Abstract
Bioinformatics and computational biology play a critical role in bioscience and biomedical research. As researchers design their experimental projects, one major challenge is to find the most relevant bioinformatics toolkits that will lead to new knowledge discovery from their data. The Bio-TDS (Bioscience Query Tool Discovery Systems, http://biotds.org/) has been developed to assist researchers in retrieving the most applicable analytic tools by allowing them to formulate their questions as free text. The Bio-TDS is a flexible retrieval system that affords users from multiple bioscience domains (e.g. genomic, proteomic, bio-imaging) the ability to query over 15 000 analytic tool descriptions integrated from well-established, community repositories. One of the primary components of the Bio-TDS is the ontology and natural language processing workflow for annotation, curation, query processing, and evaluation. The Bio-TDS’s scientific impact was evaluated using sample questions posed by researchers retrieved from Biostars, a site focusing on biological data analysis. The Bio-TDS was compared to five similar bioscience analytic tool retrieval systems with the Bio-TDS outperforming the others in terms of relevance and completeness. The Bio-TDS offers researchers the capacity to associate their bioscience question with the most relevant computational toolsets required for the data analysis in their knowledge discovery process.
INTRODUCTION
Numerous bioinformatics tool repositories or retrieval systems have been developed (1,2), but do not provide natural language processing functionality allowing users to enter their queries as free text. This problem has accelerated the growth of community based discussion platforms such as SEQAnswer Wiki (http://SEQAnswers.com/wiki/SEQAnswers), Biostars (https://www.biostars.org/) and ARAPORT (https://www.araport.org/) (3–5). The development of Natural Language Processing (NLP) applications in bioscience (6) offers an opportunity to combine ontologies (i.e. user-oriented domain information representation) (7) and free text NLP methods to minimize the gap between users and bioscience information retrieval systems. There are several examples of use of NLP for information retrieval using specified data sources, for example DNORM (8), PathNER (9) and PhenX (8–11), but there is no existing system leveraging NLP plus ontologies for general queries across biological domains.
Domain independent design to achieve broad, integrated, community usage
A bioinformatics tool is frequently used to address multiple bioscience domain needs or questions (12) even though it is initially developed for specific scientific domains such as computer science, biology, applied mathematics, plant science, microbiology, ecology, biomedical engineering, bioimaging etc. (13). The software design process used for the creation of an analytic tool generally follows the fundamental principles of the system development life cycle with the user requirements expressed using specific domain terms (14–16). Then, useful tools are applied in new scientific domains and expanded as data collection technologies spread. For example, TopHat was developed for splice junction mapping for RNA-Seq reads in mammalian-sized genomes, using bowtie aligner (17). TopHat is broadly used today in different domains and serves as a reference for new ‘read mapping’ algorithms (e.g. STAR or customized TopHat) (12,18,19). TopHat is leveraged by genome sequencing experts who know the read mapping domain vocabulary very well (e.g. genomicists, geneticists, proteomic experts, systems biologists) as well as by non-experts who just need the best, relevant, ready-to-use tool for a specific problem (e.g. biology, microbiology, plant science, health science). The query expressions posed by these potential users vary from precise questions such as ‘RNASeq read mapper for mammalian’ or ‘read mapping’, to very domain specific free text expressions like ‘best tool for microbiology genome mapping using RNASeq’ (https://liorpachter.wordpress.com/2015/11/01/what-is-a-read-mapping/).
Retrieval systems flexibility challenge: customizable open architecture across usage level
Bioinformatics is a multidisciplinary field comprising many scientific domains and sub-domains. The definition of Bioinformatics is shifting, with some communities expanding the definition to include data analytics. Computation has become an essential tool in life science research but the level of researchers’ expertise varies greatly. This diversity requires an information retrieval system to be flexible and customizable in both design (architecture) and accessibility (interoperability) (20). Common, existing systems offer web-based interfaces at the end-user level (24) and provide Application Programming Interfaces (API) (e.g. RESTful API) to allow client applications access to underlying functionality. This architecture has been adopted as the most effective approach for programmatic access following the SaaS (Software as a Service) logic (1,21). The primary challenge is to ensure the involvement of each bioscience community in terms of domain and skill level in the system development process. Systems such as ELIXIR (1) endeavor to involve the research community in their repository population and curation processes but this results in requiring user query expressions to fit the system's common standards and thus demands a relatively high level of expertise from the users. This approach thus has weaknesses in both repository content completeness (i.e. not enough information to represent the entire user domain knowledge) and user query relevance (e.g. low precision for user query results). For example, ELIXIR has based its core annotation workflow on the EDAM ontology (22). EDAM focuses only on one bioinformatics domain information representation, and does not represent some key tool development information such as version or platform which are available in the Software Ontology (SWO) (23).
Bioscience query tools discovery system (Bio-TDS)
To address these limitations, the Bio-TDS has been developed to assist researchers from diverse domains and skill levels to find the most relevant computational tools for their data analysis. This system allows end-users to retrieve tools of interest by submitting either keyword-based queries and/or free text based questions. The Bio-TDS includes functionality to gather individual community tool definitions and a ‘pipe’ that integrates these individual tool descriptions into a centralized retrieval system. By integrating analytic tool definitions from multiple repositories and providing flexible querying options, the end-user needs are addressed from the beginning to the end of the retrieval system process. For example, one of the tools repositories that has been integrated into the Bio-TDS is SEQAnswer-Wiki SEQ, the most relevant tools repository for the High Throughput Sequencing (HTS) community (e.g. NGS, RNASeq, ChipSeq) (4,12).
MATERIALS AND METHODS
System design and implementation
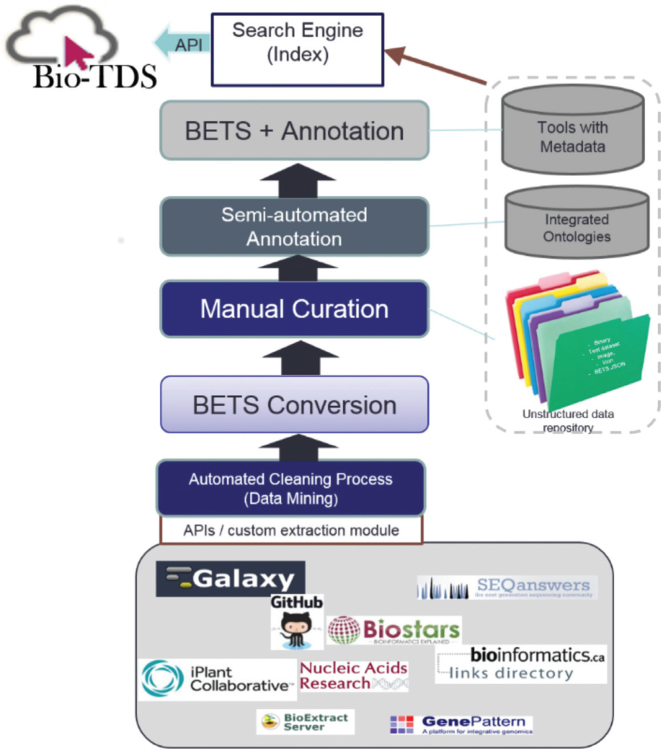
The Bio-TDS’s domain diversity has been integrated into the system through four flexible modules: (i) a tool's minimum information specification format called Bioinformatics Elaborated Tools Specifications (BETS) which provides a standard for analytic tool descriptions, (ii) a data extraction, integration and automatic curation module, (iii) an automatic ontology-based tools annotation module called TONER (Tools Ontology-based Name Entities Resolution) and (iv) an ontology and natural language driven query processing workflow for relevant tool retrieval (see Figure 1).
Figure 1.
Bio-TDS discovery system overview.
Bio-TDS module 1: tools specification and BETS construction
The Bio-TDS gathers the bioinformatics tool definitions from well-established, disparate, community-based repositories and creates a BETS document for each tool (Table 2: Supplementary Table S1). The primary purpose of BETS is to provide standard, robust representations of bioinformatics analytic tools. The BETS descriptions currently include augmented specifications from Galaxy (http://galaxy.org/), CyVerse (formerly named the iPlant Collaborative) (http://www.cyverse.org/), Bioinformatics Link Directory (BLD) (http://bioinformatics.ca/links_directory/) which contains the Nucleic Acid Research Tool List, SEQAnswers (http://SEQAnswers.com/) and Bio-Soft Net (http://en.bio-soft.net/) (24–27). The module's BETS converter, implemented in Java, allows users to convert any of the tool specification from the five listed repositories into the BETS standard in JSON (http://json.org/) format and/or revert them back into the original source repository format. This integration helps the system to maintain the community specifications and provides a lossless data extraction from these source repositories.
Table 2.
Supporting materials available at http://biotds.org/help/supporting.xhtml
| S1 | BETS Specification description and manipulation |
| S2 | Resources extraction and semi-automatics curation |
| S3 | TONER: Tools ontology-based annotation |
| S4 | BioQueryTool query processing workflow and programmatic access |
| S5 | BioQueryTool Evaluation and comparison |
‘NS’ value in a given evaluation criteria (Precision, Recall,…) indicates limited data point (missing >40% data points compare the variable dataset size) to compute an accurate meaningful criteria value. This is due to a low retrieval rate in the related repository (e.g. no result return for the query).
Bio-TDS module 2: data extraction and Bio-TDS repository population
The Bio-TDS repository currently contains over 15 000 analytic tool descriptions extracted from five repositories including Galaxy, CyVerse, Bioinformatics Link Directory, SEQAnswers and BioSoft Net. The Bio-TDS repository includes structured data stored using the open source database MySQL (https://www.mysql.com/), unstructured data stored in a files system, and indexed data using Apache Lucene (https://lucene.apache.org/core/). The MySQL relational database stores the most commonly used attributes such as ‘Name, version, description’. The unstructured data file system consists of the BETS specification in JSON format, the tool annotation data, and any images associated with a tool such as the tool icon. The analytic tool BETS documents are indexed in the Bio-TDS using the Apache Lucene indexing engine.
The process used to retrieve tool descriptions from the community repositories is dependent on each individual repository architecture. Some repositories offer access functionality through an API making tool description retrieval straightforward. Others do not provide any programmatic access and therefore require the development of custom Java scraping modules. Before an analytic tool definition is added to the Bio-TDS, it is processed through a tools checker to ensure BETS-compatibility and initial validation (Table 2: Supplementary Table S2). Once the tools pass the checker, they are moved to a BETS-converter. Each tool is curated, annotated, and stored in the Bio-TDS repository making it ready for additional annotation through our TONER module.
Bio-TDS module 3: tools ontology-based annotation: TONER
To enable accurate searching of the Bio-TDS repository, it is important to have meaningful annotations describing the tools and their features. The Bio-TDS includes an automated annotator called TONER (Tools Name Entity Resolution). TONER uses the National Center for Biomedical Ontology (NCBO) (http://www.bioontology.org/) online annotation service to provide suggested concept annotations from selected domain ontologies (28). For example, the TopHat tool description is tagged with the ‘mapping’ and ‘Linux’ concepts from the EDAM and SWO ontologies respectively. This indicates that TopHat is a tool for sequence mapping operations and is compatible with the Linux operating system. The current Bio-TDS version includes three ontologies to enable robust annotations and searching capabilities. These ontologies are: EDAM, an ontology of concepts that are prevalent within bioinformatics, including types of data, data identifiers, data formats, operations and topics (http://edamontology.org/) (22); SWO, a resource for describing analytic tools, their types, tasks, versions, data requirements and provenance (http://theswo.sourceforge.net/) (23); and NGSOnto, an ontology to describe workflows from DNA extraction to contigs in a Whole Genome Sequence experiment (http://darwin.phyloviz.net/∼msilva/NGSonto/). Attribute values within each BETS document are queried against these ontologies and the located terms are used to annotate the analytic tool.
The analytic tool descriptions (i.e. metadata) integrated into the Bio-TDS are stored in JSON format using the BETS standard. TONER processes this metadata to isolate values. If necessary, TONER splits values to provide a set of coherent strings that preserve meaningful cohesive information. These strings (also called mentions) are passed to the NCBO Annotator service, which provides a list of concept annotations for each string (fuzzy or exact-match). These annotation results and their associated domain ontology URIs are stored with the BETS document, thus adding more meaning to the tool description (Table 2: Supplementary Table S3). TONER offers an automatic process for bioinformatics tools annotation (29). TONER was evaluated by selecting 50 tools and manually checking the automatic annotation error rate. For example, the TONER annotated a specific tool with 25 ontology terms out of the 7396 terms (EDAM = 3240, SWO = 4067, NGSONTO = 89), with no term being miss-annotated, when we used 100% similarity.
Bio-TDS repository curation
In data integration, ‘perfect data’ is not practical as the corpus of information is continually updated and the lag in curation is long. The Bio-TDS curation process aims to achieve a ‘good enough’ data set (20,30) defined to be the data that allows Bio-TDS query processing to provide a relatively complete (precision) and relevant (recall) result tool set from a user query.
The Bio-TDS extraction module initially gathered a list of 16 293 tools corresponding to 10 975 from Bio-Soft Net, 2300 from BLD, 690 from SEQAnswers, 94 from CyVerse (iPlant) and 2234 from the Galaxy Tool Shed. The initial raw dataset contained duplicates, missing information, inaccurate tool descriptions, and miss-assigned attribute values. Regardless, the list of 15 989 unique tools from just five of the community repositories reveals the large number of analytic tools available in computational bioscience. With this number, it would be hard, inefficient and very expensive to consider only a manual curation approach.
A rule-based (or predicate) semi-automatic curation process has been developed for the Bio-TDS by combining human inspection and data mining methods (31,32). At the current development stage, the Bio-TDS team has already identified 10 key rules. The application of this rule set has helped to improve the repository accuracy by removing duplicates and invalid tools (from 20 000 tools to 15 000), removing inaccurate attributes values, and filling in missing information. This has achieved an overall improvement rate of ∼30%. (Table 2: Supplementary Table S2).
Ontology and natural language processing driven query processing workflow
When users query the Bio-TDS, their queries are tokenized and processed against the NCBO BioPortal (http://bioportal.bioontology.org/) (33) to extract ontology terms along with their URIs in order to enrich the query token set. NLP is applied to the query (e.g. stemming, tagging, English synonym mapping) (6) for further augmentation. The annotated query is then submitted to the Bio-TDS search engine leveraging the indexed, annotated BETS documents to retrieve the related tools ranked by relevance.
Test set design for bioscience tools repository evaluation
A test set was developed to map user queries to relevant tools. This set is a first step in developing a Gold Standard for analytic tool repository assessment. The current version of the test set consists of 229 user questions related to tool usage. These questions were extracted from Biostars and manually curated with tagging keywords (Table 2: Supplementary Table S5). Retrieval rate (RR), Precision (P), recall (R) and F-Measure (F) were calculated as P = TP/(TP + FP), R = TP/(TP+FN) and F = 2P*R/(P+R); where TP, FP and FN represent True Positive, False Positive, and False Negative in a 2×2 contingency matrix. For example, let's consider the user query Q1: ‘What Bioinformatics Methods Have Been Performed To Study DNA Methylation Data?’ BISMARK is expected to be a relevant tool (34). This query submitted to Bio-TDS returns 2,495 results with BISMARK ranked eighth. For each tool, we calculate the average of the retrieval rate (RR), Precision (P); recall (R) and F-Measure (F) among the queries (35). The average was then used to calculate the mean average of each repository, which led to the MRR (Mean Retrieval Rate), MAP (Mean Average Precision), MAR (Mean Average Recall) and Mean Average F-Measure (Table 1).
Table 1.
Bio-TDS evaluation and comparison overview
| Criteriaa | Bio-TDS | BLD | ELIXIR | GALAXY | SeqAnswer |
|---|---|---|---|---|---|
| MRR + | 1 | 0.0131 | 0.0087 | 0.0043 | 0.1484 |
| MAP + | 0.0004 | NS | NS | NS | NS |
| MAR + | 0.8755 | 0.0000 | 0.0000 | 0.0036 | 0.0339 |
| MAF + | 0.0008 | NS | NS | NS | NS |
| MRR ++ | 0.7598 | NS | 0.1441 | 0.1310 | 0.6899 |
| MAP ++ | 0.0427 | NS | NS | NS | 0.0572 |
| MAR ++ | 0.3474 | 0.0200 | 0.0696 | 0.0518 | 0.2327 |
| MAF ++ | 0.0801 | NS | NS | NS | 0.1383 |
a Evaluation Criteria: MRR = Mean Retrieval Rate; MAP = Mean Average Precision; MAR = recall; MAF = mean Average F-measure. NS: not significant result. User Query Type: +Free text Query; ++Keyword Query.
RESULTS
Flexible one-stop shop (integrated) and domain-ontology annotated bioscience tools repository
The Bio-TDS has integrated over 15 000 bioscience tools from five large community-based repositories. To provide more information, an extensive domain ontology annotation system and NLP were added to the system to enhance tool annotation. The results are that the Bio-TDS retrieves the most relevant tool description list based on user queries with minimal error (∼0.01) as compared to other like systems.
Programmatic access of Bio-TDS resources using RESTful API
To assist users with programmatic querying needs, a RESTful API module was developed to access Bio-TDS resources through URI end-points. This RESTful module allows developers to leverage Bio-TDS functionality for other bioinformatics needs. (Table 2: Supplementary Table S4).
Predicting the most relevant tools list from user questions
Frequently, users are not aware of all the existing analytic tools that are available to be applied to a specific problem. Through the Bio-TDS Web-based user interface (http://biotds.org/), users are able to enter their questions either as free text or in keyword format to obtain the list of most relevant tools. The NLP functionality integrated into the system maps the user query to the most relevant tool sets. These NLP operations allowed for the integration of an English dictionary and ontology semantic features such as synonym management to improve the completeness of user result. When a user types ‘sequence analysis’ and later ‘analyzing sequences’, the system returns very similar results. Similarly, when a user enters ‘read mapper’ or ‘mapping reads’, RNASeq mapping tools such as ‘mom’ would be include in the result set with similar precision and rank.
Web-based tools discovery and visualization
The Bio-TDS Web-based Client provides multiple views and retrieval options for the user (Table 2, Supplementary Table S4). Users are able to view query result sets retrieved from the discovery system as a list of ranked tools or in table format. Without a specific question, users can also browse the repository by domain (e.g. biochemistry, medicine), method (e.g. sequencing, imaging, visualization) or data format (e.g. SAM, image). Users can click on the tool name within a result set to view more details related to the tool. These details included information such as the association to the provenance repository, published reference paper when applicable and additional tool attributes. Users can also click to inspect the entire BETS specification of the selected tool.
Bio-TDS evaluation and comparison
The Bio-TDS query-processing module was evaluated and compared to similar systems using the retrieval rate, precision, recall and F-measure (35). The evaluation was based on a 229 user-query test set relating to 25 analytic tools available in each repository. The test set was manually created and was based on user postings from Biostars.
The Bio-TDS was evaluated by checking its ability to retrieve the user's expected tool list (i.e. completeness or recall), and its ability to sort the expected tools by the best rank (i.e. exactness or precision) (35,36). Bio-TDS always retrieves the user's expected tools (recall ∼1) with competitive exactness (precision ∼0.1). This performance is based on both free text and keyword queries. Bio-TDS was compared with other existing, like tool repository systems including the recently published ELIXIR (1). Bio-TDS outperformed these systems in precision, recall and F-measure (Table 2: Supplementary Table S5).
DISCUSSION AND CONCLUSION
Many scientists now realize that the discovery of analytic methods and the justification of the use of such methods are important, limiting factors in the data analyses and publication of research results (13,20). By integrating user thinking into the retrieval process, the Bio-TDS provides a framework to assist researchers in identifying the most relevant analytic tools to assist them in their data analysis. The Bio-TDS is open source, customizable, and provides users from multiple bioscience domains (e.g. genomic, proteomic, bioimaging) the ability to query over 15 000 analytic tools integrated from five very well established user community repositories. One of the primary components of the Bio-TDS is the ontology and natural language processing workflow for annotation, curation, query processing and evaluation. When tested with an extensive test set, the semi-automatic curation workflow demonstrates a high level of accuracy. The Bio-TDS was compared to existing analytic tool retrieval systems including the ELIXIR registry (1), Galaxy ToolShed (24), Bioinformatics Links Directory (27), SEQAnswers Wiki (4), and CyVerse (iPlant) (26). The advantages of the Bio-TDS are as follows: (i) the Bio-TDS allows users to discover relevant tools from their problem/question statement using our ontology and NLP integration, with no need to build a sophisticated query; (ii) the Bio-TDS contains more tools than any other repository to which it was compared; (iii) the Bio-TDS allows for easy integration with semi-automatic curation of new repositories achieved by creating a mapper for our BETS specification and BETS converter module; (iv) the Bio-TDS provides flexible tool annotation through the TONER module and, new ontologies can be integrated for specific applications or domains; (v) the Bio-TDS provides a test set of 229 user questions for bioscience tools repository assessment; (vi) the Bio-TDS outperformed the state-of-the-art similar systems against which it was compared in terms of retrieval rate, precision, recall and F-measure.
Overall, as bioscience tool development grows, the Bio-TDS provides users with functionality to discover relevant tools using a one-stop shop. The Bio-TDS allows users who want to explore analytic tool options to retrieve the most relevant toolkit list by submitting a question or project description as a free text description. Our expressive, cutting-edge Bio-TDS architecture makes the Bio-TDS flexible in terms of user functionality (e.g. Web interface, RESTful API programmatic access) and for user community expansion (e.g. integrating new repositories and ontologies).
FUTURE WORK
The Bio-TDS is a community need driven systems tested initially, in two bioscience research infrastructure networks. This first version constitutes a starting point for a long-term project intending to minimize the gap between bioscience analytic tools, research data, and knowledge discovery. There remains room for improvement to the Bio-TDS in order to move toward that goal. These improvements include tasks such as precision improvement (e.g. for ‘read mapper’ and ‘mapping read’ queries, the ‘mom’ tool is retrieve in both cases, but appear in different ranking positions—1st and 160th). In addition, the Bio-TDS repository will be enhanced by integrating more user domains and ontologies, such as the Bioconductor community (https://www.bioconductor.org/). The Bio-TDS test set will be expanded to contain more user questions and more tools per question. The ultimate long-term goal will be for the Bio-TDS to become a personalized tool and workflow recommending systems, which includes a customizable tools testing module for bioscience researchers.
AVAILABILITY
Bio-TDS is open source project freely available at (http://biotds.org/) under GNU Public License. The source code is available at https://bitbucket.org/USDBioinformatics/tds-v1-sources.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thanks Barb Goodman, Michael Gonzales and Ann Stapleton for their valuable feedback regarding our system.
FUNDING
National Science Foundation EAGER Award [IOS-1545596]; National Science Foundation/EPSCoR Grant [IIA-1355423]; State of South Dakota; Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health [P20GM103443]. Funding for open access charge: National Science Foundation EPSCoR Award [IIA-1355423]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and the National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ison J., Rapacki K., Ménager H., Kalaš M., Rydza E., Chmura P., Anthon C., Beard N., Berka K., Bolser D. et al. Tools and data services registry: a community effort to document bioinformatics resources. Nucleic Acids Res. 2016; 44:D38–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henry V.J., Bandrowski A.E., Pepin A.-S., Gonzalez B.J., Desfeux A.. OMICtools: an informative directory for multi-omic data analysis. Database. 2014; bau069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krishnakumar V., Hanlon M.R., Contrino S., Ferlanti E.S., Karamycheva S., Kim M., Rosen B.D., Cheng C.Y., Moreira W., Mock S.A. et al. Araport: the Arabidopsis Information Portal. Nucleic Acids Res. 2015; 43:D1003–D1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J.W., Robison K., Martin M., Sjödin A., Usadel B., Young M., Olivares E.C., Bolser D.M.. The SEQanswers wiki: A wiki database of tools for high-throughput sequencing analysis. Nucleic Acids Res. 2012; 40:D1313–D1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parnell L.D., Lindenbaum P., Shameer K., Dall’Olio G.M., Swan D.C., Jensen L.J., Cockell S.J., Pedersen B.S., Mangan M.E., Miller C.A. et al. BioStar: an online question & answer resource for the bioinformatics community. PLoS Comput. Biol. 2011; 7:e1002216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Comeau D.C., Doǧan R.I., Ciccarese P., Cohen K.B., Krallinger M., Leitner F., Lu Z., Peng Y., Rinaldi F., Torii M. et al. BioC: a minimalist approach to interoperability for biomedical text processing. Database. 2013; 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernández M., Cantador I., López V., Vallet D., Castells P., Motta E.. Semantically enhanced Information Retrieval: an ontology-based approach. J. Web Semant. 2011; 9:434–452. [Google Scholar]
- 8. Leaman R., Islamaj Dogan R., Lu Z.. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013; 29:2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu C., Schwartz J.-M., Nenadic G.. PathNER: a tool for systematic identification of biological pathway mentions in the literature. BMC Syst. Biol. 2013; 7(Suppl. 3):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zemojtel T., Köhler S., Mackenroth L., Jäger M., Hecht J., Krawitz P., Graul-Neumann L., Doelken S., Ehmke N., Spielmann M. et al. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci. Transl. Med. 2014; 6:252ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smedley D., Jacobsen J.O.B., Jäger M., Köhler S., Holtgrewe M., Schubach M., Siragusa E., Zemojtel T., Buske O.J., Washington N.L. et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc. 2015; 10:2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pabinger S., Dander A., Fischer M., Snajder R., Sperk M., Efremova M., Krabichler B., Speicher M.R., Zschocke J., Trajanoski Z.. A survey of tools for variant analysis of next-generation genome sequencing data. Brief. Bioinform. 2013; doi:10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dooley R., Vaughn M., Stanzione D., Terry S., Skidmore E.. Software-as-a-Service: The iPlant Foundation API. 5th IEEE Workshop on Many-Task Computing on Grids and Supercomputers. 2012. [Google Scholar]
- 14. Nagasaki M., Fujita A., Sekiya Y., Saito A., Ikeda E., Li C., Miyano S.. XiP: a computational environment to create, extend and share workflows. Bioinformatics. 2013; 29:137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lushbough C.M., Jennewein D.M., Brendel V.P.. The BioExtract Server: a web-based bioinformatic workflow platform. Nucleic Acids Res. 2011; 39:W528–W532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gnimpieba E.Z., Thavappiragasam M., Chango A., Conn B., Lushbough C.M.. SBMLDock: Docker Driven Systems Biology Tool Development and Usage. Computational Methods in Systems Biology. 2015; Springer International Publishing; 282–285. [Google Scholar]
- 17. Trapnell C., Pachter L., Salzberg S.L.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brueffer C., Saal L.H.. TopHat-Recondition: a post-processor for TopHat unmapped reads. BMC Bioinformatics. 2016; 17:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh S., Chan C.-K.K.. Analysis of RNA-Seq data using TopHat and Cufflinks. Methods Mol. Biol. 2016; 1374:339–361. [DOI] [PubMed] [Google Scholar]
- 20. Dooley R., Vaughn M., Terry S.. Your data, your way the iPlant Foundation API Data Services. 2012; 1–7.
- 21. Juty N., Le Noveŕe N., Laibe C.. Identifiers.org and MIRIAM registry: community resources to provide persistent identification. Nucleic Acids Res. 2012; 40:D580–D586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ison J., Kalaš M., Jonassen I., Bolser D., Uludag M., McWilliam H., Malone J., Lopez R., Pettifer S., Rice P.. EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics. 2013; 29:1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malone J., Brown A., Lister A.L., Ison J., Hull D., Parkinson H., Stevens R.. The Software Ontology (SWO): a resource for reproducibility in biomedical data analysis, curation and digital preservation. J. Biomed. Semantics. 2014; 5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goecks J., Nekrutenko A., Taylor J.. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lushbough C.M., Gnimpieba E.Z., Dooley R.. Life science data analysis workflow development using the bioextract server leveraging the iPlant collaborative cyberinfrastructure. Concurr. Comput. 2015; 27:408–419. [Google Scholar]
- 26. Goff S.A., Vaughn M., McKay S., Lyons E., Stapleton A.E., Gessler D., Matasci N., Wang L., Hanlon M., Lenards A. et al. The iPlant Collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2011; 2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brazas M.D., Yim D., Yeung W., Ouellette B.F.F.. A decade of web server updates at the bioinformatics links directory: 2003-2012. Nucleic Acids Res. 2012; 40:W3–W12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whetzel P.L. NCBO Technology: Powering semantically aware applications. J. Biomed. Semantics. 2013; 4:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang S.-Y. An ontology-supported and fully-automatic annotation technology for semantic portals. New Trends in Applied Artificial Intelligence. Springer, Berlin, Heidelberg; 1158–1168. [Google Scholar]
- 30. Buneman P. Agosti M, Borbinha J, Kapidakis S, Papatheodorou C, Tsakonas G. Curated Databases. Research and Advanced Technology for Digital Libraries: 13th European Conference, ECDL 2009, Corfu, Greece, September 27 - October 2, 2009. Proceedings. 2009; Berlin, Heidelberg: Springer; 2. [Google Scholar]
- 31. Rajasekar A., Moore R., Hou C.-Y., Lee C.A., Marciano R., de Torcy A., Wan M., Schroeder W., Chen S.-Y., Gilbert L. et al. iRODS primer: integrated rule-oriented data system. Synth. Lect. Inf. Concepts, Retrieval, Serv. 2010; 2:1–143. [Google Scholar]
- 32. Chiang G.-T., Clapham P., Qi G., Sale K., Coates G.. Implementing a genomic data management system using iRODS in the Wellcome Trust Sanger Institute. BMC Bioinformatics. 2011; 12:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noy N.F., Shah N.H., Whetzel P.L., Dai B., Dorf M., Griffith N., Jonquet C., Rubin D.L., Storey M.-A., Chute C.G. et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009; 37:W170–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bramer W.M., Giustini D., Kramer B.M.R.. Comparing the coverage, recall, and precision of searches for 120 systematic reviews in Embase, MEDLINE, and Google Scholar: a prospective study. Syst. Rev. 2016; 5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manning C.D., Raghavan P., Schütze H.. Introduction to Information Retrieval. 2008; Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.