Abstract
FINDbase (http://www.findbase.org) is a comprehensive data repository that records the prevalence of clinically relevant genomic variants in various populations worldwide, such as pathogenic variants leading mostly to monogenic disorders and pharmacogenomics biomarkers. The database also records the incidence of rare genetic diseases in various populations, all in well-distinct data modules. Here, we report extensive data content updates in all data modules, with direct implications to clinical pharmacogenomics. Also, we report significant new developments in FINDbase, namely (i) the release of a new version of the ETHNOS software that catalyzes development curation of national/ethnic genetic databases, (ii) the migration of all FINDbase data content into 90 distinct national/ethnic mutation databases, all built around Microsoft's PivotViewer (http://www.getpivot.com) software (iii) new data visualization tools and (iv) the interrelation of FINDbase with DruGeVar database with direct implications in clinical pharmacogenomics. The abovementioned updates further enhance the impact of FINDbase, as a key resource for Genomic Medicine applications.
INTRODUCTION
National and Ethnic Genomic Databases (NEGDBs), previously known as National and Ethnic Mutation Databases or NEMDBs, comprise a well-defined niche of genomic databases, aiming to record the prevalence of alleles, mainly pathogenic and of clinical relevance, but also benign, in different populations and ethnic groups worldwide in a structured manner (1). These resources can be used not only in population genetic studies, e.g. to study gene/mutation flow and admixture patterns, human demographic history but also, and most importantly, in Genomic Medicine, e.g. to stratify national molecular diagnostic services or to rationalize drug use (1). NEGDBs can nicely complement the data content of either central (or core) databases, such as ClinVar (2) or HGMD (3) and/or locus-specific databases in general (4).
The first NEGDBs were realized in a very primitive structure back in 2003, mainly to serve individual populations and ethnic groups. Couple of years later, and in an effort to maintain a homogeneous data content, a dedicated software (ETHNOS; 5) was developed, allowing the development and curation of NEGDB. In 2006, our group established FINDbase (Frequency of INherited Disorders database; http://www.findbase.org), a worldwide database with the vision to comprehensively document the prevalence of clinically relevant genomic variation allele frequencies in various populations and ethnic groups worldwide (6). FINDbase contains data only in an aggregated manner, i.e. allele frequencies stripped from any sensitive personal information of their carriers, in order to maintain anonymity (6). At the beginning, FINDbase accommodated data pertaining to the prevalence of pathogenic genomic variants, e.g. leading to monogenic disorders (6). Since 2010, FINDbase also includes prevalence data of pharmacogenomic (PGx) biomarkers (7) and since 2013, information about the various rare genetic diseases that are documented in various populations (8), all in distinct data modules. Content-wise, FINDbase is the richest database among all NEGDBs currently available and, it is considered to be one of the key resources to retrieve population-specific information for clinically relevant genomic variations, as indicated by the number and origin of visitors.
In this paper, we present data content updates in all three data modules and significant technological advances which substantially expand the existing data content and visualization tools, so that FINDbase becomes more appealing to the clinical, apart from the research community.
DATA CONTENT UPDATES
Contrary to the previous FINDbase data content updates in 2010 (9) and 2013 (10), the recent (2014–2016) update did not only include data curation, update and correction, where necessary, but also extensive data enrichment in all three data modules.
In particular, FINDbase data collection was enriched with causative genomic variants allele frequencies as far as the GJB2 and ATP7B genes are concerned, leading to non-syndromic sensorineural deafness and Wilson disease, respectively. As previously, all entries, representing 930 new records from 44 populations, were manually curated from the published literature and entered into the main FINDbase data collection, recorded against their corresponding unique PubMed ID.
Also, special attention was given to the data module documenting PGx marker allele frequencies, which was significantly enriched with the prevalence of 36 clinically actionable PGx biomarkers in 21, mostly European, populations, resulting from a large multicenter study mapping the prevalence of PGx biomarkers in Europe (11). Data enrichment was based on the contributor's unique ResearcherID (http://www.researcherid.com) and follows the microattribution approach (12), allowing unambiguous identification of curated data when data update or correction is needed and, at the same time, providing incentives to potential data contributors to share their data with the broader scientific community.
This particular data update is of clinical relevance, as it allows clinicians to assess whether a particular PGx biomarker, related with a certain drug's efficacy or toxicity, is rather prevalent in the population of interest, prior to prescribing a PGx testing to individualize drug treatment. FINDbase data compilation and representation are subject to copyright and usage principles to ensure that FINDbase and its contents remains freely available to all interested parties.
UPGRADE OF THE ETHNOS SOFTWARE
In the last FINDbase update, we have built a new module for rare genetic disease summaries (10), in which we migrated all genetic disease summary data collection from five previously developed NEGDBs (Hellenic, Lebanese, Cypriot, Egyptian and Moroccan). Based on this functionality, and given that the component services of all three modules homogeneously follow the Service Oriented Architecture (SOA; 13) and are built around the same data visualization tool (PivotViewer; http://www.microsoft.com/silverlight/pivotviewer), we have decided to (i) completely restructure and upgrade the previous version of the ETHNOS software (2), to include improved data submission, querying and curation functionalities, standing as an ‘off-the-shelf’ web application that would allow the development of new NEGDBs, (ii) exploit the entire FINDbase data content, subdivided into 3 modules, to migrate the entire FINDbase data collection to establish 90 individual ETHNOS-based NEGDBs in all 5 continents (Table 1). These databases are displayed in FINDbase home page and clustered together according to continent (ETHNOS Databases tab; Supplementary Figure S1). Resulting databases can be subsequently allocated to research groups and/or consortia in different countries for further data enrichment with published and unpublished data and expert data curation, always based on the microattribution approach (12,14). The upgraded ETHNOS software was used by the SERBORDISInn Consortium (www.serbordisinn.rs) to establish the Serbian NEGDB (Supplementary Figure S2A). In this case, the user can query the clinically relevant genomic variation allele frequency data and rare genetic disease summaries only for the Serbian population (Supplementary Figure S2B).
Table 1. Summary of FINDbase worldwide database records (assessed August 2016) within all populations. Average: Average record count per population within each continent group.
| Continents | Populations | Records | Average |
|---|---|---|---|
| Africa | 15 | 791 | 52.73 |
| Americas | 5 | 565 | 113.00 |
| Asia | 30 | 2574 | 85.80 |
| Europe | 37 | 4564 | 123.35 |
| Oceania | 3 | 94 | 31.33 |
| TOTAL | 90 | 8588 | 95.42 |
The upgraded ETHNOS software complies with the established recommendations and guidelines to develop nation-wide projects to document the genetic heterogeneity in various countries (7).
NEW FEATURES
Apart from the comprehensive data content curation and update described in the previous paragraphs, we have also incorporated new features into FINDbase, mostly of clinical relevance. These include the display of the FINDbase content in a map format, giving a visual first impression of FINDbase data content spanning across all three data modules and the interrelation of the PGx module of FINDbase with DruGeVar database, offering new data visualization and querying functionalities, as described below.
Transition to HTML5
As previously mentioned, the main visualization tool of FINDbase querying engine is PivotViewer (http://www.microsoft.com/silverlight/pivotviewer), which is based on Microsoft Silverlight technology (http://www.silverlight.net). The latter requires the installation of the Silverlight plug-in, which, although freeware, in several installations is either not available or not allowed. For this reason, we have made a transition of FINDbase to HTML5 technology, which allows the user to access the data without the need of having Silverlight installed in their workstation. Although this transition may minimally affect the processing speed in case of large data sets, our experience shows that it is overall advantageous for FINDbase users.
Integration of DruGeVar database with the pharmacogenomics module of FINDbase
To date, a large number of genomic variants have been correlated with variable drug response and severity of adverse drug reactions (15). From these, only a small fraction of these variants, referred to as PGx biomarkers, have been approved by regulatory agencies such that over 120 drugs bear PGx information in their labels. However, although comprehensive drug-gene lists exist online in both the US Food and Drug Administration (FDA; www.fda.gov) and the European Medicines Agency (EMA; www.ema.europa.eu), information related to the respective PGx biomarkers is currently missing from such lists (16). We have previously extracted information from the published literature and online resources and developed a comprehensive database, namely DruGeVar (http://drugevar.genomicmedicinealliance.org; 17), which triangulates drugs with genes and PGx biomarkers that could serve clinical PGx. DruGeVar has been developed in such way to be readily applicable as a standalone resource or a plug-in module for other databases documenting relevant information. Given the fact that the PGx biomarker data module of FINDbase, not only contains the relevant information but is also built using the same data visualization tool, namely PivotViewer and understanding the need for deploying interactive and comprehensive visualizations displaying complex and big data structures, exploiting the wealth of information in both databases that are ultimately useful for clinicians, we integrated DruGeVar and FINDbase databases.
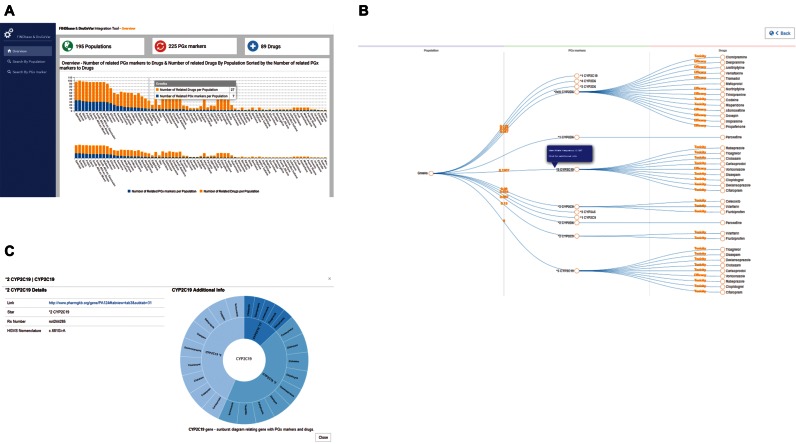
The FINDbase and DruGeVar integration tool can be accessed from the FINDbase PGx biomarkers module. The front end environment focuses on the three related components of the DruGeVar database, namely drugs, genes, PGx biomarkers and populations of the FINDbase database in purpose to highlight the relation between them. The front end environment was developed using AngularJS framework (https://angularjs.org) and the architecture of the application is based on the AngularJS components adding reusable elements to the application and the ability of the configuration of the new features. We added five different types of visualizations; (i) collapsible tree diagrams, (ii) sunburst diagrams, (iii) bar charts, (iv) stacked bar charts and (v) donut charts. The (i) and (ii) were developed using D3.js (https://d3js.org) and (iii), (iv) and (v) using C3.js (http://c3js.org). Collapsible tree diagrams are used to provide interactive visualizations using hierarchical data format. The root node of each tree defines a population or a PGx marker and leads to the related data according the selection of the user. Collapsible tree diagrams, with population as root node (Figure 1A and B), are provided selecting on an interactive world map (not shown), developed using Datamaps (http://datamaps.github.io) or by a provided list of countries. Clicking on the related nodes of the population's collapsible tree diagram, additional information for the selected node is provided. Clicking on the PGx biomarker's node, linking the selected population to the related drugs, a sunburst diagram visualization (Figure 1C), using hierarchical data format, is displayed, in addition to visualize the relation between PGx biomarker's gene, its related PGx biomarkers and drugs. The selection of a PGx biomarker, in purpose to construct a collapsible tree diagram with the PGx biomarker as root node, follows a filtering flow and begins with the selection of the related gene from a list of the available genes of the database and continues with the selection of the PGx biomarker from the related PGx biomarkers’ list according the gene's selection. In that case, the information provided includes a table, with filtering, ordering and pagination of the fields of the related drugs (Figure 2A and B) developed using Smart Table (http://lorenzofox3.github.io/smart-tablewebsite). Stacked bar charts, donut charts and bar charts are used to measure the relation between the basic components; the percentage of the related PGx biomarkers, the number of related PGx biomarkers to drugs and the number of the related drugs by population.
Figure 1.
Integration of DruGeVar with the PGx markers data module of FINDbase. (A) Bar chart showing the number of the related PGx biomarkers to drugs and the number of related drugs by population sorted by the number of related PGx biomarkers to drugs. The subchart is used for selecting specific region of the bar chart. (B) Collapsible tree diagram showing the relation between population, PGx biomarkers and drugs. The number between population and PGx biomarker defines the rare allele frequency and the label between PGx number and drug the effect on the toxicity or efficacy of the biomarker to the related drug. Clicking on the nodes of PGx biomarkers or drugs additional information for the selected element is appearing in a pop up window. (C) Pop up window showing additional information for the selected PGx biomarker. Left: Details for the selected PGx marker. Right: Sunburst visualization, showing the related additional PGx markers and drugs of the gene of the selected PGx biomarker. Clicking on the elements of the PGx biomarkers, the visualization is reconstructed focusing on the children nodes (related drugs) of the selected node, while clicking on the elements of the drugs, the visualization is reconstructed focusing on the ‘children’ of the parent node.
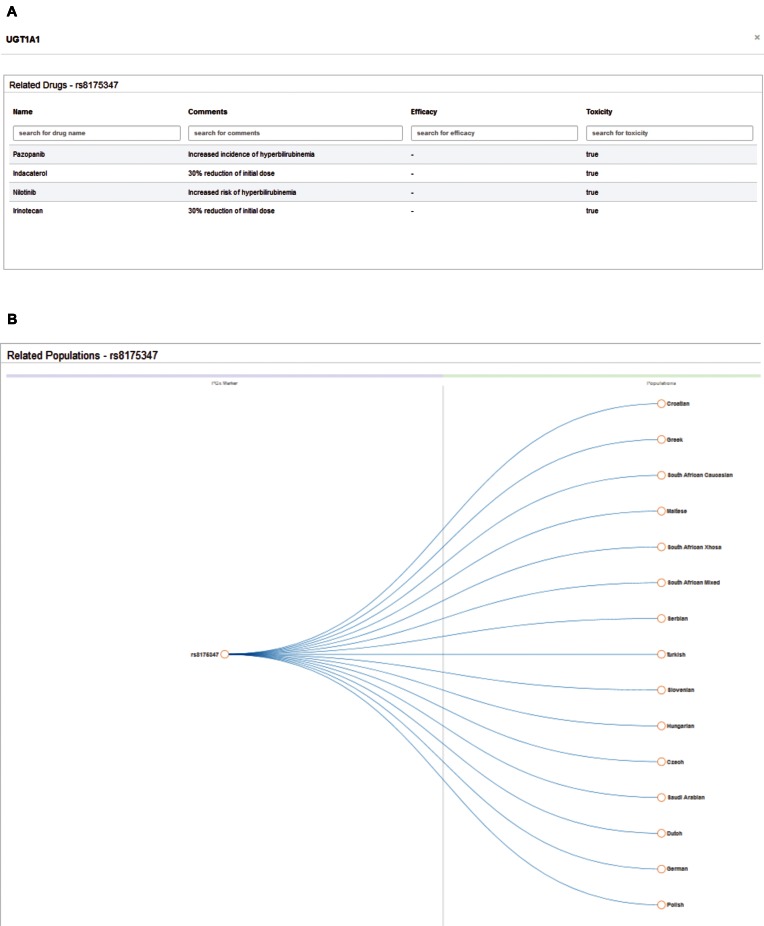
Figure 2.
Integration of DruGeVar with the PGx markers data module of FINDbase. Pop up window showing the related data of the selected PGx biomarker after filtering, displaying the related drugs to the PGx biomarker and additional information in a (A) tabular format and the related populations of the selected PGx marker in (B) collapsible tree diagram.
Establishing the minimum NEGDB information specification with VarioML
As of early 2007, immediately after its first launch, FINDbase established synergies with other related genomic databases (Human Gene Mutation Database (HGMD); http://www.hgmd.org; 3), or resources (Café Variome (http://www.cafevariome.org); 10).
In 2012, VarioML was developed within the scope of the GEN2PHEN project framework (www.gen2phen.org), as a set of tools and practices improving the availability, quality and comprehensibility of human variation information (18). VarioML enables researchers and diagnostic laboratories to easily and unambiguously share variant information, by adapting the variant specification into one's own research workflow in a straightforward and a simple ‘push-button’ manner. In other words, genomic variation data can be easily captured, federated and exchanged, while more complex data can be also described. VarioML schema is developed using RelaxNG language (http://books.xmlschemata.org/relaxng) and it consist of set of XML building blocks, which can be used for composing data exchange implementations for specific use cases. Although the VarioML is implemented using XML technologies, the usage of the schema is not limited only to XML. Translation tools have been made for example converting VarioML elements to JSON format and Java programming language classes. These open source tools and libraries are made available on GitHub (https://github.com/VarioML).
We have actively participated with the VarioML development and used FINDbase data to propose the minimum information specification for NEGDB data exchange use cases. The developed specification has necessary elements for describing population structures, genomic variants and allele frequencies in an appropriate manner needed for NEGDBs. For example, variants can have one or more frequency elements, each of which can be expressed in one of three formats: (i) decimal number, expressing frequency as a floating point value, (ii) number of cases, expressing frequency as a count and/or (iii) categorized value, expressing frequency as an ontology term, for categorized observations such as ‘exists’ or ‘less than 100’. Further context for the populations, variants and frequency values can be provided by evidence code, protocol id, database cross reference and comment elements. All VarioML elements can hold necessary attributes for unambiguously linking the elements into relevant ontologies using for example either URIs or MIRIAM identifiers (http://www.ebi.ac.uk/miriam). Further information and examples of use are available at http://www.varioml.org/freq_minspec.htm.
The map display visualization tool
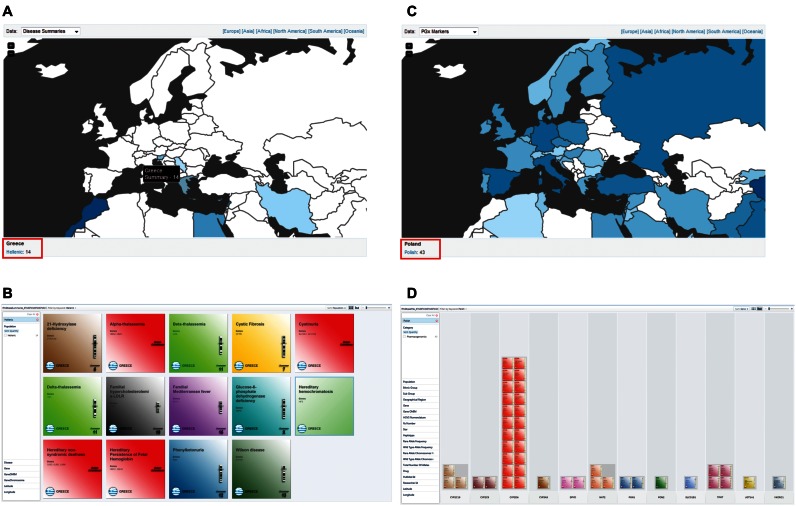
FINDbase includes a large data set of several clinically relevant genomic data allele frequencies in over 90 populations and ethnic groups worldwide. In order to give a visual impression of FINDbase data content and, at the same time, provide a quick access to the available data per population and per module, we have built a world map that is readily available at the Home page (Supplementary Figure S3). The user can zoom in and out in the map, using the + and – keys and select the data module desired, namely causative mutations, PGx markers or disease summaries from the top left corner of the map. Alternatively, the user can select the map display per continent, from the top right corner of the map. By hovering the cursor over each country, a text box display provides the exact number or records in this population for the selected data module, while a visual impression is also provided with different shades of blue, depending on the total number of entries per data module (the darker the color the more data recorded; Figure 3A and C). By clicking on the left mouse button, a hyperlink appears at the bottom left side of the map, which directs the user to the data query page, where all available records for this selection are provided (Figure 3B and D).
Figure 3.
Overview of the new FINDbase map display visualization tool. (A) Overview of the documented genetic disease summaries in European countries (part of North Africa is also shown). By hovering the cursor over Greece, a text box appears providing an overview of the number of disease summaries documented in FINDbase for the Hellenic population. By clicking on the country, a link appears in the bottom down part of the map (indicated in a red rectangle). Clicking on the link opens a new browser page, where all the different genetic disease summaries in the (B) Hellenic population appear, where the user can navigate for further information. (C) Overview of the documented PGx markers in European countries (part of North Africa is also shown). Again, by clicking on a specific country (e.g. Poland), a link appears in the bottom down part of the map (indicated in a red rectangle), which opens a new browser page, where all the different PGx markers in the Polish population appear, (D) grouped per gene. Please note that the intensity of the blue color is indicative of the number of data records that are documented in FINDbase for every country per data module, respectively (see also Supplementary Figure S4).
CONCLUSIONS AND FUTURE PERSPECTIVES
FINDbase is a comprehensive online resource documenting the prevalence of clinically relevant genomic variation allele frequency data, serving a well-defined scientific discipline. Ever since its establishment in 2006, it is considered a reference repository of such information worldwide. To maximize its utility, access of FINDbase content is kept free of charge, while both the data content and the battery of data visualization tools have been significantly enriched. This was made possible thanks to the uninterrupted funding of this database since 2005, as part of major research Consortia, such as GEN2PHEN (http://www.gen2phen.org), RD-Connect (http://www.rd-connect.eu) and the Genomic Medicine Alliance (http://www.genomicmedicinealliance.org), also ensuring fruitful interactions with the field's leading experts.
The recent update of FINDbase data modules focused primarily in the PGx data collection, not only by expanding the existing data sets with information about the prevalence of clinically actionable PGx biomarkers in many European and other populations but, most importantly, by interrelating the PGx data module with DruGeVar, hence providing the interested users with the best of both resources. Also, the upgrade of the ETHNOS software, as an ‘off-the-shelf’ web application for NEGDB development and curation, offers a unique opportunity to interested users, with minimal or even no (bio)informatics background, either to start a new NEGDB from scratch or even to ‘adopt’ an existing NEGDB from the FINDbase collection and, together with a group of expert curators from their own country, to assume responsibility of enriching it with content, with all technical support being provided centrally by FINDbase administrators. This also contributes to harmonized NEGDB development and minimizes data content heterogeneity. Also, the interrelation of DruGeVar and FINDbase PGx module can catalyze the efforts of national regulatory agencies to develop guidelines to rationalize drug use in those populations, where certain clinically actionable PGx biomarkers reach high frequencies.
To accommodate the anticipated increased data generation, especially from next-generation sequencing (NGS) and microarray experiments, FINDbase architecture will have to be further modernized. In particular, we do not only plan to design a system for microarray (e.g. for the Affymetrix DMETTM Plus platform) or NGS data uploading, but also to improvise an algorithm to automatically calculate clinically relevant genomic variants in an aggregated manner to ensure data anonymity. Such increase in data coverage would require upgraded data visualization tools, using a variety of techniques, such as the drill-down approach.
These data will be contributed with microattribution, supporting additional forms of contributor identification apart from the ResearcherID (e.g. ORCID; http://www.orcid.org), and flagged with a certain label (e.g. mA), and deposited using DOI and/or PubMed ID, hence providing minimal credit to data contributors and curators. Such data are already available for testing as far as PGx biomarkers (11) and pharmacogenes (11,19) are concerned.
Acknowledgments
Most of the work described in this paper has been supported by European Commission grants [GEN2PHEN (FP7-200754), RD-CONNECT (FP7-305444), SEE_DRUG (FP7-285950), UPGx (H2020-668353) to G.P.P. and SERBORDISInn (FP7-316088) to S.P.], by a Greek State grant (EΠAvEK 2014-2020; ELIXIR_GR) to G.P.P., by a Serbian State grant (III 41004 MESTD RS to S.P.), and by funding from the Golden Helix Foundation (UK). The authors are grateful to Milena Ugrin, Dragica Radojkovic, Mila Ljujic, Aleksandra Divac Rankov for competent data mining and curation. Also, the authors are indebted to all FINDbase users worldwide for their valuable comments and suggestions, which helped us to keep the information as updated and complete as possible and also contributed to the continuous improvement of the database profile and contents.
Footnotes
Present address: Juha Muilu, BC Platforms, Espoo, Finland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission [GEN2PHEN (FP7-200754), RD-CONNECT (FP7-305444), SEE_DRUG (FP7-285950), UPGx (H2020-668353) to G.P.P. and SERBORDISInn (FP7-316088) to S.P.]; Greek State grant [EΠAvEK 2014-2020; ELIXIR_GR) to G.P.P.]; Serbian State grant [III 41004 MESTD RS to S.P.]; Golden Helix Foundation (UK). Funding for open access charge: European Commission UPGx grant [H2020-668353].
Conflict of interest statement. None declared.
REFERENCES
- 1.Patrinos G.P. National and Ethnic Mutation databases: Documenting populations’ genography. Hum. Mutat. 2006;27:879–887. doi: 10.1002/humu.20376. [DOI] [PubMed] [Google Scholar]
- 2.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J., et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D828. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitropoulou C., Webb A.J., Mitropoulos K., Brookes A.J., Patrinos G.P. Locus-specific database domain and data content analysis: evolution and content maturation toward clinical use. Hum. Mutat. 2010;31:1109–1116. doi: 10.1002/humu.21332. [DOI] [PubMed] [Google Scholar]
- 5.van Baal S., Zlotogora J., Lagoumintzis G., Gkantouna V., Tzimas I., Poulas K., Tsakalidis A., Romeo G., Patrinos G.P. ETHNOS: a versatile electronic tool for the development and curation of National Genetic databases. Hum. Genomics. 2010;4:361–368. doi: 10.1186/1479-7364-4-5-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Baal S., Kaimakis P., Phommarinh M., Koumbi D., Cuppens H., Riccardino F., Macek M., Jr, Scriver C.R., Patrinos G.P. FINDbase: a relational database recording frequencies of genetic defects leading to inherited disorders worldwide. Nucleic Acids Res. 2007;35:D690–D695. doi: 10.1093/nar/gkl934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patrinos G.P., Al Aama J., Al Aqeel A., Al-Mulla F., Borg J., Devereux A., Felice A.E., Macrae F., Marafie M.J., Petersen M.B., et al. Recommendations for genetic variation data capture in developing countries to ensure a comprehensive worldwide data collection. Hum. Mutat. 2011;32:2–9. doi: 10.1002/humu.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgitsi M., Viennas E., Gkantouna V., Christodoulopoulou E., Zagoriti Z., Tafrali C., Ntellos F., Giannakopoulou O., Boulakou A., Vlahopoulou P., et al. Population-specific documentation of pharmacogenomic markers and their allelic frequencies in FINDbase. Pharmacogenomics. 2011;12:49–58. doi: 10.2217/pgs.10.169. [DOI] [PubMed] [Google Scholar]
- 9.Georgitsi M., Viennas E., Gkantouna V., van Baal S., Petricoin E.F., Poulas K., Tzimas G., Patrinos G.P. FINDbase: A worldwide database for genetic variation allele frequencies updated. Nucleic Acids Res. 2011;39:D926–D932. doi: 10.1093/nar/gkq1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papadopoulos P., Viennas E., Gkantouna V., Pavlidis C., Bartsakoulia M., Ioannou Z.M., Ratbi I., Sefiani A., Tsaknakis J., Poulas K., et al. Developments in FINDbase worldwide database for clinically relevant genomic variation allele frequencies. Nucleic Acids Res. 2014;42:D1020–D1026. doi: 10.1093/nar/gkt1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizzi C., Dalabira E., Kumuthini J., Dzimiri N., Balogh I., Başak N., Böhm R., Borg J., Borgiani P., Bozina P., et al. A European spectrum of pharmacogenomic biomarkers: Implications for clinical pharmacogenomics. PLoS One. 2016;11:e0162866. doi: 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patrinos G.P., Cooper D.N., van Mulligen E., Gkantouna V., Tzimas G., Tatum Z., Schultes E., Roos M., Mons B. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum. Mutat. 2012;33:1503–1512. doi: 10.1002/humu.22144. [DOI] [PubMed] [Google Scholar]
- 13.Bell M. SOA Modeling patterns for service-oriented discovery and analysis. Hoboken: Wiley & Sons; 2010. [Google Scholar]
- 14.Giardine B., Borg J., Higgs D.R., Peterson K.R., Philipsen S., Maglott D., Singleton B.K., Anstee D.J., Basak A.N., Clark B., et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat. Genet. 2011;43:295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardon L.R., Harris T. Precision medicine, genomics and drug discovery. Hum. Mol. Genet. 2016;25:R166–R172. doi: 10.1093/hmg/ddw246. [DOI] [PubMed] [Google Scholar]
- 16.Maliepaard M., Nofziger C., Papaluca M., Zineh I., Uyama Y., Prasad K., Grimstein C., Pacanowski M., Ehmann F., Dossena S., et al. Pharmacogenetics in the evaluation of new drugs: a multiregional regulatory perspective. Nat. Rev. Drug Discov. 2013;12:103–115. doi: 10.1038/nrd3931. [DOI] [PubMed] [Google Scholar]
- 17.Dalabira E., Viennas E., Daki E., Komianou A., Bartsakoulia M., Poulas K., Katsila T., Tzimas G., Patrinos G.P. DruGeVar: an online resource triangulating drugs with genes and genomic biomarkers for clinical pharmacogenomics. Public Health Genomics. 2014;17:265–271. doi: 10.1159/000365895. [DOI] [PubMed] [Google Scholar]
- 18.Byrne M., Fokkema I.F., Lancaster O., Adamusiak T., Ahonen-Bishopp A., Atlan D., Béroud C., Cornell M., Dalgleish R., Devereau A., et al. VarioML framework for comprehensive variation data representation and exchange. BMC Bioinformatics. 2012;13:254. doi: 10.1186/1471-2105-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizzi C., Peters B., Mitropoulou C., Mitropoulos K., Katsila T., Agarwal M.R., van Schaik R.H., Drmanac R., Borg J., Patrinos G.P. Personalized pharmacogenomics profiling using whole-genome sequencing. Pharmacogenomics. 2014;15:1223–1234. doi: 10.2217/pgs.14.102. [DOI] [PubMed] [Google Scholar]