Abstract
The information about the genetic basis of human diseases lies at the heart of precision medicine and drug discovery. However, to realize its full potential to support these goals, several problems, such as fragmentation, heterogeneity, availability and different conceptualization of the data must be overcome. To provide the community with a resource free of these hurdles, we have developed DisGeNET (http://www.disgenet.org), one of the largest available collections of genes and variants involved in human diseases. DisGeNET integrates data from expert curated repositories, GWAS catalogues, animal models and the scientific literature. DisGeNET data are homogeneously annotated with controlled vocabularies and community-driven ontologies. Additionally, several original metrics are provided to assist the prioritization of genotype–phenotype relationships. The information is accessible through a web interface, a Cytoscape App, an RDF SPARQL endpoint, scripts in several programming languages and an R package. DisGeNET is a versatile platform that can be used for different research purposes including the investigation of the molecular underpinnings of specific human diseases and their comorbidities, the analysis of the properties of disease genes, the generation of hypothesis on drug therapeutic action and drug adverse effects, the validation of computationally predicted disease genes and the evaluation of text-mining methods performance.
INTRODUCTION
Research on the genetic causes of disease has accelerated as a result of both the completion of the human genome and the development of the Next Generation Sequencing techniques, which has opened the promise of translating the alterations in individuals’ genomes in clinically relevant information to assist disease diagnostics and therapeutic decision-making. These efforts have generated a large volume of potentially useful information that has boosted biomedical research. Navigating and analyzing this information, however, is still cumbersome and time-consuming for researchers, due to four main hurdles. First, relevant information about the genetic causes of disease is scattered across specialized catalogues focused on specific disease classifications (i.e. Mendelian, or rare diseases), different model organisms, or on particular technological approaches (such as GWAS). Second, the fragmented nature of the processes generating this information has resulted in data of heterogeneous nature, not always annotated with controlled vocabularies and ontologies, and provided with different formats which are often non-trivial to reconcile. Third, the identification and prioritization of the relevant information from the vast quantity of data they harbor is often a challenging task for the end user. Finally, the access to most resources is usually limited to web interfaces and data downloads, hampering the usability of the information they contain. To further complicate this situation, a significant portion of the research on this field is only available as free text in scientific publications. These data are then, not amenable for computational analysis, and their inspection by researchers and clinicians, slow and tedious.
Repositories integrating and homogeneously annotating our current knowledge of the genetic causes of diseases are therefore essential to expedite translational research. To address this need we have developed DisGeNET (1,2), a discovery platform that contains a comprehensive catalogue of genes and variants associated to human diseases. DisGeNET covers the whole landscape of human diseases, including Mendelian, complex, environmental and rare diseases, and disease-related traits. DisGeNET collects data on genotype-phenotype relationships from several of the most popular resources in this area. These data are complemented with information extracted from the scientific literature using NLP-based text-mining tools. A variety of annotations and several metrics are offered to support, prioritize and facilitate the retrieval, and interpretation of the information. Furthermore, the platform offers several tools to interact with the data, including a web interface, a Cytoscape App, an R package and a SPARQL endpoint. The data are available for downloading in several formats. In this way, DisGeNET offers one of the most complete repositories on the genetic causes of human diseases to a wide range of users and purposes.
A COMPREHENSIVE CENTRALIZED REPOSITORY OF DISEASE-ASSOCIATED GENES AND VARIANTS
In order to present the most complete landscape of the extent of our knowledge of the genetic underpinnings of human diseases, DisGeNET integrates data from expert curated databases with information gathered through text-mining the scientific literature. The DisGeNET release 4.0 includes the following resources (for more details about the data processing, and versions, please check the Supplementary Data and http://disgenet.org/web/DisGeNET/menu/dbinfo):
- The Comparative Toxicogenomics Database (CTD) that provides information about chemicals and genes, and their effect in human health and disease (3)
- UniProt, a repository centred on protein sequence and function, that also include disease annotations from OMIM, and from expert curation of the scientific literature (4)
- ClinVar*, a public archive of relationships between human variants and phenotypes, including diseases (5)
- Orphanet*, a portal on rare diseases and their genes (6)
- The GWAS Catalog*, a curated collection of published GWAS results (7)
- The Rat Genome Database (RGD), a repository for the genetic, genomic and phenotypic data of the laboratory rat (8)
- The Mouse Genome Database (MGD), the main community resource for the laboratory mouse, that includes information about mouse models of disease (9)
- The Genetic Association Database (GAD), an archive of genetic association studies (10)
Additionally, DisGeNET includes two data sets obtained using different text-mining approaches
- The Literature Human Gene Derived Network (LHGDN), a data set of gene-disease associations obtained by text-mining the Entrez Gene Reference Into Function (GeneRIFs) using a Conditional Random Field approach (11)
- BeFree data, obtained using the BeFree System, which extracts gene-disease associations from MEDLINE abstracts (12,13)
* New sources with respect to the last published release
The data in DisGeNET are aggregated according to the original source in: CURATED, that includes data from UniProt, ClinVar, Orphanet, the GWAS Catalog and CTD (human data), PREDICTED, including RGD, MGD and CTD (mouse and rat data), and ALL. For further details about the data processing, see the Supplementary Material.
DisGeNET 4.0 (June, 2016) contains 429 036 gene-disease associations (GDAs), linking 17 381 genes to 15 093 diseases, disorders and clinical or abnormal human phenotypes. Ninety nine percent of the GDAs in DisGeNET are supported by at least one publication, and the information contained in DisGeNET corresponds to more than 289 000 publications. DisGeNET contains 72 870 variant-disease associations (VDAs), between 46 589 SNPs and 6356 diseases and phenotypes. The GDAs provided by the expert curated resources (DisGeNET curated) represent only the 2% of all the information (Supplementary Figure S1, panel A), highlighting the need of text-mining algorithms to unlock the information available as free text in scientific publications. The data obtained from each resource are rather unique, as evidenced by the small overlap between sources (Supplementary Figure S1, panel B). The lack of redundancy across different resources emphasizes the importance of integration, and is a consequence of the fragmented nature of data production, the different focus of individual resources and the different criteria for curation.
HOMOGENEOUS ANNOTATION OF THE DATA FOSTERS INTEROPERABILITY AND EASES INTERPRETATION
The data retrieved from the different resources are harmonized and standardized using community driven-controlled vocabularies and ontologies. Furthermore, genes, diseases, variants, GDAs and VDAs are enriched with additional information that expedites data interpretation and analysis, both manual and automatic.
Unified Medical Language System (14) (UMLS) Metathesaurus Concept Unique Identifiers are employed to homogeneously annotate diseases obtained from different sources. To allow interoperability, and to bridge the research and the clinical settings, DisGeNET provides a wide variety of disease vocabularies. These include MeSH, OMIM, Disease Ontology (15) (DO) and ICD9. Disease attributes include the MeSH disease class, the UMLS semantic type, the Human Phenotype Ontology (16) and the DO upper level classes. In version 4.0, we distinguish between abnormal phenotypes, traits, signs and symptoms from actual diseases using the UMLS semantic types followed by manual curation. In addition, disease entries referring to very general classes such as ‘Cardiovascular Diseases’, ‘Autoimmune Diseases’, ‘Neurodegenerative Diseases and ‘Lung Neoplasms’ are indicated as ‘disease group’.
Gene HGNC symbols and UniProt identifiers are mapped to Entrez gene identifiers. The genes are described with their full name, the UniProt accession, the Reactome (17) upper level pathway and the Panther (18) protein class.
DisGeNET 4.0 has made a stronger emphasis on the relationship between variants and diseases. Variant-disease information in DisGeNET originates from ClinVar, the GWAS Catalog, UniProt, GAD and from BeFree data. Variants are identified using the NCBI Short Genetic Variations database (dbSNP) identifiers, and annotated with the chromosomal position, the reference and alternative alleles and the variant class, obtained from dbSNP database (19). Additionally, variant allele frequencies computed by the Exome Aggregation Consortium, which aggregates and harmonizes exome sequencing data from a variety of large-scale sequencing projects (20), and by the 1000 Genomes Project, a public catalogue of human variation and genotype data (21), are provided. Finally, the most severe consequence type of each variant is displayed, obtained using the Ensembl Variant Effect Predictor (22).
For both, GDAs and VDAs, the publication(s) supporting the association, a representative sentence from each publication and the original source are available. In DisGeNET 4.0 the full collection of publications supporting each association has been made available (in previous releases only 10 representative papers were shown). Additionally, the associations can now be sorted or filtered by publication year. The new release also benefits from an improved disambiguation between genes and diseases implemented in the BeFree text-mining system. Furthermore, false positive GDAs have been semi-automatically removed from all text-mining data sets (BeFree, GAD and LHGDN). Lastly, each GDA is characterized with the DisGeNET gene-disease association type ontology that has been updated to include new association types (highlighted in purple in Supplementary Figure S2).
PRIORITIZATION FEATURES TO GUIDE THE EXPLORATION OF THE GENETIC BASIS OF DISEASE
To help navigating the more than 400 000 GDAs in DisGeNET, these are rated with a confidence score (the DisGeNET score) that reflects the recurrence of a GDA across all data sources. The DisGeNET score takes into account the number of sources supporting the association and the reliability of each of them (for further details see http://disgenet.org/web/DisGeNET/menu/dbinfo#score). The score is updated in each release to include the new sources incorporated in the database.
In order to facilitate ranking the genes associated with a disease, in this release we introduced two new metrics. The Disease Specificity Index (DSI) ranges from 0 to 1, and is inversely proportional to the number of diseases associated to a particular gene. A gene associated to a large number of diseases (e.g. TNF, associated to more than 1500 diseases) will have a DSI close to zero, while a gene associated to only one disease, is more ‘specific’ for that disease and has DSI of 1. The Disease Pleiotropy Index (DPI) ranges from 0 to 1 and is proportional to the number of different (MeSH) disease classes a gene is associated to. Thus, a gene associated to diseases of diverse classes (such as APOE, associated to Cardiovascular Diseases, Mental Disorders, Neoplasms, Respiratory Tract Diseases, etc), will have a DPI close to 1. Conversely, the PSCA, associated to 58 diseases, most of which are neoplasms has a relatively low DPI.
FLEXIBLE DATA ACCESS TO ANSWER DIFFERENT RESEARCH QUESTIONS
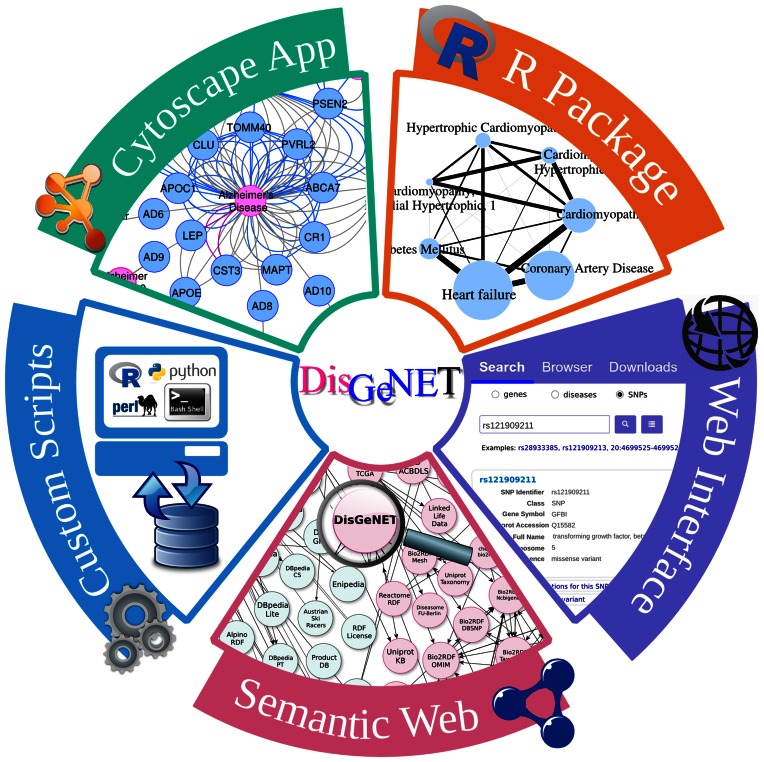
The DisGeNET platform includes several ways of accessing the data, which makes it a versatile resource to answer questions posed by different types of users, and to achieve diverse goals (Figure 1). The web interface is designed for user-friendly exploration of small portions of the data by users interested in a particular disease, gene or variant. Using customized scripts allows querying the database for large lists of genes, diseases or variants, and including DisGeNET data in computational workflows. The DisGeNET Cytoscape App is especially suited to carry out network medicine analyses and visualize their results. Accessing the data using Semantic Web technologies enables to combine DisGeNET data with other types of biological information available in the Linked Open Data (LOD) cloud (http://lod-cloud.net/). Finally, the disgenet2r R package facilitates exploring, analysing and visualizing the data using the powerful graphical and statistical capabilities of the R environment.
Figure 1.
The DisGeNET platform provides several ways to access the data.
The web interface
The DisGeNET web interface allows searching by single gene, disease and variant, using different types of identifiers. Searching by short lists of genes, diseases and variants is also possible. The user can also browse the data, using the Browser entry point. To ease the interpretation of the information, the different views of the web interface show annotations of the different biological entities to the aforementioned attributes. The interface also enables sorting and filtering the data using the DisGeNET score, the DPI, the DSI, the publication year and the DisGeNET gene-disease association type. Links to other reference resources, such as the NCBI gene, UniProt, UMLS and dbSNP, are also provided.
Programmatic access
As a result of exploring DisGeNET data using the web interface, scripts in R, Perl, Python and Bash are automatically generated. These scripts can be downloaded and customized to generate the same or similar queries, allowing to reproduce the results of the analysis performed through the web interface and to incorporate DisGeNET data as part of automatic computational workflows. Additionally, in the downloads section of the website, several exemplary scripts are provided to perform queries to the DisGeNET database, with use case examples.
The DisGeNET Cytoscape App
The DisGeNET Cytoscape plugin (1) has been updated to correspond to Cytoscape versions 3.0 or higher. The DisGeNET Cytoscape App allows visualization of the GDAs as bipartite networks, and also exploration of the data in a disease or gene-centric way. A variety of features can be used to construct the networks, such as filtering by disease class, the DisGeNET gene-disease association type, data source and score range. The networks can also be built around a particular gene, or disease. The DisGeNET Cytoscape App, and a detailed tutorial on how to use it, are available for download at http://disgenet.org/web/DisGeNET/menu/app.
DisGeNET-RDF
DisGeNET data are also published as machine-readable data through DisGeNET-RDF (23) and nanopublications (24) linked data sets, which increases the FAIRness of the data (25). Entities and properties in DisGeNET-RDF are semantically defined making extensive reuse of standard identifiers, vocabularies and ontologies such as the National Cancer Institute thesaurus, for medical vocabulary and the Semanticscience Integrated Ontology (26) for general science. DisGeNET-RDF is interlinked to other biomedical databases available in the LOD cloud that enables performing complex queries that need the interrogation of different resources to be answered. The DisGeNET SPARQL endpoint allows exploration of the DisGeNET-RDF data set and query federation to expand DisGeNET gene-disease association information with data on gene expression, drug activity and biological pathways, among others. Representative queries linking the data to other resources such as Wikipathways (27), ChEMBL (28) and the Gene Expression Atlas (29) are available at the RDF page in the website (http://www.disgenet.org/web/DisGeNET/menu/rdf#sparql-queries).
The disgenet2r R package
DisGeNET data can also be accessed via an R package, disgenet2r. The package contains a series of functions to retrieve gene-disease and variant-disease data, and to perform mappings between several biomedical vocabularies. The results of the queries can be visualized using a variety of plots, such as heatmaps, barplots and several types of networks. The package is especially well suited to explore the genetic basis of diseases, and disease comorbidities. Furthermore, the disgenet2r package permits benefiting from the Semantic Web technologies, without the need of special expertise in this area through a set of functions that use DisGeNET-RDF and other resources available in the LOD cloud. These functions include retrieving the druggable targets for a disease of interest, or obtaining the biological pathways for a list of disease genes. The disgenet2r package also expedites the integration of DisGeNET data with other R packages. The source code and documentation of disgenet2r package are available at https://bitbucket.org/ibi_group/disgenet2r.
CONCLUSIONS AND FUTURE PERSPECTIVES
New trends are starting to emerge in disease therapeutics. Some examples include immunotherapies, gene therapy, the use of siRNA and anti-sense oligonucleotides, and the more recent possibility of genome editing with CRISPR-cas9 systems. The structural constraints imposed by ‘classical’ drug development will no longer be a limitation, marking the end of the so called ‘druggable genome’ (30). With this, a new type of therapy, based on disease mechanisms is starting to emerge (31). The precise knowledge of the molecular processes underlying disease pathophysiology will become the new limiting step in drug development. In order to elucidate these mechanisms, integrative approaches that aggregate all the available information on the genetics basis of diseases are an essential step. Currently, some of the more popular resources only represent a fraction of the available knowledge. For example, OMIM (32) covers only Mendelian diseases, Orphanet (6) is a compilation of rare diseases, while the GWAS Catalog (7) is a repository for GWAS data, involving mainly complex diseases and traits (For a detailed list of available resources, see Supplementary Table S1). Conversely, DisGeNET aims at integrating information on the genetic underpinnings of all disease therapeutic areas, and in such endeavor, at being a repository of reference for closing the genotype–phenotype gap.
DisGeNET platform has been used to study a variety of biomedical problems, which include investigating the molecular basis of specific diseases (33–36), annotating lists of genes produced by different types of omics and sequencing protocols (37–39), validating disease genes prediction methods (40–42), understanding disease mechanisms in the context of protein networks (43,44), gaining insight into drug action (45) and drug adverse reactions mechanisms (46), drug repurposing (47), exploring the molecular basis of disease comorbidities (48,49), assessing the performance of text-mining algorithms (50) and as part of other resources (51–53).
DisGeNET is a well-established resource with four stable releases (as of October, 2016). It is regularly growing, fuelled and kept up-to-date by the new research, by the incorporation of new data sources, and by the interest of a growing community of users. The careful use of standards, and state of the art biomedical ontologies, the attention paid to keeping track of the provenance of the information, together with the extensive documentation of the data processing and the multiple access points, makes of DisGeNET a platform of choice to support translational research.
Acknowledgments
The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Footnotes
Present address: Núria Queralt-Rosinach, Department of Molecular and Experimental Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Instituto de Salud Carlos III-Fondo Europeo de Desarrollo Regional [CP10/00524 and PI13/00082]; Innovative Medicines Initiative Joint Undertaking [Open PHACTs No. 115191]; resources of which are composed of financial contribution from the European Union's Seventh Framework Programme [FP7/2007-2013]; EFPIA companies’ in kind contribution; European Union Horizon 2020 Programme 2014-2020 [MedBioinformatics No. 634143 and Elixir-Excelerate No. 676559]. The Research Programme on Biomedical Informatics (GRIB) is a member of the Spanish National Bioinformatics Institute (INB), PRB2-ISCIII and is supported by grant PT13/0001/0023, of the PE I+D+i 2013-2016, funded by ISCIII and FEDER. Funding for open access charge: Instituto de Salud Carlos III-Fondo Europeo de Desarrollo Regional [PI13/00082].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bauer-Mehren A., Rautschka M., Sanz F., Furlong L.I. DisGeNET: a Cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics. 2010;26:2924–2926. doi: 10.1093/bioinformatics/btq538. [DOI] [PubMed] [Google Scholar]
- 2.Piñero J., Queralt-Rosinach N., Bravo A., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis A.P., Grondin C.J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Wiegers T.C., Mattingly C.J. The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rath A., Olry A., Dhombres F., Brandt M.M., Urbero B., Ayme S. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum. Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 7.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J.F., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.-J., et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015;43:D726–D736. doi: 10.1093/nar/gku967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker K.G., Barnes K.C., Bright T.J., Wang S.A. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 11.Bundschus M., Dejori M., Stetter M., Tresp V., Kriegel H.-P. Extraction of semantic biomedical relations from text using conditional random fields. BMC Bioinformatics. 2008;9:207. doi: 10.1186/1471-2105-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bravo A., Cases M., Queralt-Rosinach N., Sanz F., Furlong L.I. A knowledge-driven approach to extract disease-related biomarkers from the literature. Biomed Res. Int. 2014;2014:253128. doi: 10.1155/2014/253128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravo À., Piñero J., Queralt-Rosinach N., Rautschka M., Furlong L.I. Extraction of relations between genes and diseases from text and large-scale data analysis: implications for translational research. BMC Bioinformatics. 2015;16:55. doi: 10.1186/s12859-015-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D2710. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D., et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groza T., Köhler S., Moldenhauer D., Vasilevsky N., Baynam G., Zemojtel T., Schriml L.M., Kibbe W.A., Schofield P.N., Beck T., et al. The human phenotype ontology: semantic unification of common and rare disease. Am. J. Hum. Genet. 2015;97:111–124. doi: 10.1016/j.ajhg.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2015;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M., Karczewski K.J., Minikel E. V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60, 706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queralt-Rosinach N., Piñero J., Bravo À., Sanz F., Furlong L.I. DisGeNET-RDF: harnessing the innovative power of the semantic web to explore the genetic basis of diseases. Bioinformatics. 2016;32:2236–2238. doi: 10.1093/bioinformatics/btw214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Queralt-Rosinach N., Kuhn T., Chichester C., Dumontier M., Sanz F., Furlong L.I. Publishing DisGeNET as nanopublications. Semant. Web. 2016;7:519–528. [Google Scholar]
- 25.Wilkinson M.D., Dumontier M., Aalbersberg Ij.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumontier M., Baker C.J., Baran J., Callahan A., Chepelev L., Cruz-Toledo J., Del Rio N.R., Duck G., Furlong L.I., Keath N., et al. The Semanticscience Integrated Ontology (SIO) for biomedical research and knowledge discovery. J. Biomed. Semantics. 2014;5:14. doi: 10.1186/2041-1480-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E.L., Bohler A., Mélius J., Waagmeester A., Sinha S.R., Miller R., et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44:D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jupp S., Malone J., Bolleman J., Brandizi M., Davies M., Garcia L., Gaulton A., Gehant S., Laibe C., Redaschi N., et al. The EBI RDF platform: linked open data for the life sciences. Bioinformatics. 2014;30:1338–1339. doi: 10.1093/bioinformatics/btt765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petryszak R., Keays M., Tang Y.A., Fonseca N.A., Barrera E., Burdett T., Füllgrabe A., Fuentes A.M.-P., Jupp S., Koskinen S., et al. Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 31.Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J., et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 32.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plaisier C.L., O'Brien S., Bernard B., Reynolds S., Simon Z., Toledo C.M., Ding Y., Reiss D.J., Paddison P.J., Baliga N.S., et al. Causal mechanistic regulatory network for glioblastoma deciphered using systems genetics network analysis. Cell Syst. 2016;3:172–186. doi: 10.1016/j.cels.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishore A., Žižková V., Kocourková L., Petrkova J., Bouros E., Nunes H., Loštáková V., Müller-Quernheim J., Zissel G., Kolek V., et al. Association study for 26 candidate loci in idiopathic pulmonary fibrosis patients from four european populations. Front. Immunol. 2016;7:274. doi: 10.3389/fimmu.2016.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatnagar M., Shorvon S. Genetic mutations associated with status epilepticus. Epilepsy Behav. 2015;49:104–110. doi: 10.1016/j.yebeh.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Abascal M.F., Besso M.J., Rosso M., Mencucci M.V., Aparicio E., Szapiro G., Furlong L.I., Vazquez-Levin M.H. CDH1/E-cadherin and solid tumors. An updated gene-disease association analysis using bioinformatics tools. Comput. Biol. Chem. 2016;60:9–20. doi: 10.1016/j.compbiolchem.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Breen M.S., Uhlmann A., Nday C.M., Glatt S.J., Mitt M., Metsalpu A., Stein D.J., Illing N. Candidate gene networks and blood biomarkers of methamphetamine-associated psychosis: an integrative RNA-sequencing report. Transl. Psychiatry. 2016;6:e802. doi: 10.1038/tp.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen M.C., Nederby L., Roug A., Villesen P., Kjeldsen E., Nyvold C.G., Hokland P. Novel scripts for improved annotation and selection of variants from whole exome sequencing in cancer research. MethodsX. 2015;2:145–153. doi: 10.1016/j.mex.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee I.-H., Lee K., Hsing M., Choe Y., Park J.-H., Kim S.H., Bohn J.M., Neu M.B., Hwang K.-B., Green R.C., et al. Prioritizing disease-linked variants, genes, and pathways with an interactive whole-genome analysis pipeline. Hum. Mutat. 2014;35:537–347. doi: 10.1002/humu.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alaimo S., Giugno R., Pulvirenti A. ncPred: ncRNA-disease association prediction through tripartite network-based inference. Front. Bioeng. Biotechnol. 2014;2:71. doi: 10.3389/fbioe.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M.-X., Chen X., Chen G., Cui Q.-H., Yan G.-Y. A computational framework to infer human disease-associated long noncoding RNAs. PLoS One. 2014;9:e84408. doi: 10.1371/journal.pone.0084408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava I., Gahlot L.K., Khurana P., Hasija Y. dbAARD & AGP: A computational pipeline for the prediction of genes associated with age related disorders. J. Biomed. Inform. 2016;60:153–161. doi: 10.1016/j.jbi.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Piñero J., Berenstein A., Gonzalez-Perez A., Chernomoretz A., Furlong L.I. Uncovering disease mechanisms through network biology in the era of Next Generation Sequencing. Sci. Rep. 2016;6:24570. doi: 10.1038/srep24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chagoyen M., Pazos F. Characterization of clinical signs in the human interactome. Bioinformatics. 2016;32:1761–1765. doi: 10.1093/bioinformatics/btw054. [DOI] [PubMed] [Google Scholar]
- 45.Vogt I., Prinz J., Worf K., Campillos M. Systematic analysis of gene properties influencing organ system phenotypes in mammalian perturbations. Bioinformatics. 2014;30:3093–3100. doi: 10.1093/bioinformatics/btu487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bauer-Mehren A., van Mullingen E.M., Avillach P., Carrascosa M.D.C., Garcia-Serna R., Piñero J., Singh B., Lopes P., Oliveira J.L., Diallo G., et al. Automatic filtering and substantiation of drug safety signals. PLoS Comput. Biol. 2012;8:e1002457. doi: 10.1371/journal.pcbi.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mullen J., Cockell S.J., Woollard P., Wipat A. An integrated data driven approach to drug repositioning using gene-disease associations. PLoS One. 2016;11:e0155811. doi: 10.1371/journal.pone.0155811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grosdidier S., Ferrer A., Faner R., Piñero J., Roca J., Cosío B., Agustí A., Gea J., Sanz F., Furlong L.I. Network medicine analysis of COPD multimorbidities. Respir. Res. 2014;15:111. doi: 10.1186/s12931-014-0111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faner R., Gutiérrez-Sacristán A., Castro-Acosta A., Grosdidier S., Gan W., Sánchez-Mayor M., Lopez-Campos J.L., Pozo-Rodriguez F., Sanz F., Mannino D., et al. Molecular and clinical diseasome of comorbidities in exacerbated COPD patients. Eur. Respir. J. 2015;46:1001–1010. doi: 10.1183/13993003.00763-2015. [DOI] [PubMed] [Google Scholar]
- 50.Xu D., Zhang M., Xie Y., Wang F., Chen M., Zhu K.Q., Wei J. DTMiner: Identification of potential disease targets through biomedical literature mining. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw503. doi:10.1093/bioinformatics/btw503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mannil D., Vogt I., Prinz J., Campillos M. Organ system heterogeneity DB: a database for the visualization of phenotypes at the organ system level. Nucleic Acids Res. 2015;43:D900–D906. doi: 10.1093/nar/gku948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamed M., Spaniol C., Nazarieh M., Helms V. TFmiR: a web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res. 2015;43:W283–W288. doi: 10.1093/nar/gkv418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornish A.J., Filippis I., David A., Sternberg M.J.E. Exploring the cellular basis of human disease through a large-scale mapping of deleterious genes to cell types. Genome Med. 2015;7:95. doi: 10.1186/s13073-015-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]