Abstract
The Human Induced Pluripotent Stem Cell Initiative (HipSci) isf establishing a large catalogue of human iPSC lines, arguably the most well characterized collection to date. The HipSci portal enables researchers to choose the right cell line for their experiment, and makes HipSci's rich catalogue of assay data easy to discover and reuse. Each cell line has genomic, transcriptomic, proteomic and cellular phenotyping data. Data are deposited in the appropriate EMBL-EBI archives, including the European Nucleotide Archive (ENA), European Genome-phenome Archive (EGA), ArrayExpress and PRoteomics IDEntifications (PRIDE) databases. The project will make 500 cell lines from healthy individuals, and from 150 patients with rare genetic diseases; these will be available through the European Collection of Authenticated Cell Cultures (ECACC). As of August 2016, 238 cell lines are available for purchase. Project data is presented through the HipSci data portal (http://www.hipsci.org/lines) and is downloadable from the associated FTP site (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp). The data portal presents a summary matrix of the HipSci cell lines, showing available data types. Each line has its own page containing descriptive metadata, quality information, and links to archived assay data. Analysis results are also available in a Track Hub, allowing visualization in the context of public genomic annotations (http://www.hipsci.org/data/trackhubs).
INTRODUCTION
The Human Induced Pluripotent Stem Cell Initiative (HipSci) was established in 2012 to address the community requirement for a large, well-characterized collection of human-induced pluripotent stem cells (iPSCs) for use in research. Human iPSCs are an invaluable system for modelling human disease (1), and are a useful tool for conducting research into the function of genetic variants both associated with complex disease and also normal human phenotypic variation. Previous research has suggested that inter-line variability may be high (2,3), making the subtle effects of common genetic variants difficult to detect. There have been previous large-scale efforts to generate and characterize pluripotent stem cells (4) (https://www.cirm.ca.gov/researchers/ipsc-initiative), but these other projects have either not systematically derived human iPSCs at the scale of HipSci or have not focused on characterizing the phenotypes related to natural genetic variation (5). The HipSci project fulfils a pressing need within the community to provide a large, well-characterized collection of human iPSCs that are systematically generated using a single experimental pipeline.
As of August 2016, the HipSci project has created 477 cell lines from healthy donors, and 86 lines from donors with a rare genetic disease. All of these lines have been characterized with a diverse range of assay data including exome-seq, RNA-seq, 450K methylation arrays and proteomic assays. The cell line catalogue, together with the extensive characterization data, provides the scientific community with a great opportunity to investigate cellular function and the impact that common genetic variation has on this function. Here we present the data access and presentation services that the HipSci project provides, which enable the community to both discover and to reuse the cell lines and data that the project has generated.
Assay data and archival strategy
The HipSci project's strategy is to derive two or three candidate iPSC cell lines from each donor, and to generate an initial set of genetic and phenotyping assays on all candidate lines (Figure 1). This initial set of data includes array-based genotyping and gene expression profiling on the iPSCs and on their progenitor cells, which are either fibroblasts or erythroblasts. The most appropriate cell line is selected for expansion, banking and further profiling using RNA-seq, exome-seq, DNA methylation profiling, proteomics in the form of mass spectrometry and high content cellular phenotyping (6).
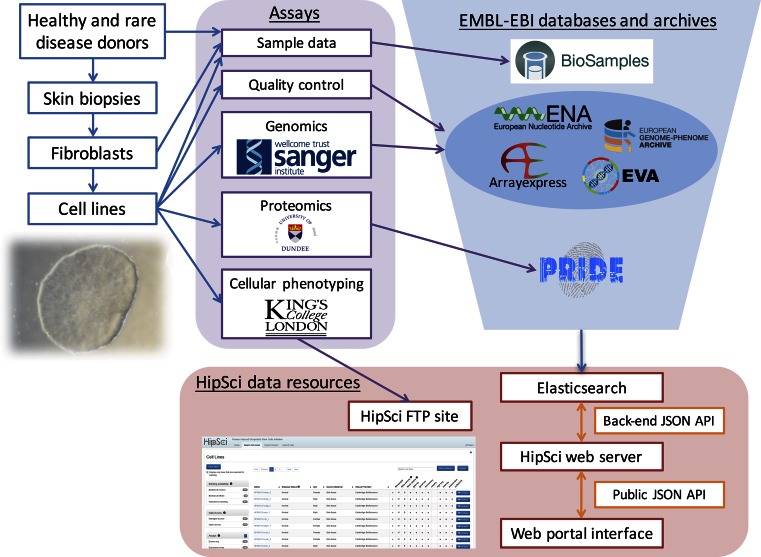
Figure 1.
HipSci data flow. Sample metadata is collected from healthy donors and donors with rare genetic disease who each provide a skin biopsy. Sample metadata is also collected on the Fibroblasts and iPSC cell lines derived from these biopsies. This sample metadata is registered immediately upon creation and before any assays are performed in EMBL-EBI's BioSamples database. For each cell line a range of data is generated including quality control and genomics assays conducted by the Wellcome Trust Sanger Institute, proteomics assays by the University of Dundee, and cellular phenotyping by Kings College London. The quality control, genomics, and proteomics data is deposited in the relevant EMBL-EBI archive, and the cellular phenotyping data is released to the HipSci public FTP site. Table 1 lists the specific archive that assay data is submitted to and shows the distinction in destination between open and managed access data. Our web portal infrastructure is based upon an Elasticsearch engine to which the sample and assay data is loaded. The public json API uses standard Elasticsearch query syntax and through an intermediary web server allows any appropriate search query to be passed through to the search engine. The web portal app displays data fed from the public API creating searchable and filterable views of all of HipSci's rich data.
The HipSci project follows a rapid pre-publication data release strategy, through submission to EMBL-EBI data archives and, in the case of preliminary data and files not appropriate for an official archive, through release via the project FTP site. EMBL-EBI's BioSamples (7) database is used to register sample metadata for all tissue donors, tissue samples, and iPSC lines generated by the project. The samples are registered immediately upon creation and before any assays are performed. The project follows this registration strategy so that all assay data produced for every iPSC line is linked to a single stable cell line identifier. The use of BioSamples ensures that these lines are well described as it supports ontology annotation, which allows the project to unambiguously annotate attributes such as disease and cell type. BioSamples also supports rich relationships between samples, which allows us to track the connection between cell lines and the tissue sample and donor from which they were derived. When assay data is submitted to other EMBL-EBI archives, the BioSamples accession is used to link to the appropriate sample record, ensuring that all archived cell line data is discoverable using this single identifier.
Table 1 lists the assay data that are generated for each HipSci cell line. The assay data is submitted to the appropriate EMBL-EBI assay archive, which ensures that data is stored using well-established infrastructure, is distributed to users efficiently, and can be easily found in the most commonly used data repositories. Our data portal (http://www.hipsci.org/lines/#/lines) links to the assay datasets in each archive, removing the need for replicating files between the HipSci resource and the individual archives.
Table 1. HipSci assay data archival strategy.
| Assay | Data type | Consent | Archive |
|---|---|---|---|
| Sample collection | Sample descriptions | Open and Managed | BioSamples |
| Genotyping array | Genotypes and imputed genotypes | Open | EVA |
| Genotyping array | Genotypes and imputed genotypes | Managed | EGA |
| Expression array | Array Signal intensity data | Open | ArrayExpress |
| Expression array | Array Signal intensity data | Managed | EGA |
| Exome-seq | Aligned reads | Open | ENA |
| Exome-seq | Aligned reads | Managed | EGA |
| Exome-seq | Variant calls and imputed genotypes | Open | EVA |
| Exome-seq | Variant calls and imputed genotypes | Managed | EGA |
| RNA-seq | Aligned reads | Open | ENA |
| RNA-seq | Aligned reads | Managed | EGA |
| RNA-seq | Abundance of transcripts | Open | ENA |
| RNA-seq | Abundance of transcripts | Managed | EGA |
| Methylation array | Array Signal intensity data | Open | ArrayExpress |
| Methylation array | Array Signal intensity data | Managed | EGA |
| Proteomics | Mass spectrometry | Open and Managed | PRIDE |
| Cellular phenotyping | Morphology and DAPI/Edu staining intensity data | Open and Managed | HipSci FTP site |
This table describes the archive used for each assay and consent type combination. Consent for data is either ‘Open’ or ‘Managed’, corresponding to the terms agreed by the donor at the time of sample donation.
The HipSci tissue samples were collected under different participant consents. This mixed-consent mode was both a consequence of timing and the health status of the individuals participating. Both the ‘healthy’ samples that were collected early in the project, and all of the rare disease samples, were collected using managed-access consent. In order to facilitate broad re-use, the consent policy for the healthy samples was updated to allow for a more open mode of data release.
The types of consent affect our strategy for archiving and distributing the data. For example, data from openly consented healthy samples that is uniquely associated with the sample donor, such as genotype or genomic sequence data, can be made freely available to all users through the European Nucleotide Archive (ENA) (8) and ArrayExpress (9). In order to meet the conditions of the managed-access consent signed by other donors, the equivalent data types are submitted to the European Genome Phenome Archive (EGA) (10), where the data is only accessible to bona fide researchers who have been granted permission to access the data by the HipSci Data Access Committee (http://www.sanger.ac.uk/about/who-we-are/policies/open-access-science).
Proteomics and cellular phenotyping data are not uniquely linkable to the sample donor, so for all cell lines these data are distributed openly without managed-access restrictions. Proteomics mass spectrometry data is deposited in the PRoteomics IDEntifications (PRIDE) database (11). HipSci's high content cellular phenotyping data is a relatively new data type, for which there is no well-established public archive. We therefore distribute these data via our project FTP site (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp). We are working with the archives hosted at EMBL-EBI to establish the best location for this data.
Cell line availability
HipSci cell lines are made available to purchase from the European Collection of Authenticated Cell Cultures (ECACC) (https://www.phe-culturecollections.org.uk/collections/ecacc.aspx). The HipSci portal provides direct links to ECACC from individual purchase buttons located both beside each cell line on the cell line summary matrix and from the individual cell line pages (Figure 2). ECACC supports the purchase of multiple cell lines in a single transaction and is able to ship cell lines worldwide. For each cell line, a user must complete a material transfer agreement that sets out the terms of use for the cell line prior to the cell line shipment. Each cell line is provided with a batch-specific certificate of analysis, which documents the results of the quality control (QC) testing criteria performed upon the banked line and includes extensive guidance for the handling of HipSci iPSCs. HipSci cell lines are released under a not-for-profit material transfer agreement for academic non-commercial use; commercial entities can purchase a significant subset of HipSci cell lines through the European Bank of induced Pluripotent Stem Cells (EBiSC; https://ebisc.org/).
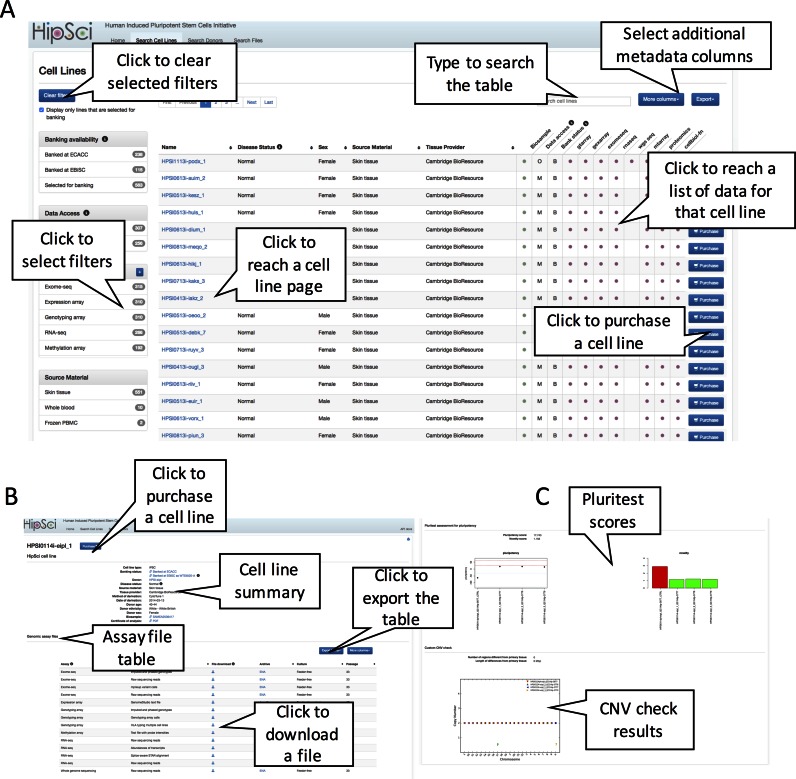
Figure 2.
Using the HipSci cell line and data browser. The HipSci data browser provides views to explore the complex data the project has collected. (A) The main page of the browser is a table listing all the available cell lines with both a search box and specific filters to restrict the table by attributes like disease state, assay availability, banking availability, and source material. The table itself contains the cell line links, which take the user to cell line summary pages and assay links that take the user to pages listing all the files available for that line and assay type. (B) The cell line summary page itself contains descriptive information about a line, including disease state, derivation method, donor sex and tissue provider. Below the cell line summary, the table lists all files associated with a line, the assay that produced them, and what culture conditions and passage number they were produced under. (C) After the Assay file table are the line QC results for Pluritest and the HipSci copy number variation (CNV) check. These graphs present the results for the given line, its clones and the control data generated using the donor tissue sample.
Data access
The HipSci cell lines and data can be explored and downloaded through several different mechanisms.
Website
The HipSci website (http://www.hipsci.org) provides summary information about the HipSci project. Information includes assay descriptions, our data reuse policy and announcements about the project. The website also acts as the entry point to our data portal. The content is designed to provide users with context about the project, the data we hold, and to help them find and use our data. All news announcements are also made on our twitter feed (@hipsci).
Cell lines and data browser
The browser presented in HipSci's dedicated web portal is designed for users to browse cell lines and their related data (http://www.hipsci.org/lines) (Figure 2). Our primary user group is biologists who want to select cell lines meeting certain criteria, such as donor disease, age, iPSC derivation method. This user group will ultimately want to either purchase the lines through ECACC, or download the associated assay data for use in their own experiments.
EMBL-EBI is currently working on projects to unify both archival submission and display; in the meantime, there is no central solution to provide the community with a coherent view of all the HipSci project data and cell lines. The HipSci data portal serves this need, by providing the iPSC community with a unified view of the HipSci collection, enabling them to discover cell lines they want to purchase and by giving them a consolidated view of all the data associated with a particular cell line. The portal also hosts data that does not have a current home in any of EMBL-EBI's archives, such as the cellular phenotyping and cell line QC data. The portal points users to the archive location for all other data types.
The cell line and data browser is the primary point of access for users wishing to query the cell lines and assay data that HipSci has produced. As of August 2016, there are 2668 cell lines and 7589 individually archived assay results displayed in the data browser.
There are three main entry points into the data from the browser's navigation bar: ‘search cell lines’, ‘donors’ or ‘files’ (Figure 2). Each of these entry points shows a table with high-level information about the cell line, sample donor or archived file, respectively. Users can customize their view by adding specific columns to the table display, by sorting the row order by clicking on the columns, or by exporting the table as a text file that can be loaded into a spreadsheet. A search box allows users to restrict the table to lines that match particular criteria, for example, cell line name ‘HPSI0114i-eipl_1′, or assay type, for example ‘proteomics’ or ‘RNA-seq’, or disease state, for example ‘Bardet-Biedl’. A list of filters allows the user to refine their search for cell lines or assay files that meet their criteria, such as disease, banking availability, and available assay data. The search box and filters reduce the table contents to lines that match the user's selection. The cell line table has columns for each of the assay types, with clickable dots, which take the user to more information about the available assay data (Figure 2).
The browser also has a cell line detail view, which displays comprehensive information about a single chosen cell line, for example http://www.hipsci.org/lines/#/lines/HPSI0114i-eipl_1. This detail can be reached by clicking on the cell line name in the cell line table. Each cell line page presents disease and demographic information about the sample donor; the clinic who provided the tissue; the method of iPSC derivation; characterization of the line; archived assay data for that cell line; and a link to ECACC in order to purchase a line, if it has been released to the bank.
The cell line characterization section includes results of the pluritest assay (12) and an analysis of copy number variations from genotyping array data (13). This section provides the user with a view of the summary statistics the project used to select which lines should be expanded, banked, and distributed. The cell line detail view contains a link to the certificate of analysis that a customer will receive when purchasing a cell line. These documents demonstrate to a recipient that the batch has passed all the HipSci project QC tests. It lists the tests that the cell line has passed to ensure sterility, viability, pluripotency, identity, morphology and clearance of reprogramming factors.
Application programming interface (API)
The data presented in the HipSci browser is pulled into the website using a RESTful Application Programming Interface (API). This API is publicly accessible and allows searching, filtering, and sorting of cell lines, of sample donors, and of archived data files. Programmatic users access the same information as a user of the web interface. This API is used by ECACC to acquire and update cell line data on commercially available lines.
The API (Figure 1) is powered by Elasticsearch, a popular search engine with extensive online documentation describing many different types of search query over http using a JSON syntax. A web server sits between the user and the search engine, to protect the search engine from inappropriate actions (such as deleting a record), but for all other cases a user's search request is forwarded directly to the Elasticsearch infrastructure. This allows users to compose many complex queries with no restrictions; for example, to search for cell lines matching a query string ‘diabetes’, whilst filtering for lines from female donors in the age range 35–39. The HipSci website has documentation describing use of the API (http://www.hipsci.org/lines/#/api).
FTP site
The HipSci project is committed to early data access, and our rapid archiving strategy ensures that data are available for download by FTP directly from the relevant EMBL-EBI archive. HipSci also has its own FTP site (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp) to facilitate the download of preliminary data and of new data types, such as high content cellular phenotyping, for which there is no well-established assay archive (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp/data/). In providing the raw data via FTP we ensure that all users can access specific data types and support those who wish to use data from many different cell lines and assay types at once.
The HipSci FTP site provides index files that list all of the assay data available from EMBL-EBI archives (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp/archive_datasets/). These tab-delimited index files enable users to locate all the data quickly and to understand the relationship of all data files to their respective cell line. These index files also make it much easier for users to download in bulk all of the HipSci project data.
Browsing the HipSci analysis results
Visualizing the analysis results associated with the HipSci lines alongside a broader range of genomic annotation allows our users to put the HipSci results in genomic context alongside information such as the GENCODE annotation (14), Ensembl regulatory annotation (15) or genomic sequence variants from projects such as the 1000 Genomes Project (16) or COSMIC (17). We enable this contextual visualization by using Track Hub technology (18); these hubs allow users to attach analysis files to either the UCSC (19) or Ensembl (20) genome browsers with a single URL, and to explore the HipSci results. The hub files themselves are text files which contain pointers to the publicly mounted location of the results files and to the associated metadata such as the cell line name, disease state and analysis type. By default, results from only a few lines will be displayed when the hub is first attached to the browser. Both UCSC and Ensembl have powerful configuration menus that allow users to change the currently viewed selection of cell lines and analysis file tracks within the hub. Our hub is registered in the Track Hub registry (http://trackhubregistry.org/) to enable broad discovery and reuse. The hub itself is hosted from our FTP site (ftp://ftp.hipsci.ebi.ac.uk/vol1/ftp/track_hub/hipsci_hub/) and we provide users with direct links to the UCSC and Ensembl genome browsers along with instructions for use from our website. The trackhub published in August 2016 only contains exome sequencing alignment data. As more analysis results are released by the consortium, in a trackhub suitable format, they will be added to the hub.
Data reuse
The HipSci project data is released early, prior to publication, in the expectation that it will be valuable for many researchers. In keeping with Fort Lauderdale principles, the public community of users may utilise the data for their own research, but are expected to allow the HipSci consortium to make the first presentations and to publish the first papers with global analyses of the data. After the HipSci consortium has published analyses, then researchers outside the project are free to present and publish using the project data for their analyses.
Prior to the first major HipSci paper, other researchers may still present methods development posters that include small amounts of HipSci data, provided the quantity of HipSci data is sufficiently small, and the project is properly acknowledged. A more thorough description of these data use conditions is available on the website (http://www.hipsci.org/data/policy).
Future
Moving forward we will continue to gather together all of the data generated by the HipSci project, and to enable the research community to discover data of interest to them. The depth of genomic sequence variation data available for these cell lines means that the data sets can be challenging to search because of the volume of samples and genomic variant sites which need to be queried: currently, the only solution to this requires a user to download all of the VCF files we present and to query across all of them. We are working with the European Variation Archive (EVA) to take advantage of their ‘variant search’ API (http://www.ebi.ac.uk/eva/?API), and once the openly consented HipSci genotype data is loaded into the EVA variation database, we will build the relevant search tools into the HipSci data portal. This will allow users to query across all the openly consented HipSci sample genomic variants and genotypes, allowing users to issue queries such as ‘which HipSci cell line is homozygous for allele X at genome position Y?’
iPSCs provide a compelling model for human biology because it is possible to differentiate them into many types of human cells. HipSci is now generating assay data on differentiated cells, such as macrophages and sensory neurons. These data will be integrated into the HipSci data portal, and we will work to build tools that allow users to intuitively search across sample, cell, and tissue type. The HipSci project has produced a valuable catalogue of cell lines. This article presents the current tools, which can be used to explore and visualize the HipSci data. As the project moves forward, we expect to continue extending and expanding these tools to ensure that the HipSci data remains useful to the community.
Contact details
General questions about the HipSci project and its resources should be addressed to hipsci@ebi.ac.uk. Specific questions about purchasing HipSci cell lines should be sent to ECACC using culturecollections.technical@phe.gov.uk. The source code supporting our data portal and website can be found on Github https://github.com/hipsci/hipsci.github.io. You can also follow us on twitter @hipsci.
Acknowledgments
We thank all the members of the HipSci consortium and Sophie Janacek for a critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust [WT-098503/D/12]; European Molecular Biology Laboratory. Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sterneckert J.L., Reinhardt P., Schöler H.R. Investigating human disease using stem cell models. Nat. Rev. Genet. 2014;15:625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- 2.Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S., et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nazor K.L., Altun G., Lynch C., Tran H., Harness J.V., Slavin I., Garitaonandia I., Müller F.J., Wang Y.C., Boscolo F.S., et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solomon S.L. The New York stem cell foundation: accelerating cures through stem cell research. Stem Cells Transl. Med. 2012;1:263–265. doi: 10.5966/sctm.2012-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The International Stem Cell Initiative. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat. Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leha A., Moens N., Meleckyte R., Culley O.J., Gervasio M.K., Kerz M., Reimer A., Cain S.A., Streeter I., Folarin A., et al. A high-content platform to characterise human induced pluripotent stem cell lines. Methods. 2016;96:85–96. doi: 10.1016/j.ymeth.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faulconbridge A., Burdett T., Brandizi M., Gostev M., Pereira R., Vasant D., Sarkans U., Brazma A., Parkinson H. Updates to BioSamples database at European Bioinformatics Institute. Nucleic Acids Res. 2014;42:D50–D52. doi: 10.1093/nar/gkt1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silvester N., Alako B., Amid C., Cerdeño-Tárraga A., Cleland I., Gibson R., Goodgame N., Ten Hoopen P., Kay S., Leinonen R., et al. Content discovery and retrieval services at the European Nucleotide Archive. Nucleic Acids Res. 2015;43:D23–D29. doi: 10.1093/nar/gku1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T., et al. ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015;43:D1113–D1116. doi: 10.1093/nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lappalainen I., Almeida-King J., Kumanduri V., Senf A., Spalding J.D., Ur-Rehman S., Saunders G., Kandasamy J., Caccamo M., Leinonen R., et al. The European Genome-phenome Archive of human data consented for biomedical research. Nat. Genet. 2015;47:692–695. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller F.J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O., et al. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danecek P., McCarthy S.A., HipSci C., Durbin R. A method for checking genomic integrity in cultured cell lines from SNP genotyping data. PLoS One. 2016;11:e0155014. doi: 10.1371/journal.pone.0155014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino D.R., Wilder S.P., Johnson N., Juettemann T., Flicek P.R. The Ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S., et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44:D717–D725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates A., Akanni W., Amode M.R., Barrell D., Billis K., Carvalho-Silva D., Cummins C., Clapham P., Fitzgerald S., Gil L., et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]