Abstract
In the last decade, miRNAs and their regulatory mechanisms have been intensively studied and many tools for the analysis of miRNAs and their targets have been developed. We previously presented a dictionary on single miRNAs and their putative target pathways. Since then, the number of miRNAs has tripled and the knowledge on miRNAs and targets has grown substantially. This, along with changes in pathway resources such as KEGG, leads to an improved understanding of miRNAs, their target genes and related pathways. Here, we introduce the miRNA Pathway Dictionary Database (miRPathDB), freely accessible at https://mpd.bioinf.uni-sb.de/. With the database we aim to complement available target pathway web-servers by providing researchers easy access to the information which pathways are regulated by a miRNA, which miRNAs target a pathway and how specific these regulations are. The database contains a large number of miRNAs (2595 human miRNAs), different miRNA target sets (14 773 experimentally validated target genes as well as 19 281 predicted targets genes) and a broad selection of functional biochemical categories (KEGG-, WikiPathways-, BioCarta-, SMPDB-, PID-, Reactome pathways, functional categories from gene ontology (GO), protein families from Pfam and chromosomal locations totaling 12 875 categories). In addition to Homo sapiens, also Mus musculus data are stored and can be compared to human target pathways.
INTRODUCTION
The understanding of regulatory mechanisms of non-coding RNAs is growing rapidly. Small non-coding RNAs, so called miRNAs or microRNAs play a central role in the regulation of molecular pathways. Already in 2001, miRNAs were described as ‘tiny regulators with great potential (1). The most comprehensive collection of miRNAs is the miRBase that can be considered as reference database and central repository. First published in 2004 with 506 miRNAs from six organisms (2), the 10th release published in 2008 already contained 5071 miRNA precursors from 58 species (3). These correspond to 5922 mature miRNA sequences.
For the analysis of miRNAs a wide variety of computational tools has been developed in the last decade. Akhtar et al. published a comprehensive review containing the description of 129 stand-alone and web-based analysis packages (4). One important task in miRNA research is to understand which pathways are regulated either by single miRNAs or miRNA sets. A selection of tools that provide solution for this task includes miRNApath (5) a Bioconductor package for enrichment of miRNA expression data, miTALOS (6) a web-server for the analysis of tissue specific regulation in signaling pathways or DIANA-miRPath (7,8), a broad target pathway analysis tool that is regularly updated. Similar functionality is also included in general ‘omics’ enrichment toolboxes such as GeneTrail2 (9) and in miRNA analysis pipelines such as Oasis (10).
Similar to the approaches implemented in the tools mentioned above, we performed an in silico enrichment analysis for single miRNAs in 2010. We asked whether predicted target genes of miRNAs accumulate in certain KEGG pathways or gene ontologies and how specific the regulation is. The result was the dictionary of miRNAs and their putative target pathways (11). In the past six years, the knowledge on miRNAs has improved tremendously. The most recent version 21 of miRBase (12)—available since June 2014—lists 28 645 precursor miRNAs, expressing 35 828 mature miRNA products, in 223 different species. For Homo sapiens alone, 2600 miRNAs are annotated and the number has tripled compared to the data contained in our miRNA to target pathway dictionary. In addition to miRNA resources, also pathway databases, gene ontologies and bioinformatics methods have been extended significantly in the past six years. With GeneTrail2 (9), we developed a comprehensive gene set analysis toolbox containing substantially increased functionality as compared to the original version GeneTrail (13).
Since it has become evident that in silico target prediction bears substantial challenges (14), increased experimental effort has led to a comprehensive collection of miRNAs and validated target genes. Among the most comprehensive databases storing miRNA-target interactions (MTIs) are TarBase (15) and miRTarBase (16). The latter contains 2599 human miRNAs and 14 773 genes targeted by at least a single miRNA. Here, MTIs are classified either as ‘strong evidence’ or ‘weak evidence’. Strong evidence targets include interactions validated by reporter assay, Western blot and qPCR. Weak evidence interactions are based on microarrays, next-generation sequencing, pSILAC and other experiments.
The increased knowledge on miRNAs, the availability of large sets of experimentally validated miRNA targets and changes in gene set enrichment and pathway analyses call for the incorporation of this state-of-the-art information in an updated version of miRNA target pathway dictionary. For each miRNA we created three sets of experimentally validated and two sets of predicted MTIs. For the experimentally validated sets, we extracted for each miRNA all targets from miRTarBase and used them to create three test sets: (i) MTIs validated by any experimental method, (ii) MTIs validated by methods with strong evidence and (iii) MTIs validated with weak experimental evidence. In order to build the predicted MTI sets, we used three well established miRNA target prediction frameworks: DIANA-microT (24), miRDB (25) and TargetScan (26). From each of those, we extracted all precomputed MTIs and created two further sets containing the intersection and the union of the three predicted data sets. Using the five MTI sets we computed for each human miRNA enrichments based on 280 KEGG pathways (17), 1300 pathways from Reactome (18), 310 pathways from BioCarta (19), 6169 gene ontology (GO) (20) categories (molecular function, cellular components and biological processes), 617 categories from the Small Molecule Pathway Database (22), as well as enrichments based on the 14 chromosomes, 806 cytogenetic bands, 560 Pfam protein families (28) and on 221 categories from the National Cancer Institute (NCI) Pathway Interaction Database (21). In sum, 12 875 different functional categories were investigated. All results have been stored in the miRNA Pathway Dictionary Database (miRPathDB), freely accessible at https://mpd.bioinf.uni-sb.de/. Altogether, our database stores significant interactions for 2571 miRNAs and 7565 functional categories for Homo sapiens. Besides human, we also incorporated significant interactions for 1933 miRNAs and 8201 functional categories for Mus musculus and enable comparison of these to understand whether miRNA pathway regulations are conserved between organisms.
Data sources and enrichment analyses for miRPathDB
Data sources
With miRPathDB, we strive to provide a new database resource that covers a broad range of miRNAs, target genes, as well as potentially enriched pathways and functional categories. The results included in miRPathDB generally rely on three data sources: human or mouse miRNAs, targets of miRNAs and functional categories. With respect to the miRNAs, we use information from the miRBase version 21 (2). Predicted MTIs are extracted from precomputed data sets provided by DIANA-microT (24), miRDB (25) and TargetScan (26). Based on these data sets, we created two test sets containing the intersection and the union of the provided predictions for each miRNA. Experimentally validated MTIs were retrieved from miRTarBase version 6 (16). The respective MTIs were then used to create three test sets with different experimental evidence levels (any, weak and strong). The third resource, functional categories, are obtained from the GeneTrail2 (9) data warehouse. This data warehouse includes functional categories from various third party resources including KEGG (17), Reactome (18), BioCarta (19), GO (20), cytogenetic bands, disease categories from the NCI Pathway Interaction Database (21) and the Small Molecule Pathway Database (22). Altogether, we considered 12 875 functional categories for human and 9741 for mouse in the current statistical analysis.
Enrichment analysis and clustering
In order to identify if the targets of a certain miRNA are enriched in a biological category, we use the hypergeometric test implemented in the GeneTrail2 C++ library (9). This test checks if this category contains more targets of the analyzed miRNA then expected by chance. In order to calculate this chance, the hypergeometric test relies on a reference set (background). In our case, this is the list of all miRNA targets in the corresponding target sets (weak experimental evidence, strong experimental evidence, any experimental evidence, intersection of predicted targets, union of predicted targets).
For each miRNA and the associated test sets, the following analysis is carried out: We used the hypergeometric test to compute a P-value for the given test set, reference set and biological category. Multiple testing corrections were performed by controlling the false discovery rate (Benjamini–Hochberg adjustment). The significance level was set to 0.05. We only focused on significantly enriched categories rather than depleted ones. P-values for depleted categories were set to 1. The minimal category size was set to 2, the maximal category size to 1000.
To process the results of the enrichment analyses and to enable the integrative visualization as heat maps, we used the freely available statistical programming environment R, version 3.0.2. In order to build heat maps for each database and target set category, we used the following methodology. We used the GeneTrail2 enrichment results to build a pathway x miRNA P-value matrix for each database and target set category. The respective P-values were log10 transformed and discretized. We then performed complete linkage hierarchical clustering with the Euclidian distance, using the hclust method from the stats package, in order to group similar signatures.
Especially for GO categories and miRNA sets, the hypergeometric distribution is known to be biased, as described by Bleazard et al. (23). In their paper, the authors argue that this bias might be caused by the many-to-many relationship between miRNA sets and associated targets. They even emphasize that the respective bias increases with the number of considered miRNAs in the test set. This means that for our considerations of single miRNAs the respective bias seems to have a less strong influence although it cannot be ruled out completely. Additionally, the representation of results as heat map helps to discover specific miRNA – pathway regulations.
Database implementation and functionality
Database implementation and updates
miRPathDB is designed as a document-oriented NoSQL database that provides a RESTful API and is connected to a user friendly web interface. The user interface is based on HTML5 and JavaEE technology using the Thymeleaf template engine, JQuery and AJAX. The database information is visualized using the DataTables plug-in for JQuery and the Highcharts JavaScript library.
The database is implemented in a manner that semi-automated update routines can be used to incorporate new results in regular intervals. Each new version of miRPathDB will get a new version number (currently 1.0), all pathway resources will be updated and all enrichment results will be recomputed. The update routines currently require 900 CPU hours of computing time. All compute intensive tasks are performed using the GeneTrail2 C++ library (https://github.com/unisb-bioinf/genetrail2) and GNU Parallel (27).
Database functionality
With the database we want to enable researchers to identify which miRNAs target a specific pathway, which pathways are regulated by a specific miRNA and how specific these interactions are. miRPathDB contains data about molecular pathways and biological processes that are significantly more targeted by certain miRNAs than expected by chance. For each miRNA, we extracted five target sets (experimental evidence, strong experimental evidence, weak experimental evidence, intersection of predicted target data sets, union of predicted target data set) and pre-computed enrichment analyses for the categories mentioned above.
Following the goal described above we generated several representations of our database: a miRNA centric representation, a pathway centric representation and a detailed representation for each miRNA and pathway.
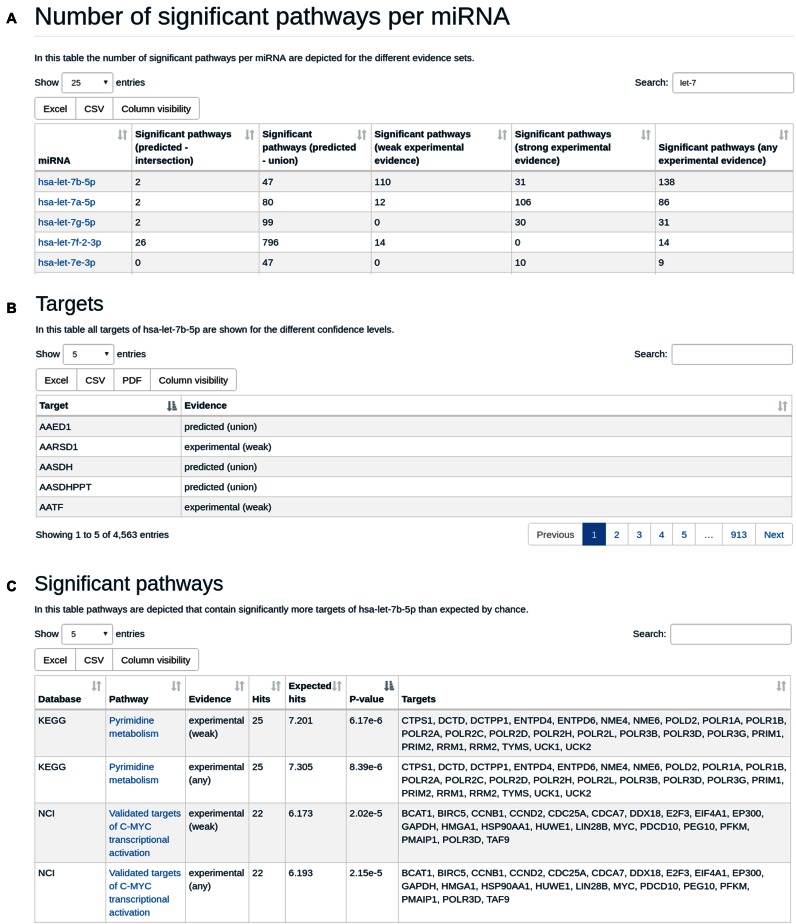
In the miRNA centric representation, all miRNAs are listed along with the number of significantly targeted categories with respect to the five target sets per miRNA. The user can sort miRNAs in alphabetic order or according to the number of targeted categories. Also, the results can be filtered by, e.g. typing ‘let-7’ in the search field, effectively, making it possible to inspect only results of the let-7 family. A typical result of the miRNA centric view is shown in Figure 1A. From here, users can select a miRNA of interest and obtain detailed information about its targets and regulated pathways. First, the target genes for this miRNA are listed and for each target the evidence(s) of this interaction is shown. The respective result is presented in Figure 1B. Per default, five entries are shown but the lists can be expanded to show, e.g. 50, 100, 250 or all target genes. Again, a gene name can be queried in the search field to see whether this gene is contained in the target gene list of the current miRNA. In addition to the target genes, the target pathways are listed. The basic set-up is similar to the target gene representation, however, in addition expected and actual number of target genes on the pathway are included, the respective adjusted significance value as well as the set of all genes that are targeted by this miRNA on that pathway. The representation of target pathways for hsa-let-7b-5p is presented in Figure 1C.
Figure 1.
(A) Result representation of the human miRNA centric view, restricted to miRNAs containing ‘let’. (B) Target gene results for hsa-let-7b-5p, the first miRNA from the results presented in Figure 1A. (C) Target pathway results for hsa-let-7b-5p, the first miRNA from the results presented in Figure 1A.
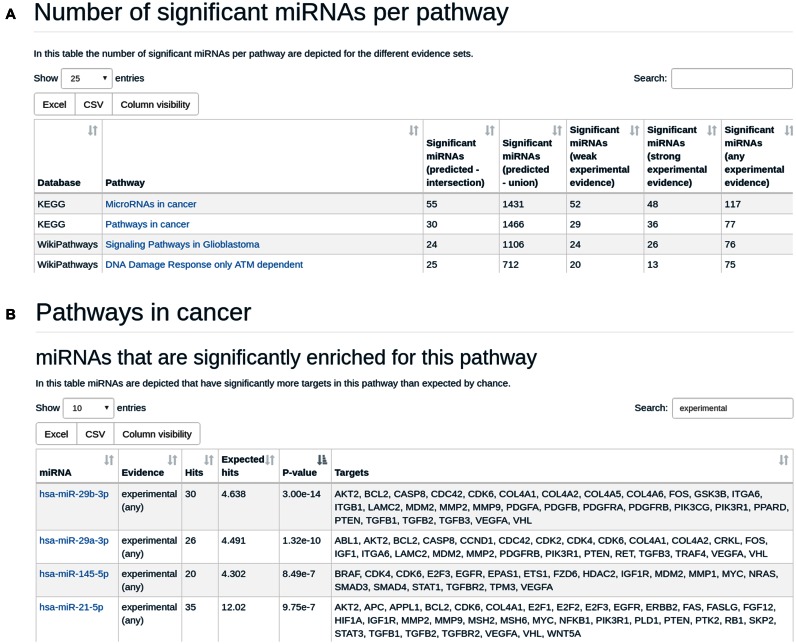
The pathway centric representation lists all pathways that are significantly enriched for at least one miRNA and one of the target sets. It follows generally the same scheme as the miRNA centric one: per pathway the number of miRNAs significantly targeting this pathway dependent on the different target set categories is listed. Again, the representation can be restricted to pathways containing a certain name, in the example in Figure 2A, only pathways with ‘cancer’ are listed. By selecting one pathway from the list the details are presented for this pathway. miRNAs with enriched number of target genes on that pathway are listed with the expected and actual number of target genes and the adjusted P-value followed by target gene names. An example is presented in Figure 2B. The detailed representation can be directly accessed for certain miRNAs or pathways by the search button in the upper right corner of the miRPathDB web page.
Figure 2.
(A) pathway centric view for human miRNAs and pathways. (B) Results of the pathway centric view for the second pathway from Figure 2A.
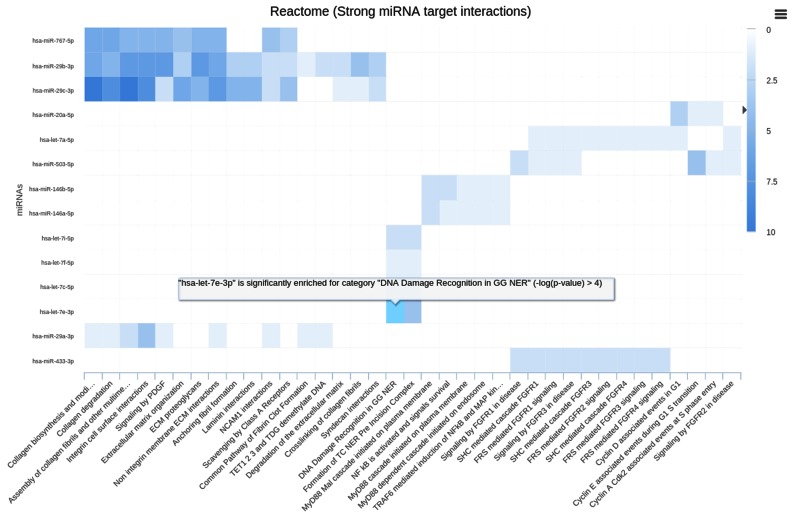
In order to complement this functionality, we also provide a graphic visualization of miRNA-pathway interactions as interactive heat maps that provide a comprehensive overview of pathways targeted by the different (single) miRNAs.
For each category and the five different target sets significance values are presented for miRNAs and the targeted pathways. The significance values are color-coded on a logarithmic scale. On mouse over, the P-value for the miRNA in the row and the pathway in that column is highlighted. An example for human target pathways of miRNAs with strong evidence target genes from Reactome is available in Figure 3. Using this representation, researchers can immediately see whether a miRNA is targeting only few pathways and is rather specific or whether a miRNA is targeting almost all categories.
Figure 3.
Interactive map for strong validated human target pathways in Reactome.
Since miRNAs and their targets are conserved between organisms, we implemented the functionality described above for Mus musculus and Homo sapiens. Users can switch between the organisms by clicking on the respective organism logo located besides the search function in the upper right corner of the miRPathDB home page. Thereby, the degree of conservation of pathways between mouse and human can be assessed.
Download of results and data availability
In each of the miRNA and pathway centric representations the user can select the columns of interest and selectively hide information. The results can then be downloaded in common formats, including flat files (comma separated) that can be used as input for other tools or Excel lists. Beyond that, we also offer a download of the complete result tables from the miRPathDB homepage.
CONCLUSION
The increasing number of human miRNAs, availability of experimentally validated targets and updates in pathway resources lead to an altered picture of miRNAs targeting pathways. The complexity of the analyses calls for a concise and easy to use data repository storing the most recent interactions between miRNAs and target pathways. In the present study, we systematically analyzed target gene sets of miRNAs as well as the regulatory influence of miRNAs on pathways. With the miRNA Pathway Dictionary Database (miRPathDB), which is freely accessible at https://mpd.bioinf.uni-sb.de/, we provide a comprehensive collection of single miRNAs that regulate pathways, gene ontologies and other categories, hence complementing the hitherto available miRNA target enrichment programs, tailored for miRNA sets.
FUNDING
Funding for open access charge: Saarland University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhtar M.M., Micolucci L., Islam M.S., Olivieri F., Procopio A.D. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016;44:24–44. doi: 10.1093/nar/gkv1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiromatzo A.O., Oliveira T.Y., Pereira G., Costa A.Y., Montesco C.A., Gras D.E., Yosetake F., Vilar J.B., Cervato M., Prado P.R., et al. miRNApath: A database of miRNAs, target genes and metabolic pathways. Genet. Mol. Res. 2007;6:859–865. [PubMed] [Google Scholar]
- 6.Kowarsch A., Preusse M., Marr C., Theis F.J. miTALOS: Analyzing the tissue-specific regulation of signaling pathways by human and mouse microRNAs. RNA. 2011;17:809–819. doi: 10.1261/rna.2474511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlachos I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., Hatzigeorgiou A.G. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stockel D., Kehl T., Trampert P., Schneider L., Backes C., Ludwig N., Gerasch A., Kaufmann M., Gessler M., Graf N., et al. Multi-omics enrichment analysis using the GeneTrail2 web service. Bioinformatics. 2016;32:1502–1508. doi: 10.1093/bioinformatics/btv770. [DOI] [PubMed] [Google Scholar]
- 10.Capece V., Garcia Vizcaino J.C., Vidal R., Rahman R.U., Pena Centeno T., Shomroni O., Suberviola I., Fischer A., Bonn S. Oasis: online analysis of small RNA deep sequencing data. Bioinformatics. 2015;31:2205–2207. doi: 10.1093/bioinformatics/btv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backes C., Meese E., Lenhof H.P., Keller A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010;38:4476–4486. doi: 10.1093/nar/gkq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y.A., Muller R., Meese E., Lenhof H.P. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das N. MicroRNA Targets - How to predict? Bioinformation. 2012;8:841–845. doi: 10.6026/97320630008841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., Anastasopoulos I.L., Maniou S., Karathanou K., Kalfakakou D., et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou C.H., Chang N.W., Shrestha S., Hsu S.D., Lin Y.L., Lee W.H., Yang C.D., Hong H.C., Wei T.Y., Tu S.J., et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein L.D. Using the Reactome database. Curr. Protoc. Bioinformatics. 2004 doi: 10.1002/0471250953.bi0807s7. doi:10.1002/0471250953.bi0807s7. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura D. BioCarta. Biotech Software Internet Report. 2004;2:117–120. [Google Scholar]
- 20.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jewison T., Su Y., Disfany F.M., Liang Y., Knox C., Maciejewski A., Poelzer J., Huynh J., Zhou Y., Arndt D., et al. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42:D478–D484. doi: 10.1093/nar/gkt1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bleazard T., Lamb J.A., Griffiths-Jones S. Bias in microRNA functional enrichment analysis. Bioinformatics. 2015;31:1592–1598. doi: 10.1093/bioinformatics/btv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paraskevopoulou M. D., Georgakilas G., Kostoulas N., Vlachos I. S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong N., Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2014;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal V., Bell G. W., Nam J. W., Bartel D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tange, O. GNU Parallel - The Command-Line Power Tool. login: The USENIX Magazine. 2011;2011:42–47. [Google Scholar]
- 28.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]