Abstract
Plant Reactome (http://plantreactome.gramene.org/) is a free, open-source, curated plant pathway database portal, provided as part of the Gramene project. The database provides intuitive bioinformatics tools for the visualization, analysis and interpretation of pathway knowledge to support genome annotation, genome analysis, modeling, systems biology, basic research and education. Plant Reactome employs the structural framework of a plant cell to show metabolic, transport, genetic, developmental and signaling pathways. We manually curate molecular details of pathways in these domains for reference species Oryza sativa (rice) supported by published literature and annotation of well-characterized genes. Two hundred twenty-two rice pathways, 1025 reactions associated with 1173 proteins, 907 small molecules and 256 literature references have been curated to date. These reference annotations were used to project pathways for 62 model, crop and evolutionarily significant plant species based on gene homology. Database users can search and browse various components of the database, visualize curated baseline expression of pathway-associated genes provided by the Expression Atlas and upload and analyze their Omics datasets. The database also offers data access via Application Programming Interfaces (APIs) and in various standardized pathway formats, such as SBML and BioPAX.
INTRODUCTION
Multi-faceted bioinformatic resources and databases can facilitate understanding of a plant's genetic complexity, structure, development, evolution and its response to environmental stress conditions. It is a prerequisite for developing new and improved varieties of crops to meet the growing demand for production and yield and challenges posed by population growth, global climate change, and biotic and abiotic stresses. The Gramene database (http://gramene.org/) (1) provides online resources to plant researchers for conducting a comparative analysis of plant genomes and pathways (1). The Plant Reactome database (http://plantreactome.gramene.org), the Pathways portal of Gramene, hosts metabolic, transport, genetic, signaling and developmental pathways for 63 plant species including various crops, experimental models and other species important to understand the evolution of pathway networks (Table 1). The Plant Reactome (Supplementary Figure S1) was developed in collaboration with the Human Reactome project (2) and utilizes their data model to organize proteins, protein complexes, small molecules and macromolecular interactions into reactions and pathways in the context of their subcellular location and interactions to build a systems-level pathway network of a plant cell.
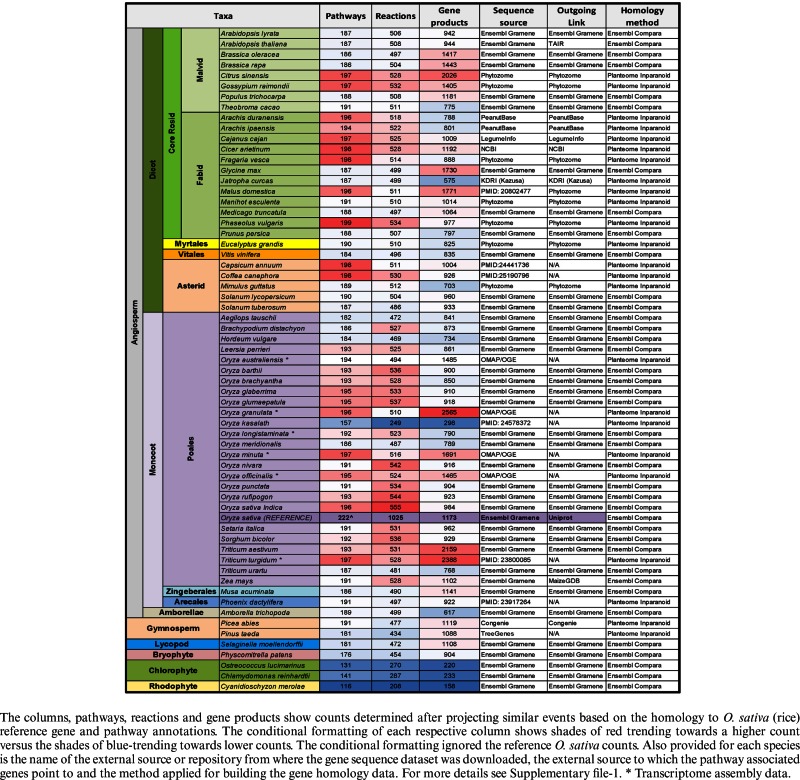
Table 1. Taxonomic distribution of the 63 plant species represented in the Plant Reactome pathway database.
|
The Plant Reactome features Oryza sativa (rice) as a reference species. To construct the reference pathway network, metabolic pathways from RiceCyc (3) were bulk imported into the Reactome data structure using BioPAX Level-2 conversion tools. Subsequently, these pathways and their various components were curated, and pathway diagrams were manually drawn to meet the Reactome database model standards. The dataset for 12 rice metabolic pathways was also inferred based on gene homology from the human Reactome (2) that includes evolutionary conserved regulatory processes, such as cell growth, cell cycle, DNA replication/repair, etc. As protein functions have not been systematically and experimentally characterized for large number of genes from many plant species, we use an integrated approach for curating the rice and non-rice literature to build the reference datasets. These reference data sets are then used to project homologous events in 62 model, crop and evolutionary significant plant species. The combination of pathway analysis tools, annotations, gene expression data and the search and browse functionalities enable plant researchers to review, reuse, discover and build stochastic models for further studies in their species of interest. Achieved by investigating the molecular interactions, gene expression under various conditions, and evolution of pathways, reactions and small molecules, thus moving the research beyond the realms of genome and transcriptome sequencing. The database extensively collaborates with various plant genomics projects such as Gramene's Ensembl genome portal (1), Phytozome (4), PeanutBase (5), TreeGenes (6), Genome Database for Rosaceae (7), SolGenomics Network (8), MaizeGDB (9), TAIR (10), AraPort (11), Legume Information System (12) and Planteome (13) (Table 1). The collaboration extends to large online resource providers such as Gene Ontology (14), EMBL-EBI's Gene Expression Atlas (15), ChEBI (16), PubMed, UniProt (17) and NCBI, in order to share data and extend its linkages (Supplementary Figure S2). The database also provides access to data in various community-wide standardized pathway formats and via Application Programming Interfaces (APIs).
PLANT REACTOME RESOURCE
Since its first beta release in January 2013, Plant Reactome has grown to include primary reference annotations in the form of 222 pathways and 1025 reactions associated with 1173 rice gene products. We continue to curate rice pathways manually and also use automated methods and scripts that integrate data related to structure and function of proteins, their subcellular locations, the expression of protein-coding genes, and diversity of small molecule and metabolite from the external resources mentioned above. In addition to these reference rice pathways, the database provides gene homology-based pathway projections for 62 other plant species (Table 1) as described later. The Plant Reactome allows researchers to query and visualize pathways in species of their choice and analyze their genome-scale expression data for pathway enrichment analysis to identify differentially expressed pathways and genes. Users can also compare projected pathways with reference rice pathways and visualize curated baseline and differential expression data for all pathway-associated genes fetched programmatically from EMBL-EBI's Expression Atlas (15).
The database homepage, accessible from http://plantreactome.gramene.org, provides quick links to the pathway browser, data analysis tools and download options, user guide, video tutorials, database release summary, news and documentation about the project, data model, schema and APIs. (Supplementary Figure S1). The homepage also provides direct access to the primary database search feature.
We will use an example of the jasmonic acid (JA) signaling pathway and the COI1 (CORONATINE INSENSITIVE1) gene from the reference species rice and its projections on other angiosperm species like soybean, tomato, maize and non-angiosperms lycopod Selaginella and unicellular green algae Chlamydomonas. We will use this example to describe the development and various functionalities of the Plant Reactome resource including analysis tools for use in research, education, and training. Some studies on Plant COI1 gene family members are reported. The A. thaliana COI gene encodes an F-Box domain containing protein that binds to jasmonate-isoleucine (JA-Ile)/coronatine (COR) and acts as a jasmonic acid (JA) receptor, thus playing a role in the JA signaling pathway (18,19). COR is a phytotoxin produced by the fungus Pseudomonas syringae a well-known pathogen of crop plants (19). The rice COI1 gene is known for its role in immunity, plant development and senescence (20,21). Rice has three gene family members and the COI1b mutant stayed green in the dark-induced natural senescence and showed a decrease in spikelet fertility and grain weight leading to lower yield (22). The tomato homolog is essential for seed maturation, immunity, and glandular trichome development (23,24), whereas soybean COI homolog complements and restores Arabidopsis COI function (25). Studies also showed that COI1 plays an interactive role with abscisic acid signaling in regulating the stomatal closure and ion channel activation in response to drought (26,27). To investigate if COI1 expression is affected by drought conditions, we selected a Glycine max (soybean) RNA-seq based drought transcriptome study as an example for OMICs data analysis.
PLANT REACTOME SEARCH
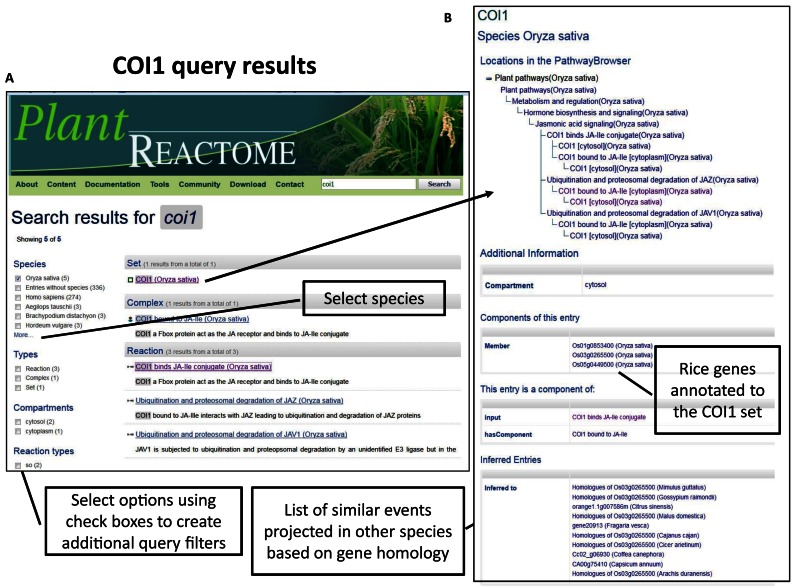
The quick search box is powered by the Solr search platform (http://lucene.apache.org/solr/), designed for highly scalable, load-balanced, and dynamically indexed full-text searches. The Plant Reactome implementation makes particular use of the auto-completion feature and faceted search result filters, which aid the user with search suggestions and the ability to narrow down results with precision. The quick search tool on the right-most upper side of Plant Reactome homepage (http://plantreactome.gramene.org/) allows users to search for any small molecules including metabolites, proteins, protein complexes, enzymes, reactions and pathways in all 63 species, including the reference species Oryza sativa (rice). For example, querying for the COI1 (Figure 1A) gene returns results showing that, if selected for reference O. sativa, there are five entities in the database that either bears the name COI1 or associated with the gene set called COI1. The list of facets with check boxes in the left-hand menu allows additional query filters. One has an option to go further for a detailed view (Figure 1B) of the search on the COI1 set from rice. The top panel shows event hierarchy and provides links to the Pathway Browser (Figure 1B), and the bottom panel suggests associated cellular component locations, three rice gene products that are part of the COI1 set, reaction components, and a list of similar events projected in other species inferred by gene homology-based projection. In this query result, users also learn that three rice gene products are part of the COI1 set.
Figure 1.
Plant Reactome search results for the query ‘COI1’. COI1 is the CORONATINE INSENSITIVE1 gene homolog. (A) The search results selected for rice, showing entities that either bears the name COI1 or associated with the gene set called COI1. (B) Detailed view of the search on COI1 set from rice showing event hierarchy, links to the Pathway Browser and additional information about the gene set.
The additional small molecule search available from http://plantreactome.gramene.org/cgi-bin/small_molecule_search allows searches of various chemical compound entities. The query form allows the user to illustrate a molecule and submit searches to the remote ChEBI database of small molecules (16). Resulting matches connect to the same molecule in the Plant Reactome database. For example, a search for C2H4 finds a match to Ethene or Ethylene (CHEBI:18153) and a link to the pathways involving that molecule in the database.
PATHWAY BROWSER
The Plant Reactome utilizes the conceptual framework of a eukaryotic plant cell to annotate the interactions of small molecules including metabolites, enzymes, and regulators as reactions and pathways. The pathways, in turn, are organized around a hierarchical classification schema similar to the Gene Ontology (GO) that allows building relationships between pathways (e.g. grouping of similar pathways, parent–child relationship, connections between interacting pathways, etc.). Users can reach the pathway browser directly from the database homepage or from query results, such as those for COI1 (Figure 1).
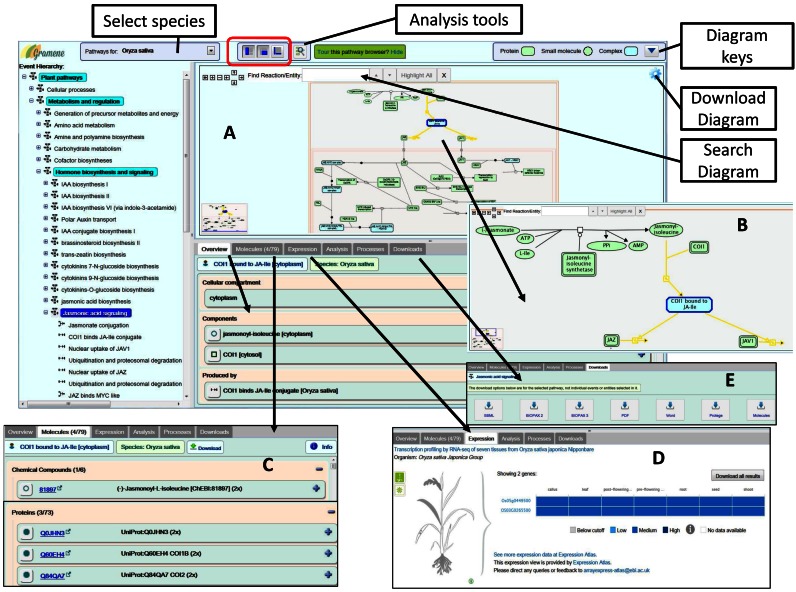
By default, the pathway browser window provides an overview of all available rice reference pathways, but users can choose another plant species from a pull-down list. An example screen shot of the pathway browser window is shown in Figure 2A displaying overview of the O. sativa (rice) jasmonic acid (JA) signaling pathway. An event hierarchy located on the left-hand panel allows selection of pathways, sub-pathways and reactions. Clicking the hyperlinked name of an event in the hierarchy will open the respective pathway diagram in the top right-hand side panel, known as the pathway diagram viewer. The pathway diagram viewer graphically depicts connected reactions and events associated with a pathway in the context of their subcellular locations and allows zooming to facilitate easy access to the details about each entity. The zoom feature allows adjustment of the diagram views, such as the zoomed-in view (Figure 2B) of the reaction involving jasmonic acid and the COI1 gene products, in addition to its upstream and downstream events. Users can search various entities on the given pathway diagram and can click on individual entities to get associated data and external links in the pathway and entity detail panel found below the pathway diagram. Users can also access various other features, such as the summation of the pathway and the Molecules tab (Figure 1C) listing interacting molecules depending on the selected event.
Figure 2.
A view of the Plant Reactome Pathway Browser details. (A) An overview of the selected O. sativa (rice) jasmonic acid (JA) signaling pathway. Zoom in and out feature (red outlined section) allows adjusting the diagram views, such as the zoomed-in view (B) of the reaction involving jasmonic acid. (C) The Molecules tab provides a list or interacting molecules. (D) Depending on the selected entities in the diagram, the expression tab displays baseline (RNA-seq based) gene expression data analyzed and served by the Gene Expression Atlas. (E) Download the selected entities data in various standard formats, including SBML and BioPAX.
Contingent upon the availability of data accessed programmatically from the EMBL-EBI Gene Expression Atlas (15), the Expression tab (Figure 1D) displays a baseline expression profile of the genes associated with the selected reference rice pathway. Similar baseline data is also available for nine other species. By choosing the data cells in the expression view, users can find the actual data value as well as the highlighted anatomogram image (such as parts of a rice plant) shown on the left-hand side. Once in the expression view, clicking on the ‘see more expression data’ hyperlink at the bottom of that view takes the user to an external Expression Atlas page of the collaborator database that may show available differential expression data for the genes associated with the selected reaction or pathway view. The 22 June 2016, release of the EMBL-EBI Gene Expression Atlas includes 671 gene expression dataset from fourteen plant species. Baseline expression data generated from the RNA-seq transcriptome studies provide estimates of transcript abundance for each gene across various experimental conditions, e.g. tissues, cell types and development stages. The differential expression data scored for each experimental study provides information on the differential expression of a given gene or a gene set between two conditions including the treatments as previously described (15).
All three panels in the pathway browser window are tightly linked and clicking on any of the panels changes the view of other panels displaying associated features and data. The icon pointing to the Analysis tools options is also available on top of the pathway browser page (Figure 2A) as well as the homepage of the project website and described in the respective section (Figure 3).
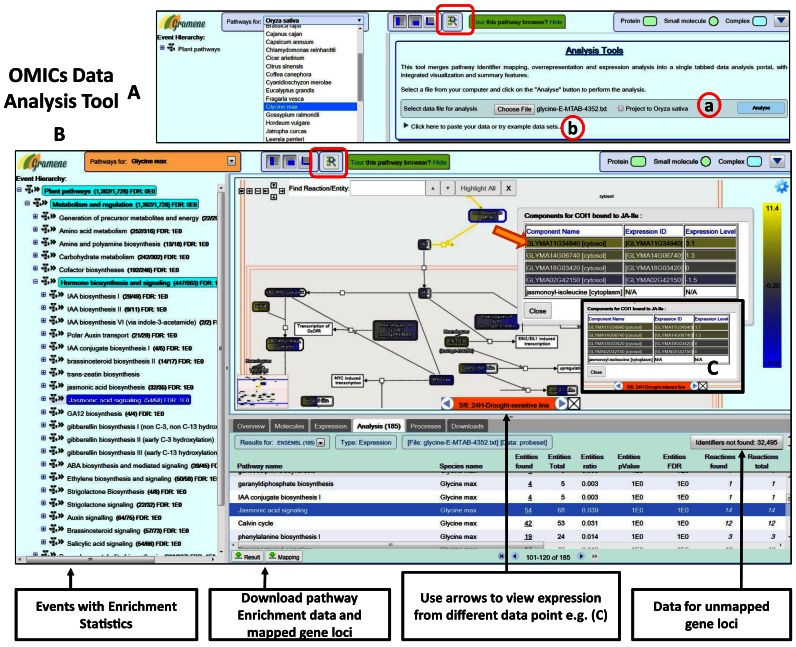
Figure 3.
Plant Reactome pathway analysis tools interface: OMICs data analysis. (A) The analysis tool is accessible from the project website's homepage (http://plantreactome.gramene.org) by clicking on the Analyze Data icon or if the users are on the pathway browser click on the Analyze your data icon (red box). The analysis tools interface replaces the Pathway diagram view in the pathway browser. Users can input their data either by, (a) uploading the data file (this figure) or (b) by copying the data into the text box (Supplementary Figure S3). After selecting the data, click on the Analyze button and the pathway, enrichment analysis data is returned in the tabulated form (B) in the Analysis tab. Select the event from the left-hand side panel to view the diagram with expression data. By hovering the computer mouse over the colored genes sets and clicking the blue info icon a list of mapped entities with painted expression values is displayed. Left or the right arrow next to the orange text box displays expression from a different data column (C).
The information on selected entities data is downloadable in various standard formats, including SBML and BioPAX, underneath the Download tab (Figure 2E).
PATHWAY PROJECTIONS FOR PLANT GENOMES
The human Reactome database (2) allows the construction of gene homology-based pre-computed pathway network projections from a single reference species to other species. We used a similar approach, to construct pathway projections for 62 plant species (http://plantreactome.gramene.org/stats.html), which were generated using rice as a reference species (Table 1). Homology data for 38 of these species were accessed from Gramene's Ensembl Compara (1,28), while the remaining 24 species were generated in-house by comparing them with O. sativa reference annotations via the InParanoid homology clustering method developed by us (29–31). We extract these species homologs for O. sativa from a local InParanoid MySQL database. Both sets of homology data, formatted as flat files containing one-to-many pairs of homologous gene locus identifiers, are then fed to a Reactome pathway projection Perl script that generates new gene product, reaction, and pathway data in the Plant Reactome database. Table 1 provides a complete summary of the projected pathways, reactions, and associated homologous gene counts. The counts and projections exclude the original 12 pathways initially projected for rice using the human Reactome annotations as described above. The availability of projected pathways and reactions are unlike the KEGG (32) or BioCyc-based (33) projections in that Plant Reactome will only display the individual reactions of a pathway associated with gene product activity. The projected data for any of these species is accessible to users with full functionalities described in this publication.
PATHWAY ANALYSIS TOOLS
Moving beyond differential gene expression, ontology-based enrichment and genome annotations, researchers are often looking for ways to analyze their data for making inferences and building a hypothesis based on molecular interactions and pathways. Therefore, Plant Reactome provides two types of analysis tools for its users: (i) pathway enrichment and overlay visualization of user-provided data from OMICs experiments (Figure 3 and Supplementary Figure S3) and (ii) interspecies pathway comparison (Figure 4). Both tools described in the following sections provide data analysis, visual insights and data downloads. Users can access the Analysis Tools from the Plant Reactome homepage (Supplementary Figure S1) by clicking on the Analyze Data icon or from the icon in the header of the Pathway Browser (Figure 2).
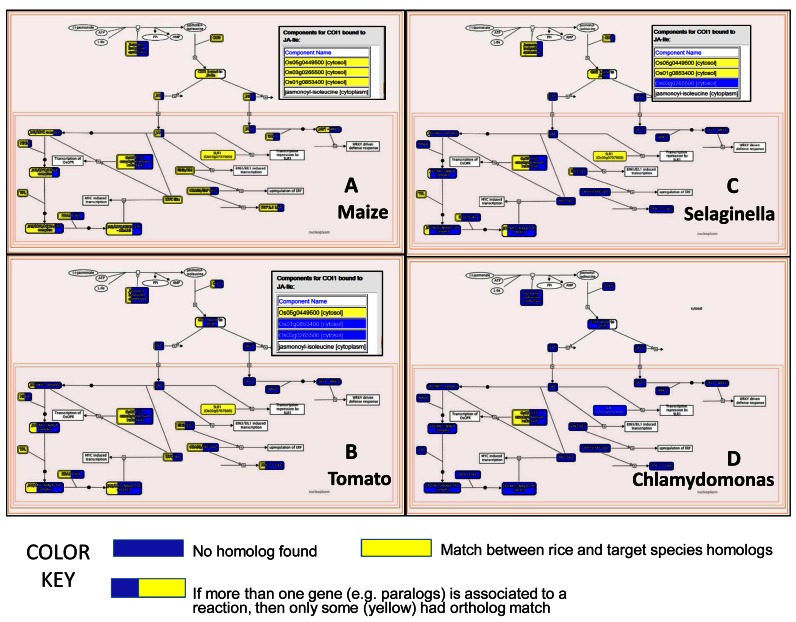
Figure 4.
Plant Reactome pathway analysis tools interface: Interspecific pathway comparative views of the jasmonic acid signaling pathway when compared to angiosperms maize (A) and tomato (B), lycopod Selaginella (C) and unicellular green algae Chlamydomonas (D) based on the respective gene homology matches with the rice reference. Colored boxes suggest that either a respective rice gene homolog was found (yellow color) or not (blue). The reaction associated with multiple rice genes may display both yellow and blue colors depending on the number of homolog matches which also corresponds to the proportion of the yellow or blue color. The inset showing Components for COI1 bound to JA-Ile suggests which of the rice genes have homologs mapped from the respective species.
Pathway analysis using OMICs Data
The Analysis Tools allow uploading, visualization and analysis of user-defined high-throughput OMICs data (e.g. transcriptome, proteome, metabolome, etc.) in the context of Plant Reactome pathways. The analysis tool does not accept raw data, and users need to process their data in a tab-delimited file format before uploading the data file to conduct pathway enrichment analysis (Figure 3A). The users can either upload the data file or copy the data in the text box (Supplementary Figure S3) and may choose to project their data against only the reference O. sativa (rice) or against another species. Before opting for the latter select the appropriate species from the drop-down menu, in this case, Glycine max. To illustrate the utility of the OMICs analysis tools, we used the pre-analyzed and publicly available RNA-seq based transcriptomic data (E-MTAB-4352; source: Gene Expression Atlas (15)) generated from the two accessions of G. max (soybean) that show different phenotype response to drought treatment (SUPPLEMENTARY FILE-3). After selecting the data, clicking on the Analyze button initiates the analysis/process. Depending on the size of the data, and mapped entities in the database (e.g. Gene IDs provided by the Source database (Table 1) and UniProt IDs for rice only, metabolite name or IDs, etc.), the pathway enrichment analysis data is returned in the tabulated form (Figure 3B). Other tabs continue to provide the data mentioned in Figure 2. Users would also notice that the event hierarchy panel on the left-hand side now shows the data statistics next to the pathway event because all the events recorded in the Plant Reactome database for the selected species, in this case, G. max are included in the analysis. Users can choose the pathway event from the left-hand side panel and the tabulated data in the Analysis tab. For example, the jasmonic acid signaling pathway showing 54 genes from the user input data mapped to the projected soybean JA signaling pathway with a total of 68 genes and 14 reactions. Users also get options to download the mapped, unmapped and analyzed data for later use in publications. If the input file has multiple data columns, by default, it displays data from the first data column in the overlaid views. Change it by clicking the left or the right arrows on either side of the orange highlighted text box at the bottom of the pathway diagram.
Visualize the expression data corresponding to the various genes mapped to a given pathway in the pathway diagram panel by selecting the gene set or an event and clicking the blue info icon. A dialogue box opens listing the gene homologs or gene family members mapped to the given event and overlays their expression profile (inset: Components for COI1 bound to JA-Ile). For example, at the 24-h time point after drought treatment in sensitive soybean accession (main figure) compared to the expression of same genes at the same time point after drought treatment in the tolerant soybean line (Figure 3C). Although the gene duplicates are likely to encode similar proteins, their expression and functions may differ across the tissues, organ and cells during plant development as well as in response to stimuli and environment (9,14,16–20).
Our analysis results suggest that the soybean COI1 homolog GLYMA11G34940 mapped to the COI1-JA-Ile complex shows about two-fold increased expression in response to the drought treatment in the tolerant line. The susceptible line shows downregulation of the other homolog GLYMA0-2G42150, whereas, GLYMA14G06740 and GLYMA18G03420 do not indicate any change in their expression when compared between the two lines. Although transcript abundance scored in this data may not provide a direct correlation to the translated protein function and abundance, the observation suggests that at least two COI1 gene family members in soybean show altered expression at transcript level under drought response.
OMICs data analysis views like these are a simple way to begin building hypotheses searching for candidate genes that show altered transcript abundance in phenotypically different accessions in response to the same treatment. Such analyses have the potential to become powerful tools for deciphering and discovering candidate genes regulating the important agronomic traits like abiotic and biotic stress resistance or tolerance, development, anatomy, physiology, yield and nutrition quality. It applies in particular when these inferences are combined with ontology-based gene function annotations, overlapping genetic markers, SNPs, QTLs and syntenic alignments to closely related genomes, thus helping researchers formulate new hypotheses using a systems biology approach and narrowing down the scale of experimental tests.
Interspecific pathway comparison
The second type of analysis tool provided by the Plant Reactome allows comparison of events such as pathways and reactions between the reference species rice and any of the other species by way of mapped gene entities. See analysis steps shown in Supplementary Figure S4 using rice to grape comparison. In another example, b selecting the species of choice, for example, Zea mays (maize) and clicking on the Compare button, the analyzed results are displayed and available for download from the Analysis tab of the pathway browser window described earlier. In this analysis, clicking on the hyperlinked event name in the event hierarchy panel or those in the analysis results table, for example, the jasmonic acid signaling pathway, will render a reference rice pathway diagram overlaid with homolog mappings from the selected species (Figure 4), in this case Z. mays (maize) with colored gene set boxes—suggesting either a respective rice gene homolog was found (yellow color) or not found (blue). In the case of multiple rice genes associated with the reaction, users may see both yellow and blue colors depending on the number of homolog matches between the species, which also correspond to the proportion of the yellow or blue color. Hovering the computer mouse over the colored box and clicking the blue info icon pops an inset showing the detailed gene list with color as shown in the Figure 4. Views such as these provide insights into the evolution of gene families, pathways, and reactions. For example, we observe that the jasmonic acid signaling pathway show likely conservation for all the curated events with at least one or more rice gene homologs present (yellow color) in the closely related maize, also a monocot (Figure 4A), including the components for COI1 bound to JA-Ile. A similar comparison with tomato (Figure 4B), a dicot and a non-angiosperm basal vascular plant Selaginella (Figure 4C), a lycopod, show some degree of conservation, but also no homologs (all blue boxes) for certain reactions downstream of the JA-Ile complex formation stage. Similarly, Chlamydomonas (Figure 4D), green algae from an aquatic habitat, on the other hand, is probably missing homologs for a majority of the genes. These views provide clues for building a hypothesis that either certain parts of the JA-signaling pathway may lack support in tomato and Selaginella or need better annotation. A similar observation for Chlamydomonas suggests that the pathway may not be required for the unicellular algae to sustain in the aquatic habitat, even though the precursors for the biosynthesis of JA may exist in Chlamydomonas (34) and are known to be induced by salt stress. Though these inferences need to be tested, we strongly recommend researchers perform a second set of data mining and gene homology analysis elsewhere. Depending on the version and quality of the species-specific genome annotation and the gene sets acquired from the source archive (Table 1) as well as the applied stringency of the gene homology prediction methods (Supplementary file-1), data may show variation in the projection of homologous events.
DATA INTEGRATION, CURATION AND DATABASE DEVELOPMENT
The Plant Reactome was initiated in 2012 using rice as a reference species. All the curation is accomplished using manual and semi-automated methods. We integrate curated data in the central pathway curation database maintained by the human Reactome project (2). Thus we share the same small molecule data archive, data models, review process, database servers, curation and database production tools. The reference rice pathways were first created by adopting metabolic pathways from RiceCyc (3), which were then imported in bulk into the central pathway curation database in the BioPAX level-2 format using import tools provided by the Reactome project. Subsequently, various other types of pathways involved in hormone signaling, development, transport, and stress response were curated by mining the published literature and using the Reactome Curator Tool. At time of release, the reference pathways checked into the central pathway database and marked for release are sliced out. These rice reference pathways serve as templates for computationally projecting systems-level pathway networks for 62 other species by identifying corresponding homologs of rice genes (Table 1) using the method described in the Supplementary File-1.
It is noteworthy that projected pathways based on the transcriptomes and some sequenced plant genomes are not available in any of the online annotated genomics resources including the Gramene Ensembl, Phytozome and NCBI databases (Table 1). For example, annotated leaf transcriptomes of wild Oryza species (provided by the OMAP project) (35) and the Kasalath rice genome (36) a representative of O. sativa AUS clade (Table 1), were used for deducing the projected pathways in the corresponding species using our in-house InParanoid clustered analysis (29–31). We managed to project pathways for these representative species because the Reactome data model allows inclusion of sequenced entities from a sequenced transcriptome, genome or for that matter a proteome. We included all the transcript isoforms from the assembled transcriptomes and did not make an effort to find the canonical longest isoform or the unigene. Similarly, polyploid gene annotations were not excluded and thus researchers may see an increased number of associated genes or transcript isoforms (Table 1) like the associated gene family members and paralogs for diploid species. Species such as O. sativa Aus Kasalath show lower projection counts, perhaps due to the lower coverage of the sequenced genome (36) or artifacts of the genome annotation quality. However, lower pathway counts for non-Angiosperm species are reflective of the evolutionary tree of life for plants.
DATA DOWNLOAD AND WEBSERVICES
We facilitate download of various data sets associated with the plant pathways (Figure 2). Users can download pathway data in community standard formats such as SBML, BioPAX level-2 and level-3, as well as in the Protégé OWL, PDF and Word formats. Users can also access a hierarchical list of pathways and reactions in a particular species, and download a list of small molecules and gene products participating in a particular pathway. Full downloads of the latest released database (in MySQL) are also available, along with tab-delimited files containing (i) all curated and projected gene product identifiers grouped by pathway and species, and (ii) all projected gene products grouped by pathway and reaction and indexed to curated Oryza sativa gene products (Supplementary Figure S1). These dataset are helpful if an external source is interested in establishing reciprocal links to the Plant Reactome entities as well as using the embedded identifiers for building Application Programming Interface (API)-based queries.
Application developers and advanced users may access Plant Reactome data via its API deployed with each release. This API is built on the work of the human Reactome project. It is used internally to provide data for the web application and is also provided externally as a service to the community. Many methods can be called over the HTTP protocol from a web browser, or directly from the command line; others can be invoked using client-side Javascript. Documentation for the API is available in the Reactome Developer Guide. Follow the same instructions for forming URLs, but replace the domain name ‘reactome.org’ suggested in the Reactome documentation with ‘plantreactome.gramene.org’, and then invoke the RESTful web service methods. For details consult Supplementary file-2.
OUTREACH AND TUTORIALS
To assist the plant research community in exploring resources and tools available at Plant Reactome, we regularly organize online webinars. Using case studies from a variety of crops and model plant species, we show how to use the available resources, data, and tools for plant comparative genomics and pathway analysis. The recorded webinars also serve as video tutorials and can be found on Gramene's YouTube channel (https://goo.gl/qQ2Pjn).
CONCLUSION
The Plant Reactome is a network of plant proteins linked to their molecular functions and interactors in the context of their locations within the plant cell. It serves as an archive of plant pathways (with all associated components) as well as a conceptual framework that can facilitate the analysis of genome-scale expression data. Thus, it enables researchers to discover previously unknown interactions that drive a plant's essential developmental genetic program and/or mitigate stress signals from the environment. The homology-based projections cover a broad taxonomic range within the plant kingdom including models and crop species from monocots and dicots represented by various grasses, fruits, legumes and bioenergy feedstock model crops, namely Brachypodium, Setaria, Sorghum, Poplar and Jatropha, as well as the basal angiosperm Amborella trichopoda. The database extends its pathway projections for non-Angiosperm species represented by gymnosperms Pinus taeda (Loblolly pine) and Pinus abies (Norway spruce), lycopod Selaginella moellendorffii, moss Physcomitrella patens, green algae Chlamydomonas reinhardtii and Ostreococcus lucimarinus and red algae Cyanidioschyzon merolae, which are included for their evolutionary importance (Table 1). Therefore, the Plant Reactome is a unique resource and provider of pathway collection and analysis tools.
Currently, the curation of reference pathways is ongoing, and we continue to develop projections for more plant species with each database release. We update our database 3–5 times per year, and post the current statistics and notes on new or updated data, tools and website content at http://plantreactome.gramene.org/pages/content/release-summary. Our plans include adopting the Reactome 3.5 or later platform, staying synchronized with future Reactome code releases, projections for new plant species, and adding new pathways for response to biotic and abiotic stresses, development, transport, and metabolism.
We encourage feedback from the community of plant genomic researchers about the Plant Reactome data, our website, downloads, online webinars and recorded tutorials. We are especially looking for help in reviewing our curated pathways and identifying new plant species with genome and transcriptome data for which plant biology researchers would like to have pathway projections.
Acknowledgments
We would like to thank Cold Spring Harbor Laboratory's Gramene project members Dr. Marcela Tello-Ruiz, Joe Mulvaney, Andrew Olson and Kapeel Chougle for coordinating the updates to the Gramene database searches and its Ensembl Plant genome browser and the Dolan DNA Learning Center for help with the organization of Plant Reactome webinars. We also acknowledge Ontario Institute for Cancer Research for providing access to the Reactome central curation database and hosting our development and live infrastructure support. Christopher Sullivan from the Center for Genome Research and Biocomputing at Oregon State University provided timely support for local infrastructure needs. We also thank Robin Haw, Sheldon McKay and David Croft from the Reactome project for providing timely technical assistance on Plant Reactome development. The pathway curation help provided by Oregon State University undergraduate students Dylan Beorchia, Teague Green, Kindra Amoss and Christina Partipilo is greatly appreciated. The authors are grateful to our users, researchers and numerous collaborators for sharing datasets generated in their projects and for valuable suggestions and feedback on improving the overall quality of the database.
Authors contributions: P.J., D.W, P.D., L.S. envisioned and proposed the project. P.J. leads the Plant Reactome project as Co-PI of the Gramene Project led by the P.I. D.W. P.J., S.N., P.G., V.A., P.D.D. curated the pathways, reviewed by P.D. P.D., chief editor of the Human Reactome, also provided training for Plant Reactome curators. J.P. is the lead Bioinformatics software and database developer for the project. G.M. carried out the initial RiceCyc import and provided timely support on curation fixes, data standards, and data model. J.W. and A.F. provided the timely support on running the Plant Reactome functionalities and database build. L.S. provided the infrastructure support from OICR and human Reactome project. M.K., A.M.P.F., R.P. curated the Gene Expression Atlas data and provided programmatic access to the data visualization widget. J.E. provided local infrastructure support and contributed the InParanoid gene homology dataset. The manuscript was written by S.N., P.G., J.P., P.D., J.E. and P.J. and reviewed by everyone. Funding agencies had no role in the study design, data analysis, or preparation of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Gramene database Award [NSF IOS-1127112] supported the project with in-kind infrastructure and intellectual support from the human Reactome database project [NIH: P41 HG003751, ENFIN LSHG-CT-2005-518254, Ontario Research Fund, and EBI Industry Programme); The Planteome Project [NSF IOS:1340112] provided InParanoid gene homology dataset. Funding for open access charge: The Gramene database Award [NSF IOS-1127112].
Conflict of interest statement. None declared.
REFERENCES
- 1.Tello-Ruiz M.K., Stein J., Wei S., Youens-Clark K., Jaiswal P., Ware D. Gramene: a resource for comparative analysis of plants genomes and pathways. Methods Mol. Biol. 2016;1374:141–163. doi: 10.1007/978-1-4939-3167-5_7. [DOI] [PubMed] [Google Scholar]
- 2.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dharmawardhana P., Ren L., Amarasinghe V., Monaco M., Thomason J., Ravenscroft D., McCouch S., Ware D., Jaiswal P. A genome scale metabolic network for rice and accompanying analysis of tryptophan, auxin and serotonin biosynthesis regulation under biotic stress. Rice (N. Y.) 2013;6:15. doi: 10.1186/1939-8433-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dash S., Cannon E.K.S. Chapter 8 - PeanutBase and Other Bioinformatic Resources for Peanut A2 - Stalker, H. Thomas. In: Wilson R.F., editor. Peanuts. AOCS Press; 2016. pp. 241–252. [Google Scholar]
- 6.Wegrzyn J.L., Lee J.M., Tearse B.R., Neale D.B. TreeGenes: a forest tree genome database. Int. J. Plant Genomics. 2008:412875. doi: 10.1155/2008/412875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung S., Ficklin S.P., Lee T., Cheng C.-H., Blenda A., Zheng P., Yu J., Bombarely A., Cho I., Ru S., et al. The Genome Database for Rosaceae (GDR): year 10 update. Nucleic Acids Res. 2014;42:D1237–D1244. doi: 10.1093/nar/gkt1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H., et al. The Sol Genomics Network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 2015;43:D1036–D1041. doi: 10.1093/nar/gku1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andorf C.M., Cannon E.K., Portwood J.L. II, Gardiner J.M., Harper L.C., Schaeffer M.L., Braun B.L., Campbell D.A., Vinnakota A.G., Sribalusu V.V., et al. MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res. 2016;44:D1195–D1201. doi: 10.1093/nar/gkv1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M., et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnakumar V., Hanlon M.R., Contrino S., Ferlanti E.S., Karamycheva S., Kim M., Rosen B.D., Cheng C.Y., Moreira W., Mock S.A., et al. Araport: the Arabidopsis information portal. Nucleic Acids Res. 2015;43:D1003–D1009. doi: 10.1093/nar/gku1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dash S., Campbell J.D., Cannon E.K., Cleary A.M., Huang W., Kalberer S.R., Karingula V., Rice A.G., Singh J., Umale P.E., et al. Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Res. 2016;44:D1181–D1188. doi: 10.1093/nar/gkv1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper L., Walls R.L., Elser J., Gandolfo M.A., Stevenson D.W., Smith B., Preece J., Athreya B., Mungall C.J., Rensing S., et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gene Ontology, C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petryszak R., Keays M., Tang Y.A., Fonseca N.A., Barrera E., Burdett T., Füllgrabe A., Fuentes A.M., Jupp S., Koskinen S., et al. Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan J., Li H., Li S., Yao R., Deng H., Xie Q., Xie D. The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell. 2013;25:486–498. doi: 10.1105/tpc.112.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H.Y., Seo J.S., Cho J.H., Jung H., Kim J.K., Lee J.S., Rhee S., Do Choi Y. Oryza sativa COI homologues restore jasmonate signal transduction in Arabidopsis coi1-1 mutants. PLoS One. 2013;8:e52802. doi: 10.1371/journal.pone.0052802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye M., Luo S.M., Xie J.F., Li Y.F., Xu T., Liu Y., Song Y.Y., Zhu-Salzman K., Zeng R.S. silencing COI1 in rice increases susceptibility to chewing insects and impairs inducible defense. PLoS One. 2012;7:e36214. doi: 10.1371/journal.pone.0036214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S.H., Sakuraba Y., Lee T., Kim K.W., An G., Lee H.Y., Paek N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015;57:562–576. doi: 10.1111/jipb.12276. [DOI] [PubMed] [Google Scholar]
- 23.Li C., Zhao J., Jiang H., Wu X., Sun J., Zhang C., Wang X., Lou Y., Li C. The wound response mutant suppressor of prosystemin-mediated responses6 (spr6) is a weak allele of the tomato homolog of CORONATINE-INSENSITIVE1 (COI1) Plant Cell Physiol. 2006;47:653–663. doi: 10.1093/pcp/pcj034. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Dai L., Jiang Z., Peng W., Zhang L., Wang G., Xie D. GmCOI1, a soybean F-box protein gene, shows ability to mediate jasmonate-regulated plant defense and fertility in Arabidopsis. Mol. Plant Microbe Interact. 2005;18:1285–1295. doi: 10.1094/MPMI-18-1285. [DOI] [PubMed] [Google Scholar]
- 26.Harb A., Krishnan A., Ambavaram M.M., Pereira A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010;154:1254–1271. doi: 10.1104/pp.110.161752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munemasa S., Oda K., Watanabe-Sugimoto M., Nakamura Y., Shimoishi Y., Murata Y. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 2007;143:1398–1407. doi: 10.1104/pp.106.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myburg A.A., Grattapaglia D., Tuskan G.A., Hellsten U., Hayes R.D., Grimwood J., Jenkins J., Lindquist E., Tice H., Bauer D., et al. The genome of Eucalyptus grandis. Nature. 2014;510:356–362. doi: 10.1038/nature13308. [DOI] [PubMed] [Google Scholar]
- 30.Shulaev V., Sargent D.J., Crowhurst R.N., Mockler T.C., Folkerts O., Delcher A.L., Jaiswal P., Mockaitis K., Liston A., Mane S.P., et al. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonnhammer E.L., Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–D239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M. KEGG Bioinformatics Resource for Plant Genomics and Metabolomics. Methods Mol. Biol. 2016;1374:55–70. doi: 10.1007/978-1-4939-3167-5_3. [DOI] [PubMed] [Google Scholar]
- 33.Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A., et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arisz S.A., Munnik T. The salt stress-induced LPA response in Chlamydomonas is produced via PLA2 hydrolysis of DGK-generated phosphatidic acid. J. Lipid Res. 2011;52:2012–2020. doi: 10.1194/jlr.M016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wing R.A., Ammiraju J.S., Luo M., Kim H., Yu Y., Kudrna D., Goicoechea J.L., Wang W., Nelson W., Rao K., et al. The oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- 36.Sakai H., Kanamori H., Arai-Kichise Y., Shibata-Hatta M., Ebana K., Oono Y., Kurita K., Fujisawa H., Katagiri S., Mukai Y., et al. Construction of pseudomolecule sequences of the aus rice cultivar Kasalath for comparative genomics of Asian cultivated rice. DNA Res. 2014;21:397–405. doi: 10.1093/dnares/dsu006. [DOI] [PMC free article] [PubMed] [Google Scholar]