Abstract
A system-wide understanding of cellular function requires knowledge of all functional interactions between the expressed proteins. The STRING database aims to collect and integrate this information, by consolidating known and predicted protein–protein association data for a large number of organisms. The associations in STRING include direct (physical) interactions, as well as indirect (functional) interactions, as long as both are specific and biologically meaningful. Apart from collecting and reassessing available experimental data on protein–protein interactions, and importing known pathways and protein complexes from curated databases, interaction predictions are derived from the following sources: (i) systematic co-expression analysis, (ii) detection of shared selective signals across genomes, (iii) automated text-mining of the scientific literature and (iv) computational transfer of interaction knowledge between organisms based on gene orthology. In the latest version 10.5 of STRING, the biggest changes are concerned with data dissemination: the web frontend has been completely redesigned to reduce dependency on outdated browser technologies, and the database can now also be queried from inside the popular Cytoscape software framework. Further improvements include automated background analysis of user inputs for functional enrichments, and streamlined download options. The STRING resource is available online, at http://string-db.org/.
INTRODUCTION
The flow of information and energy through the cell proceeds along specific and evolved interfaces: across and between nucleotides, proteins, lipids, metabolites and other small molecules. Among these interfaces, those between proteins are arguably among the most important, being biochemically diverse and information-rich, and showing exquisite specificity (1–3). Apart from direct physical binding, proteins also have many other, indirect ways of cooperation and mutual regulation: they can influence each other's production and half-life transcriptionally and post-transcriptionally, exchange reaction products, participate in signal relay mechanisms, or jointly contribute toward specific organismal functions. Together, these direct and indirect interactions constitute ‘functional association’, a useful operational umbrella-term for specific and functionally productive interactions of any type (4–9).
Assembling all known and predicted protein functional associations for a given organism results in a protein network of genome-wide functional connectivity. These networks represent a crucial, intermediate level of information aggregation: they are placed between pathway databases at one extreme (which provide mechanistic detail but often have low coverage), and high-throughput experimental interaction discovery and ad hoc predictions at the other extreme (which have high coverage but usually also high levels of false positives). As such, protein networks are ideally suited to serve as scaffolds or filters for further data integration, for visualization and for molecular discovery. They are essential for modern life sciences: protein networks are used to increase discovery power for noisy data sets by ‘network smoothing’ (10,11), help define drug efficiency by network-based ‘drug-disease proximity measures’ (12), help to interpret the results of genome-wide association screens (13–17) and enable the discovery of new molecular players through the ‘guilt by association’ concept (18,19).
A number of databases and online resources are dedicated to protein networks, at various levels of abstraction and each with a somewhat different focus/scope. First, individual well-supported protein–protein interactions are curated manually from the published literature, through dedicated efforts by members of the IMEx consortium (20,21), but also as part of more general annotation workflows such as within the UniProt consortium (22). Second, a number of databases assemble larger, genome-wide protein networks that are nevertheless still restricted to experimentally observed interactions only; examples include BioGRID (23), HINT (24), iRefWeb (25) and APID (26). Lastly, resources such as STRING include indirect and predicted interactions on top, aiming for inclusiveness in scope and for maximal coverage. Apart from STRING, this latter group includes GeneMANIA (27), Integrated Multi-species Prediction (28), Integrated Interactions Database (29), HumanNet (17), FunCoup (30) and others. For this group of data resources, it is particularly important to provide interaction weights (such as quality scores or confidence estimates), to allow the users to prune down these inclusive networks, as needed.
Within the spectrum of the above resources, STRING aims to set itself apart in three ways: (i) comprehensiveness – it covers the largest number of organisms and uses the widest breadth of input sources, including automated text-mining and computational predictions, (ii) usability – in terms of an intuitive web interface, Cytoscape integration and programmatic access options, and (iii) quality control and traceability – each interaction is annotated with benchmarked confidence scores, separately per evidence type, and the underlying evidence can be tracked to its source. STRING has been maintained continuously since the year 2000, and has already been described in several publications (31–34). Below, we provide a brief overview of the main features, and describe recent technical developments.
DATABASE CONTENT
For each protein–protein association stored in STRING, a score is provided. These scores (i.e., the ‘edge weights’ in each network) represent confidence scores, and are scaled between zero and one. They indicate the estimated likelihood that a given interaction is biologically meaningful, specific and reproducible, given the supporting evidence. For each interaction, the supporting evidence is divided into one or more ‘evidence channels’, depending on the origin and type of the evidence. There are seven channels, and they are assembled, scored and benchmarked separately. In the network visualization on the web frontend, the evidence channels are usually delineated by edges of different color, and each of the channels can be disabled individually by the user, in case some types of evidence might not be considered suitable for a particular question that is being studied. Based on the seven channels, a combined and final confidence score is computed for each interaction, and it is this ‘combined score’ that is typically used as the final measure when building networks or when sorting and filtering interactions. For a given interaction, it is generally a good sign of support when not only the combined score is high, but when there is also more than one evidence channel contributing to the score. Furthermore, it is important to note that the interactions in STRING have gene-locus resolution only: we do not discriminate between different splice isoforms or post-translationally modified forms. Hence, the interacting units in STRING are actually the protein-coding gene loci (represented by their main, canonical protein isoform).
Briefly, the seven evidence channels in STRING are (i) The experiments channel: Here, evidence comes from actual experiments in the lab (including biochemical, biophysical, as well as genetic experiments). This channel is populated mainly from the primary interaction databases organized in the IMEx consortium, plus BioGRID. (ii) The database channel: In this channel, STRING collects evidence that has been asserted by a human expert curator; this information is imported from pathway databases. (iii) The textmining channel: Here, STRING searches for mentions of protein names in all PubMed abstracts, in an in-house collection of more than three million fulltext articles, and in other text collections (35,36). Pairs of proteins are given an association score when they are frequently mentioned together in the same paper, abstract or even sentence (relative to how often they are mentioned separately). This score is raised further when it has been possible to parse one or more sentences through Natural Language Processing, and a concept connecting the two proteins was encountered (such as ‘binding’ or ‘phosphorylation by’). (iv) The coexpression channel: For this channel, gene expression data originating from a variety of expression experiments are normalized, pruned and then correlated (34). Pairs of proteins that are consistently similar in their expression patterns, under a variety of conditions, will receive a high association score. In addition to large-scale microarray data, in version 10.5 of STRING, RNAseq expression data are now also processed; this results in the inclusion of 16 previously non-covered organisms into this channel. (v) The neighborhood channel: This channel, and the next two, are genome-based prediction channels, whose functionality is generally most relevant for Bacteria and Archaea. In the neighborhood channel, genes are given an association score where they are consistently observed in each other's genome neighborhood (such as in the case of conserved, co-transcribed ‘operons’). (vi) The fusion channel: Pairs of proteins are given an association score when there is at least one organism where their respective orthologs have fused into a single, protein-coding gene. Finally, (vii) The co-occurrence channel: In this channel, STRING evaluates the phylogenetic distribution of orthologs of all proteins in a given organism. If two proteins show a high similarity in this distribution, i.e. if their orthologs tend to be observed as ‘present’ or ‘absent’ in the same subsets of organisms, then an association score is assigned. For this channel, the details of the STRING implementation have recently been described, separately (37).
Apart from direct evidence collected in the seven evidence channels, another important contribution of interactions in STRING comes from the transfer of evidence from one organism to another. This so-called ‘interolog’ transfer (38,39) is based on the observation that orthologs of interacting proteins in one organism are often also interacting in another organism – this inference is the more confident the better the orthology relationships can be established. STRING relies on hierarchical orthology relations imported from the eggNOG database (40), and conducts an all-against-all transfer of interactions, benchmarked separately for each evidence channel. Transfers between closely related organisms are made more confidently, whereas the existence of paralogs (i.e., implied gene duplications) will lower the transfer score. Overall, the biggest benefit of the transfers can be seen for poorly studied organisms, where the fraction of interactions supported by transfers only can be as high as 99%. In contrast, in well-studied model organisms such as Escherichia coli, the corresponding fraction is below 20%.
USER INTERFACE
The protein networks stored in STRING can be accessed in a number of ways. Programmatic access is provided via a REST-API (41), via an R/Bioconductor package (34) and via a mechanism to add additional user-provided interactions, as well as protein-centric information, onto the website (‘data payload’) (32). Studies that require genome-wide networks can refer to the STRING download pages, where the complete interaction scores, as well as accessory information, are available (the downloads are free for academics; commercial users need a license for some of the files). As of version 10.5, the downloads can now be pruned down, prior to receiving the files, by organism (or by groups of organisms), which greatly facilitates subsequent data processing. The most important interface to STRING, however, remains the web frontend (Figure 1). In 2016, it has been completely redesigned from the ground up; this was done in order to remove dependencies on deprecated web technologies such as Adobe Flash. The new website allows easier and more intuitive browsing of the networks and the underlying evidence, and it is tightly integrated with the database backend to provide speedy responses. Users can make search results and gene sets persistent by logging in, and stable URLs are provided on each page to facilitate sharing of results.
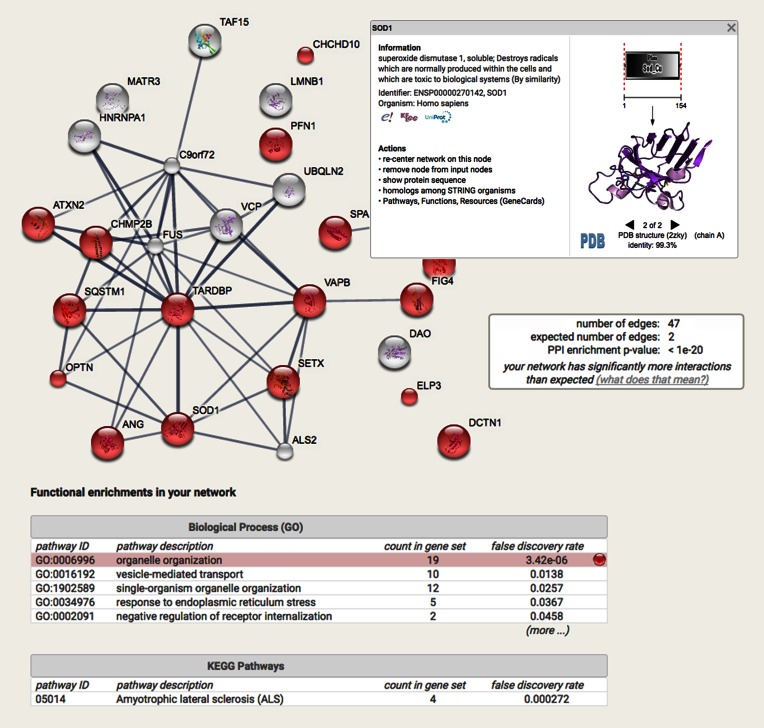
Figure 1.
Network and Enrichment Analysis. Combined screenshots from the STRING website, showing results obtained upon entering a set of 31 proteins suspected to be involved in Amyotrophic Lateral Sclerosis (55). The insets are showing (from top to bottom): the accessory information available for a single protein, a reported enrichment of functional connections among the set of proteins, and statistical enrichments detected in functional subsystems. In the bottom inset, one enriched function has been selected, and the corresponding protein nodes in the network are automatically highlighted in color.
Importantly, users are now—by default—provided with statistical analysis results for each network. The analysis is done server-side, in the background, so as not to slow down the user experience, and it produces alerts when a network is enriched in certain known functions, or has more interactions (edges) than expected. This is particularly meaningful when users arrive to the website with a set of proteins instead of just a single query protein, as it provides a functional characterization of the set (this feature is increasingly used by STRING users). The enrichment tests are done for a variety of classification systems (Gene Ontology, KEGG, Pfam and InterPro), and employ a Fisher's exact test followed by a correction for multiple testing (42,43).
CYTOSCAPE APP INTEGRATION
The web interface of STRING is designed primarily for users interested in small- to medium-scale networks, whereas the API, R package and download files are mainly intended for bioinformaticians who want to integrate STRING with other resources or perform large-scale network analyses. To bridge the gap between the two, we have developed a so-called App for the Cytoscape software framework (44,45), which allows users to easily retrieve, visualize and analyze networks of hundreds to thousands of proteins via a GUI.
The App allows users to query STRING in three different ways from within Cytoscape: by protein names, by disease or by PubMed query. The first of these mirrors the ‘Multiple proteins’ query in the STRING web interface and allows users to retrieve a network for a list of up to 2000 protein names or identifiers from, for example, a proteomics or transcriptomics study. The second option is to retrieve a network for a disease of interest; it first retrieves a list of the top-N human proteins associated with the disease from the DISEASES database (46) and subsequently loads the STRING network for these proteins into Cytoscape. The third option, PubMed query, allows users to retrieve a STRING network pertaining to any topic of interest based on text mining of PubMed abstracts. The app fetches the abstracts for a user-specified query via NCBI E-utilities, counts how many of these mention each protein from the organism of interest, ranks the proteins by comparing these counts to precomputed background counts over entire PubMed and retrieves a STRING network for the top-N proteins. The underlying text mining is performed by the software also used for the text-mining channel in STRING.
When a network is retrieved by the App, it comes associated with a large number of node attributes for each protein and edge attributes for each interaction, which can subsequently be used within Cytoscape. These include STRING and UniProt accessions to facilitate cross-linking with other resources, a human-readable name for display purposes and the protein sequence. If a protein was retrieved through a protein name query, we store also the exact query term with which the protein was found. This is helpful when querying for proteins identified in a proteomics or transcriptomics study, since it facilitates subsequent import of tabular data from the study (Figure 2). If available for the organism in question, the App also fetches information on the subcellular localization and tissue expression of each protein from the COMPARTMENTS (47) and TISSUES (48) databases as well as drug target information from Pharos (http://pharos.nih.gov/). For each interaction, the edge attributes include the overall confidence score and the subscores from each individual evidence channel.
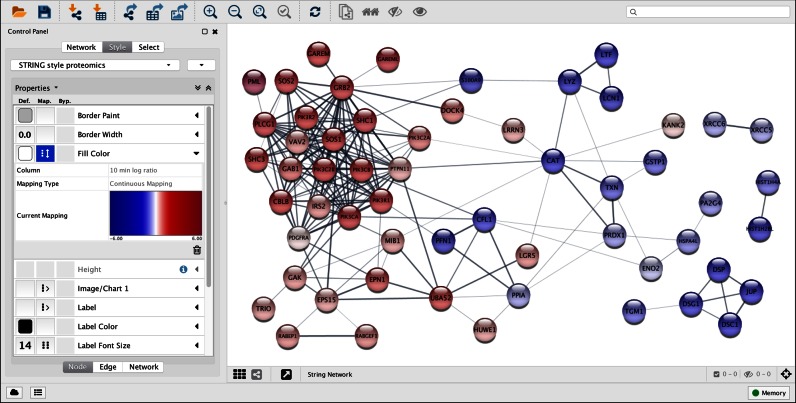
Figure 2.
STRING network visualization within Cytoscape. Using the Cytoscape STRING app, a network was retrieved for 78 proteins interacting with TrkA (tropomyosin-related kinase A) 10 min after stimulating neuroblastoma cells with NGF (nerve growth factor) (56). With a confidence cutoff of 0.4, the resulting network contains 182 functional associations between 57 of the proteins; the 21 proteins with no associations to other proteins in the network were removed. Nodes are colored according to the protein abundance (log ratio) compared to the cells before NGF treatment. The confidence score of each interaction is mapped to the edge thickness and opacity.
Cytoscape and its hundreds of apps provide numerous ways for users to interact with, visualize and analyze STRING networks (49), including integrating additional data from public repositories or their own experiments, changing visual styles and applying algorithms for network layout, clustering (50), enrichment analysis (51,52) and network analysis (53). In addition to these, the STRING App allows users to modify an already retrieved network in three different ways. First, the confidence cutoff for the imported evidence channels can be increased or decreased, which in the latter case involves fetching additional interactions from STRING. Second, users can expand the network by a user-specified number of interactors that are most closely associated with all network nodes or a selected subset of them. Third, any number of additional nodes can be queried by name and added to the existing network. Furthermore, the App provides a results panel with links to related databases such as UniProt (22), GeneCards (54), Pharos, COMPARTMENTS, TISSUES and DISEASES.
OUTLOOK
The availability of completely sequenced genomes, and of protein–protein interaction data, continues to grow quickly. Hence, the data importing and processing for STRING will be further streamlined in order to accommodate this. The upcoming version 11 of STRING will cover more than 4000 organisms, and will contain pre-computed protein networks for all of them. We are also developing a separate and distinctive interface specifically for the investigation of virus-host protein–protein interactions, which will incorporate many of the evidence channels present in STRING. This specialized database will enable querying for a whole virus or for specific viral proteins and will superimpose the viral interaction network onto that of the host.
Furthermore, we plan to extend the analysis options for user-provided gene set input, addressing a frequently expressed user need. This will include the possibility to report statistical enrichments for ranked genes lists, even genome-wide rankings. Together with the up-to-date network information, this will allow users to extract the maximum functional information from their queries, for any organism of interest.
Acknowledgments
The authors are indebted to Yan P. Yuan (EMBL Heidelberg) for IT support, and to Dr. Thomas Rattei (University of Vienna) for producing and sharing systematic, all-against-all protein-protein similarity data.
FUNDING
Core funding for STRING comes from the Swiss Institute of Bioinformatics (Lausanne), the Novo Nordisk Foundation (Copenhagen, NNF14CC0001), and the European Molecular Biology Laboratory (EMBL Heidelberg). J.H.M. has been funded by NIHGMS grant P41-GM103311. Funding for Open Access charges: University of Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Aloy P., Russell R.B. Ten thousand interactions for the molecular biologist. Nat. Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- 2.Gao M., Skolnick J. Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22517–22522. doi: 10.1073/pnas.1012820107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garma L., Mukherjee S., Mitra P., Zhang Y. How many protein-protein interactions types exist in nature? PLoS One. 2012;7:e38913. doi: 10.1371/journal.pone.0038913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright A.J., Ouzounis C.A. Functional associations of proteins in entire genomes by means of exhaustive detection of gene fusions. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snel B., Bork P., Huynen M.A. The identification of functional modules from the genomic association of genes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rives A.W., Galitski T. Modular organization of cellular networks. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1128–1133. doi: 10.1073/pnas.0237338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Las Rivas J, de Luis A. Interactome data and databases: different types of protein interaction. Comp. Funct. Genomics. 2004;5:173–178. doi: 10.1002/cfg.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannenfelser R., Clark N.R., Ma'ayan A. Genes2FANs: connecting genes through functional association networks. BMC Bioinformatics. 2012;13:156–168. doi: 10.1186/1471-2105-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Studham M.E., Tjärnberg A., Nordling T.E., Nelander S., Sonnhammer E.L. Functional association networks as priors for gene regulatory network inference. Bioinformatics. 2014;30:i130–i138. doi: 10.1093/bioinformatics/btu285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cun Y., Frohlich H. Network and data integration for biomarker signature discovery via network smoothed T-statistics. PLoS One. 2013;8:e73074. doi: 10.1371/journal.pone.0073074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofree M., Shen J.P., Carter H., Gross A., Ideker T. Network-based stratification of tumor mutations. Nat. Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guney E., Menche J., Vidal M., Barábasi A.L. Network-based in silico drug efficacy screening. Nat. Commun. 2016;7:10331–10343. doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillenmeyer S., Davis L.K., Gamazon E.R., Cook E.H., Cox N.J., Altman R.B. STAMS: STRING-Assisted Module Search for Genome Wide Association Studies and Application to Autism. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw530. doi:10.1093/bioinformatics/btw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiserson M.D., Eldridge J.V., Ramachandran S., Raphael B.J. Network analysis of GWAS data. Curr. Opin. Genet. Dev. 2013;23:602–610. doi: 10.1016/j.gde.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia P., Zhao Z. Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum. Genet. 2014;133:125–138. doi: 10.1007/s00439-013-1377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tasan M., Musso G., Hao T., Vidal M., MacRae C.A., Roth F.P. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat. Methods. 2015;12:154–159. doi: 10.1038/nmeth.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee I., Blom U.M., Wang P.I., Shim J.E., Marcotte E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furlong L.I. Human diseases through the lens of network biology. Trends Genet. 2013;29:150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Tian W., Zhang L.V., Taşan M., Gibbons F.D., King O.D., Park J., Wunderlich Z., Cherry J.M., Roth F.P. Combining guilt-by-association and guilt-by-profiling to predict Saccharomyces cerevisiae gene function. Genome Biol. 2008;9(Suppl. 1):S7. doi: 10.1186/gb-2008-9-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orchard S., Kerrien S., Abbani S., Aranda B., Bhate J., Bidwell S., Bridge A., Briganti L., Brinkman F.S., Cesareni G., et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods. 2012;9:345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatr-Aryamontri A., Breitkreutz B.J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das J., Yu H. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst. Biol. 2012;6:92–103. doi: 10.1186/1752-0509-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner B., Razick S., Turinsky A.L., Vlasblom J., Crowdy E.K., Cho E., Morrison K., Donaldson I.M., Wodak S.J. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database (Oxford) 2010 doi: 10.1093/database/baq023. doi:10.1093/database/baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso-Lopez D., Gutiérrez M.A., Lopes K.P., Prieto C., Santamaría R., De Las Rivas J. APID interactomes: providing proteome-based interactomes with controlled quality for multiple species and derived networks. Nucleic Acids Res. 2016;44:W529–W535. doi: 10.1093/nar/gkw363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuberi K., Franz M., Rodriguez H., Montojo J., Lopes C.T., Bader G.D., Morris Q. GeneMANIA prediction server update. Nucleic Acids Res. 2013;41:W115–W122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong A.K., Krishnan A., Yao V., Tadych A., Troyanskaya O.G. IMP 2.0: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2015;43:W128–W133. doi: 10.1093/nar/gkv486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotlyar M., Pastrello C., Sheahan N., Jurisica I. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016;44:D536–D541. doi: 10.1093/nar/gkv1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt T., Ogris C., Sonnhammer E.L. FunCoup 3.0: database of genome-wide functional coupling networks. Nucleic Acids Res. 2014;42:D380–D388. doi: 10.1093/nar/gkt984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschini A., Lin J., von Mering C., Jensen L.J. SVD-phy: improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics. 2016;32:1085–1087. doi: 10.1093/bioinformatics/btv696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walhout A.J., Sordella R., Lu X., Hartley J.L., Temple G.F., Brasch M.A., Thierry-Mieg N., Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 39.Yu H., Luscombe N.M., Lu H.X., Zhu X., Xia Y., Han J.D., Bertin N., Chung S., Vidal M., Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M., et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivals I., Personnaz L., Taing L., Potier M.C. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics. 2007;23:401–407. doi: 10.1093/bioinformatics/btl633. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 44.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pletscher-Frankild S., Pallejà A., Tsafou K., Binder J.X., Jensen L.J. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015;74:83–89. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 47.Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O'Donoghue S.I., Schneider R., Jensen L.J. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014 doi: 10.1093/database/bau012. doi:10.1093/database/bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santos A., Tsafou K., Stolte C., Pletscher-Frankild S., O'Donoghue S.I., Jensen L.J. Comprehensive comparison of large-scale tissue expression datasets. PeerJ. 2015;3:e1054. doi: 10.7717/peerj.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Lotia S., Pico A.R., Bader G.D., Ideker T. A travel guide to Cytoscape plugins. Nat. Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris J.H., Apeltsin L., Newman A.M., Baumbach J., Wittkop T., Su G., Bader G.D., Ferrin T.E. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. 2011;12:436–449. doi: 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 52.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scardoni G., Tosadori G., Faizan M., Spoto F., Fabbri F., Laudanna C. Biological network analysis with CentiScaPe: centralities and experimental dataset integration. F1000Res. 2014;3:139–146. doi: 10.12688/f1000research.4477.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fishilevich S., Zimmerman S., Kohn A., Iny Stein T., Olender T., Kolker E., Safran M., Lancet D. Genic insights from integrated human proteomics in GeneCards. Database (Oxford) 2016 doi: 10.1093/database/baw030. doi:10.1093/database/baw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abel O., Powell J.F., Andersen P.M., Al-Chalabi A. ALSoD: A user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012;33:1345–1351. doi: 10.1002/humu.22157. [DOI] [PubMed] [Google Scholar]
- 56.Emdal K.B., Pedersen A.K., Bekker-Jensen D.B., Tsafou K.P., Horn H., Lindner S., Schulte J.H., Eggert A., Jensen L.J., Francavilla C., et al. Temporal proteomics of NGF-TrkA signaling identifies an inhibitory role for the E3 ligase Cbl-b in neuroblastoma cell differentiation. Sci. Signal. 2015;8:ra40. doi: 10.1126/scisignal.2005769. [DOI] [PubMed] [Google Scholar]