around them. InParanoid has proven to be successful in finding high quality orthologs (13) but provides only orthology relations for two species at the time.
An extension has been developed called Hieranoid (14), which uses InParanoid combined with a fully bifurcated species tree. Hieranoid progresses through the species tree from the leaves toward the root. At each internal node, earlier found ortholog groups are turned into representative consensus sequences. Those are re-used like a pseudo-species as input to InParanoid at the next node. As the algorithm progresses toward the root, new species are added to the respective clusters and an ortholog tree is built.
HieranoiDB is a web-based tool for exploring and visualizing hierarchical groups of orthologs from Hieranoid. The current release contains pre-computed orthologs for 66 representative species, which circumvents the need for users to install and run Hieranoid on these, and allows them to retrieve relevant data promptly. The HieranoiDB website features online searching and browsing, as well as graphical visualization of the orthologs in a tree with marked speciations and duplications, which is valuable for interpreting evolutionary scenarios.
Furthermore, finding a tree with a sequence of interest might be problematic if the user knows only a rough description of the protein function, or the sought protein is not present in the used data set. HieranoiDB helps the user to find orthologs to genes of interest, or similar ones, by offering a range of web-based search modes. We here describe how HieranoiDB was constructed and how to use its different capabilities.
MATERIALS AND METHODS
Hieranoid
Hieranoid 2.0 was executed with BLAST (2.2.18) for similarity searching (15) and with MUSCLE (3.8.31) (16) for building alignments, on the 66 species (http://www.ebi.ac.uk/reference_proteomes) that make up the 2011_04 Quest for Orthologs (QfO) reference proteomes (17). The QfO is a joint community effort to improve the quality and standards for methods inferring orthologs. The fully bifurcated species guide tree required by the Hieranoid algorithm was based on the NCBI taxonomy (http://www.ncbi.nlm.nih.gov/taxonomy) (18). When possible, accession numbers in the QfO proteomes were mapped to entry names in UniProt (2016_07) (19) to allow for searching with both.
Processing ortholog trees
Hieranoid ortholog trees were slightly modified as required for input to the Environment for Tree Exploration (ETE) package (3.0.0b32) (http://etetoolkit.org/) (20). ETE is used for most tree manipulations such as extracting protein identifiers, adding duplication nodes, and editing node labels. Species names were prepended to protein identifiers in the trees. Sequence descriptions were parsed from the reference proteomes FASTA files with Biopython (1.67) (21) and stored in the database for further use in word searching.
Inferring evolutionary events
Hieranoid stores the InParanoid results at each node in the species guide tree in the way they are reported by InParanoid. These correspond to speciation nodes. However, InParanoid does not connect inparalogs to each other via duplication nodes. Instead they are just listed, and will in Hieranoid appear as directly connected to a multifurcating speciation node. The duplication nodes must therefore be reconstructed in HieranoiDB, using the species tree. This is done by first grouping the sequences by their species into two inparalog groups that correspond to the two species groups below that node in the species tree, turning it into a bifurcating speciation node. Secondly, a duplication node is created to join the sequences in each inparalog group, if it contains more than one. This duplication node is placed between the speciation node and the inparalogs it joins.
Website
The interactive web system is implemented using the Django (1.9.6) (22) web-framework for Python (2.7) using the standard concept of the three layers model-view-controller (MVC). The layer ‘model’ defines the data structure and uses Django's Object-Relational-Mapping (ORM) for accessing the database through objects. The layer ‘controller’ defines the logic how to react to user requests and returns data in the JavaScript Object Notation (JSON) format. Finally, the layer ‘view’ defines the website frontend, describes which data are to be displayed, and passes that data to templates that are unfolded into dynamic HTML pages. For simplicity to store searchable data, the flat file SQLite (3.13) relational database was used (23).
To build a website layout that runs equally well on mobile devices and personal computers as well as wide range of browsers, CSS, HTML, and JavaScript components of the Materialize (0.97.6) library were used. For the rendering of the trees, a TnT Tree (0.1.7) library was used. Fetching of search queries is done using AJAX methods of JQuery (1.12.0). Because of the large data volume, the responsiveness of the website had to be optimised. The use of AJAX allows data to be downloaded sequentially and on user action such as navigating to the next page, or scrolling results further. Thanks to sequential fetching, the website is rendered immediately without any delay.
FUNCTIONALITY
On the front page of the website, the user can either search for a protein of interest or browse the database via the species tree that was initially used to guide Hieranoid. The purpose of this browsing feature is to categorize species, and allow selection of how deep the displayed trees should be. In the download section, flat files of all orthologs in the release are made available in the Newick tree, SQL, and pairwise formats.
Searching
The search box on every page can be used for both identifiers and for free-text searching. If the sought accession number or identifier is not present in database, information from the entry descriptions are used to help identifying the protein. In this case, a list of proteins with descriptions matching the query word(s) is presented. Here, information about species, full protein description, and both accession number and identifier are listed. A new view with the tree containing the protein of interest can be opened by clicking on an identifier in this list.
We also cover the case when a protein of interest is not featured in our database, but the user is still interested in orthologs to the best matching protein among the organisms present in the current release. For this purpose, calling BLAST that executes on the web server was implemented. The user provides a sequence in FASTA format, and default BLAST parameters are used for searching. Such a query returns a table with matches sorted according to their bit-score. Output from the BLAST search is handled in similar way as output from the text search; by clicking on an identifier the user is presented the tree containing that sequence.
Browsing
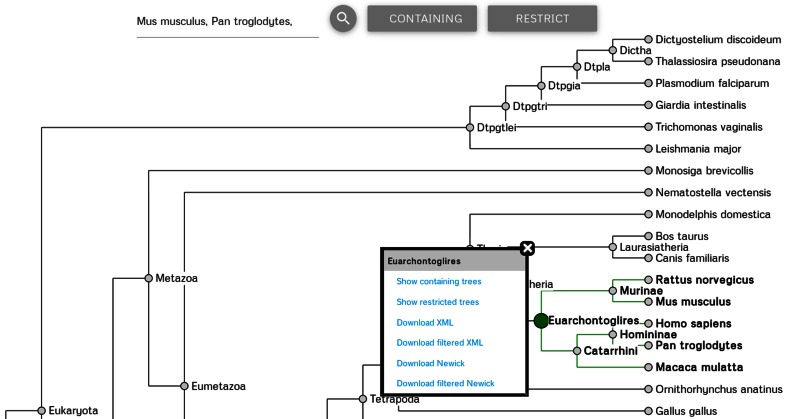
The website also allows the user to browse the database by navigating a species tree. After opening the browse page, the user is presented with the guide species tree of all featured organisms, see Figure 1.
Figure 1.
Browsing of ortholog trees in HieranoiDB uses the guide species tree, partially shown here. By asking for mouse and chimpanzee, the clade Euarchontoglires was selected.
Clicking on a tree node opens a menu for accessing relevant trees. The user can select either all trees that contain proteins from (some of) the species below the node (and potentially also from other species), or trees restricted to only proteins from (some of) the species below the node. One can also export these trees in OrthoXML or Newick format. Alternatively, organisms of interest can be typed in the search box. Also clade names can be used. The most recent common ancestor for the thus selected organisms, as well as its subtree, will then be highlighted in the guide tree. The buttons next to the search box or the links in the tree node menu will generate tabular lists of the trees for the selected species.
Ortholog tree view
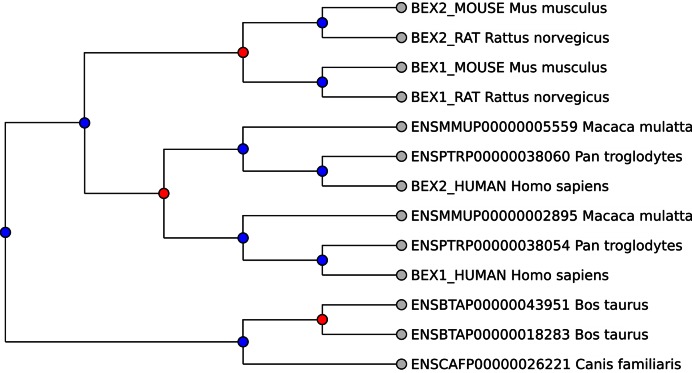
The previously described entry points lead to the ortholog tree view, shown in Figure 2. We demonstrate it with the ortholog tree containing BEX1 and BEX2 (Brain-Expressed X-linked proteins 1 and 2). Many ortholog assignments are the same as in InParanoid, but HieranoiDB adds evolutionary information by showing them in the context of a larger tree, rooted at the Eutheria clade. This tree includes gene duplications in the Murinae (mouse/rat) and Catarrhini (human/chimp/macaque) lineages, and allows the user to see orthologs and paralogs that would not be shown in one view in InParanoid. In this case, it appears that the ancestral BEX gene was independently duplicated in the ancestors of the Murinae and Catarrhini lineages, as well as in cow. This evolutionary scenario is corroborated by other databases such as Ensembl Compara (24), TreeFam (25) and PhylomeDB (26). However, the trees in the two latter databases also contain other proteins beyond BEX1 and BEX2. TreeFam adds BEX4, BEX5, BEX6, and NGFRAP1 as outparalogs to the BEX1/BEX2 group, while PhylomeDB adds NGFRAP1 either as outparalog or inparalog depending on which phylome is selected. This difference can be explained by the fact that Hieranoid will not include outparalogs that are not orthologous to any other protein in the tree and therefore produces tighter ortholog groups.
Figure 2.
Example of HieranoiDB ortholog tree with BEX1 and BEX2 proteins. Blue nodes are speciations and red nodes are duplications.
The tree view in HieranoiDB has interactive features on the shown tree and allows searching either for proteins of interest or species. Search results are highlighted in the tree. To facilitate manipulation of the tree such that only interesting nodes are left, the user can collapse or expand any internal node. From information that is displayed on the tree, the user can choose to show or hide inparalog scores computed by InParanoid or switch between identifiers and accession numbers. Clicking on a leaf brings a pop-up box which shows information about protein link to database. Each tree can be exported as a high resolution image in scalable vector graphics (SVG) or as data in the Newick or OrthoXML formats. It is also possible to export all sequences in the tree in FASTA format, and all pairs of orthologs obtained from the inferred evolutionary events. We do not provide a web services API yet, but all trees can be downloaded in bulk or for each clade separately.
DISCUSSION
HieranoiDB is an on-line database that extends Hieranoid by providing a web-based interface to explore its hierarchical groups of orthologs in an efficient and convenient manner. HieranoiDB not only allows the user to search and navigate the data, but also enriches the hierarchical groups of orthologs with information about species and inferred duplication nodes not provided in the Hieranoid output. Added value of HieranoiDB is the interactive ortholog tree view with marked speciation and duplication events, which can be used to obtain pairs of orthologs. Another important aspect is a web design that is suited for both mobile and desktop devices. It was made responsive, despite the large data volume, through the use of AJAX. This new website makes it easy for non-bioinformaticians to search and visualize information about Hieranoid orthologs and paralogs without the need for any custom processing or coding. Finally, the user can also download orthology data in multiple formats, for use in external software. Future plans for the HieranoiDB database are to extend it to more species, add a web-services API, and integrate information on protein domains.
FUNDING
Vetenskapsrådet [2015-05342]; The Swedish Research Council; the Erasmus Mobility Programme. Funding for open access charge: The Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gabaldon T., Koonin E.V. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 3.Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Biol. 1970;19:99–113. [PubMed] [Google Scholar]
- 4.Sonnhammer E.L.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 5.Altenhoff A.M., Škunca N., Glover N., Train C.-M., Sueki A., Piližota I., Gori K., Tomiczek B., Müller S., Redestig H., et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linard B., Allot A., Schneider R., Morel C., Ripp R., Bigler M., Thompson J.D., Poch O., Lecompte O. OrthoInspector 2.0: Software and database updates. Bioinformatics. 2015;31:447–448. doi: 10.1093/bioinformatics/btu642. [DOI] [PubMed] [Google Scholar]
- 7.DeLuca T.F., Cui J., Jung J.-Y., St Gabriel K.C., Wall D.P. Roundup 2.0: enabling comparative genomics for over 1800 genomes. Bioinformatics. 2012;28:715–716. doi: 10.1093/bioinformatics/bts006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M., et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakaya A., Katayama T., Itoh M., Hiranuka K., Kawashima S., Moriya Y., Okuda S., Tanaka M., Tokimatsu T., Yamanishi Y., et al. KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2013;41:D353–D357. doi: 10.1093/nar/gks1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriventseva E.V., Tegenfeldt F., Petty T.J., Waterhouse R.M., Simão F.A., Pozdnyakov I.A., Ioannidis P., Zdobnov E.M. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnhammer E.L.L., Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remm M., Storm C.E.V., Sonnhammer E.L.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons1. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 13.Altenhoff A.M., Boeckmann B., Capella-Gutierrez S., Dalquen D.A., DeLuca T., Forslund K., Huerta-Cepas J., Linard B., Pereira C., Pryszcz L.P., et al. Standardized benchmarking in the quest for orthologs. Nat. Methods. 2016;13:425–430. doi: 10.1038/nmeth.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber F., Sonnhammer E.L.L. Hieranoid: Hierarchical Orthology Inference. J. Mol. Biol. 2013;425:2072–2081. doi: 10.1016/j.jmb.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 15.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnhammer E.L.L., Gabaldón T., Sousa da Silva A.W., Martin M., Robinson-Rechavi M., Boeckmann B., Thomas P.D., Dessimoz C., consortium, the Q. for O. Big data and other challenges in the quest for orthologs. Bioinformatics. 2014;30:2993–2998. doi: 10.1093/bioinformatics/btu492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2015;43:D6–D17. doi: 10.1093/nar/gku1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huerta-Cepas J., Dopazo J., Gabaldón T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:1–7. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal S., Kumar R. Learning Django Web Development Packt Publishing. 2015. [Google Scholar]
- 23.Van der Lans R.F. The SQL Guide to SQLite Lulu. 2009. [Google Scholar]
- 24.Herrero J., Muffato M., Beal K., Fitzgerald S., Gordon L., Pignatelli M., Vilella A.J., Searle S.M.J., Amode R., Brent S. Ensembl comparative genomics resources. Database. 2016;2016:bav096. doi: 10.1093/database/bav096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber F., Patricio M., Muffato M., Pignatelli M., Bateman A. TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 2014;42:D922–D925. doi: 10.1093/nar/gkt1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huerta-Cepas J., Capella-Gutiérrez S., Pryszcz L.P., Marcet-Houben M., Gabaldón T. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]