Abstract
The Protein Data Bank Japan (PDBj, http://pdbj.org), a member of the worldwide Protein Data Bank (wwPDB), accepts and processes the deposited data of experimentally determined macromolecular structures. While maintaining the archive in collaboration with other wwPDB partners, PDBj also provides a wide range of services and tools for analyzing structures and functions of proteins. We herein outline the updated web user interfaces together with RESTful web services and the backend relational database that support the former. To enhance the interoperability of the PDB data, we have previously developed PDB/RDF, PDB data in the Resource Description Framework (RDF) format, which is now a wwPDB standard called wwPDB/RDF. We have enhanced the connectivity of the wwPDB/RDF data by incorporating various external data resources. Services for searching, comparing and analyzing the ever-increasing large structures determined by hybrid methods are also described.
INTRODUCTION
The Protein Data Bank Japan (PDBj) (1) accepts and annotates macromolecular structure data in collaboration with other worldwide Protein Data Bank (wwPDB) (2,3) partners, PDBe (4), RCSB PDB (5) and BMRB (6). The wwPDB partners recently started a new, unified deposition and annotation system called OneDep. Accordingly, depositions from Asia and the Middle East are directed to the PDBj deposition site (http://deposit-pdbj.wwpdb.org) and are annotated by the annotators at PDBj.
In addition to processing depositions and maintaining the archive, PDBj provides a wide range of analysis tools and derived databases to facilitate structural biology and bioinformatics research. Since the last publication of our NAR DB article (1), PDBj has undergone a number of major changes regarding user interfaces and analysis tools as well as additional data provided. The previously described Resource Description Framework (RDF) format, PDB/RDF, is now one of the wwPDB standard formats called wwPDB/RDF and is enhanced with supplementary information in order to connect PDB data with other biological data resources.
USER INTERFACES
User interfaces include interactive (and graphical) web interfaces for humans and RESTful web services for computer programs. We also expose our backend database in the forms of web services or dump files for enabling very complex queries. These are described in turn.
Web interface
The web interface of PDBj was updated to provide a uniform integrated interface for the available services as well as to provide a scalable interface for devices ranging from smartphones to workstations. This update incorporates several innovative/renovative features as described below.
We have implemented various functionalities to ease novice as well as expert users to interact with inherently complex macromolecular structural information. For the novice user, we have created a series of interactive tutorials for PDBj's search services (http://pdbj.org/help/tutorials). In addition, the service finder on the PDBj top page helps the user to find particular services provided by PDBj. The omni-search service, which is present on the top of nearly every page at the PDBj web site, provides faceted navigation of search results categorized into PDB entries, general information pages, status search and chemical components (Chemie, see below). In fact, the omni-search provides candidate search results in these categories while the user is typing (Figure 1).
Figure 1.
Basic web user interface. (A) On the PDBj home page, the user finds the main menu for all the services (on the left side of the page), interactive tutorials (top center) and service finder (in the middle). The omni-search bar (top left) guides the user to input keywords that are categorized into service types. (B) A search result is shown in several facets (PDB entries, ‘Info pages’, release status and chemical components) where the given keywords are found. (C) In the summary page of each PDB entry are shown basic information of the entry such as the title, authors, citation and validation report. Interactive molecular graphics are available for asymmetric units as well as biological units using Molmil (8) or JSmol (asymmetric units only) (http://wiki.jmol.org/index.php/JSmol) (right bottom).
One of the most basic utilities for structural analysis is the molecular viewer. We previously developed the Java applet-based molecular viewer jV (7). However, modern devices such as smartphones and tablets don't support Java applets. In order to offer a high performance, high quality molecular viewer on these platforms as well as on regular workstation platforms where the usage of Java applets has also diminished in recent years, we have developed a new molecular viewer named Molmil and have deployed this for various services on our web site (8), including the database of electrostatic molecular surfaces (eF-site) (7) even for very large structures. During the development of Molmil, we also developed a new JSON based format derived from PDBx/mmCIF called PDBx/mmJSON (8). Molmil, as well as PDBj's Mine PDB Explorer service use this format. The user can download the mmJSON file of PDB entries via a REST service (see http://pdbj.org/help/file-formats). The summary page of each entry is semantically annotated with schema.org vocabulary (http://schema.org/) in a microformat. The Sagace search engine (http://sagace.nibiohn.go.jp/en/) (9), for example, exploits these annotations to display PDB-specific search results with thumbnails of macromolecular structures.
Along with the graphical interfaces on Web browsers, we have also renewed our RESTful web services. In fact, all services provided by the main PDBj web site are also available as a REST service as described on http://pdbj.org/help/rest-interface.
Chemie (http://pdbj.org/chemie-search) is a new service offered by PDBj. Much like its counterparts, RCSB PDB's Ligand Expo (http://ligand-expo.rcsb.org/) and PDBe's PDBeChem (http://www.ebi.ac.uk/pdbe-srv/pdbechem/), PDBj's Chemie offers users an interface to search and explore the Chemical Component Dictionary of the wwPDB via text search of various fields, as well as sub-structure search via SMILES with Tanimoto scoring using fingerprints (10) generated by the Open Babel software (11).
PDBj Mine RDB
The most basic infrastructure underlying PDBj's web services is the PDBj Mine relational database (RDB) which has been completely redesigned in recent years. When the old version of the PDBj Mine RDB was developed (12), PDBj's backend system depended on the ‘PDBMLplus’ files that were based on PDBj's own extension of the standard PDBML files (13). Since then, the PDBx/mmCIF format (14) has been adopted as the canonical file format of the wwPDB. Accordingly, we have separated the additional components of the PDBMLplus files from the core PDBML parts. This separation made it possible to design an easier-to-use RDB of the entire PDB data, hence PDBj Mine RDB version 2 offering a leaner database structure as well as better scalability for the recently released large structures. In a nutshell, each table in the PDBj Mine RDB schema corresponds to a category as defined in the PDBx/mmCIF dictionary and different tables are cross-referenced via foreign keys when they are defined in the dictionary and make sense in the RDB. A complete documentation of the schema is provided at http://pdbj.org/mine-rdb-docs. The RDB is implemented in PostgreSQL (version 9.3) (http://www.postgresql.org/) and the complete database dump and weekly updates are available at our ftp site (ftp://ftp.pdbj.org/mine2/ and see also http://pdbj.org/help/mine2-rdb-local-install for installation instruction) along with an extensive list of example SQL queries (http://pdbj.org/help/mine2-sql). SQL queries can be executed on the web browser (http://pdbj.org/mine/sql/) or via the REST interfaces with several options for the output file formats.
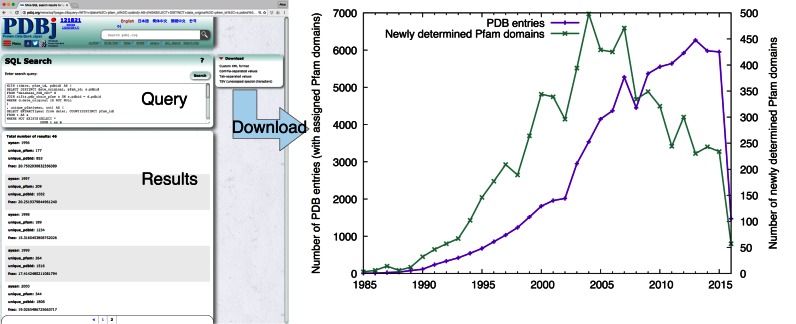
One weakness of the PDB data is the scarcity of up-to-date cross-references to other biological databases, which makes it difficult to interpret the biological relevance of macromolecular structures in a wider context. In order to solve this problem at least partially, we have also integrated the SIFTS resource developed by PDBe and UniProt (15) into the PDBj Mine RDB. Currently, the SIFTS summary files provided for ‘quick access’ (https://www.ebi.ac.uk/pdbe/docs/sifts/quick.html) are incorporated. Example queries that combine the PDB and SIFTS data are also provided at http://pdbj.org/help/mine2-sql. Using this resource, the user can analyze various trends in structural biology as reflected in the PDB. As an example, we show in Figure 2 the yearly change of newly identified Pfam (16) structures, which was obtained by a single SQL query using the web interface.
Figure 2.
An example of PDBj Mine SQL queries. Queries can be executed on the web browser as well as via the RESTful web service (the database dump for local installation is also available). The result of a query can be saved in such formats as a custom XML, comma-separated values or tab-separated values. The plot on the right shows the yearly change of newly determined Pfam domains in the PDB, as obtained from a single SQL query against the PDBj Mine RDB. (Note that the data in the years 2015 and 2016 are incomplete because not all the PDB entries deposited in these years have been released yet.)
WWPDB/RDF: PDB ON THE SEMANTIC WEB
Basic structure of wwPDB/RDF
Partly as a product of the BioHackathon (17), we started distributing PDB/RDF, the PDB data in the Resource Description Framework (RDF) format, in 2011 (1). The PDB/RDF data has become one of the standard formats of the wwPDB and is now called wwPDB/RDF provided at http://rdf.wwpdb.org/pdb/ on the web as well as on our FTP site at ftp://ftp.pdbj.org/RDF. We also provide the Chemical Component Dictionary as RDF (http://rdf.wwpdb.org/cc/). These RDF resources are referenced by some of the major data providers such as UniProt (18) and PubChem (19). More recently, we, in collaboration with the BMRB team, have also developed BMRB/RDF which translates nuclear magnetic resonance data in the BMRB into the RDF format accompanied with many links to external biological data resources (20).
The structure of the wwPDB/RDF is mostly unchanged from that which was previously described (1) except for the base URI (now http://rdf.wwpdb.org/ rather than http://pdbj.org/rdf/). That is, the ‘ontology’ of the wwPDB/RDF is based on the PDBx/mmCIF dictionary (http://mmcif.wwpdb.org/) (14) so that each PDBx/mmCIF category corresponds to an OWL class and each PDBx/mmCIF item is a data property in the RDF version. However, there are a few major enhancements in the current version for increased interoperability with other data resources. First, some descriptive attributes were added in the datablock layer of each entry. Second, many external resources are referred to by the URI's based on the Identifiers.org (http://identifiers.org/) hosted at European Bioinformatics Institute (21). These changes were recommended in the guideline proposed by the Database Center for Life Science (DBCLS) (http://wiki.lifesciencedb.jp/mw/RDFizingDatabaseGuideline [in Japanese]) that coordinates a database integration project in Japan. In the previous PDB/RDF, we excluded some categories such as entity_poly_seq in order to reduce the data volume. In the current version, however, we have added entity_poly_seq, pdbx_poly_seq_scheme and pdbx_nonpoly_scheme categories to wwPDB/RDF so that we can make residue-by-residue references to external resources based on, for example, the SIFTS PDB-UniProt sequence mapping. On the other hand, the atom_site category is still excluded from the distribution due to its large volume.
SIFTS enhancement
Similarly to the PDBj Mine RDB, wwPDB/RDF is also supplemented with the SIFTS resource (15) based on the SIFTS ‘Quick access’ files (https://www.ebi.ac.uk/pdbe/docs/sifts/quick.html). Our SIFTS RDF data are distributed as a separate release from the wwPDB/RDF at our FTP site (ftp://ftp.pdbj.org/RDF/sifts/). Nevertheless, thanks to the very design of the Semantic Web technologies of which RDF constitutes the core, integration of the SIFTS resource and wwPDB/RDF as well as other data resources should be trivial. To ensure the connectivity of various data, the links to external resources described in the SIFTS data are also based on the URI's provided by the Identifiers.org (http://identifiers.org/). In addition, we also provide sequence alignments between PDB and UniProt sequences using the FALDO ontology (22).
SPARQL endpoint at National Bioscience Database Center (NBDC RDF Portal)
The National Bioscience Database Center of Japan (NBDC) provides the RDF Portal (http://integbio.jp/rdf/) where they publish various resources (mostly developed in Japan) in the RDF format and provide a SPARQL endpoint to query these resources. The RDF Portal includes the entire wwPDB/RDF data with the SIFTS data together with other data resources. The aforementioned BMRB/RDF (20) is also a part of this portal. Currently, the wwPDB/RDF data are updated quarterly in January, April, July and October. Although the RDF Portal is not necessarily up-to-date, it provides a convenient access to the wwPDB/RDF data. Some example SPARQL queries are also provided at http://integbio.jp/rdf/?view=detail&id=pdbj.
ANALYSIS TOOLS FOR LARGE AND/OR COMPLEX STRUCTURES
Most of the services offered by the PDBj are summarized in Table 1 (35–49). Here, we focus on those related to analysis of large structures and complexes.
Table 1. Analysis tools and derived databases.
| Servicea | Description | URLb | Reference |
|---|---|---|---|
| PDBj Minec | Simple, advanced and SQL search for PDB data | pdbj.org/mine | |
| Chemie searchd | Searching the Chemical Component Dictionary | pdbj.org/chemie-search | |
| Sequence Navigatore | BLAST (27) search against PDB | pdbj.org/seq-navi | |
| Structure Navigatore | ASH structure similarity search | pdbj.org/struc-navi | (35) |
| SeSAW | Functional site prediction | pdbj.org/sesaw | (36) |
| GIRAF | Interaction interface similarity search | pdbj.org/giraf | (37,47–49) |
| Molmild | WebGL-based Molecular graphics viewer | pdbj.org/molmil | (8) |
| Yorodumie | Integrated PDB/EMDB data browser | pdbj.org/yorodumi | |
| ASH | Pairwise structure alignment | pdbj.org/ash | (35,38–39) |
| MAFTTash | Integrated multiple sequence/structure alignment | pdbj.org/mafftash | (39) |
| CRNPRED | One-dimensional structure prediction | pdbj.org/crnpred | (40,41) |
| Spanner | Homology modeling | pdbj.org/spanner | (42) |
| SFAS | Automated function annotation | pdbj.org/sfas | |
| eF-sitef | Electrostatic surface database | pdbj.org/ef-site | (7,43) |
| eF-seek | Search against eF-site | pdbj.org/ef-seek | (44) |
| HOMCOSd | Searching and modeling of protein complex | homcos.pdbj.org | (26) |
| ProMode Elasticg | ProMode with elastic network model | pdbj.org/promode-elastic | (31,32) |
| Molecule of the Monthe | Japanese translation of RCSB PDB MoM | pdbj.org/mom | (45) |
| PDBj Gamese | Simple and fun games | pdbj.org/pdbj-games |
aNew or largely updated services are in boldface.
bThe ‘http://’ prefix is omitted.
cSimplified RDB schema for the SQL search integrated with SIFTS resources(15), more powerful search engine and updated web interface.
dNew service.
eUpdated web interface.
fExtended database with large structures.
gUpdated web interface and extended database.
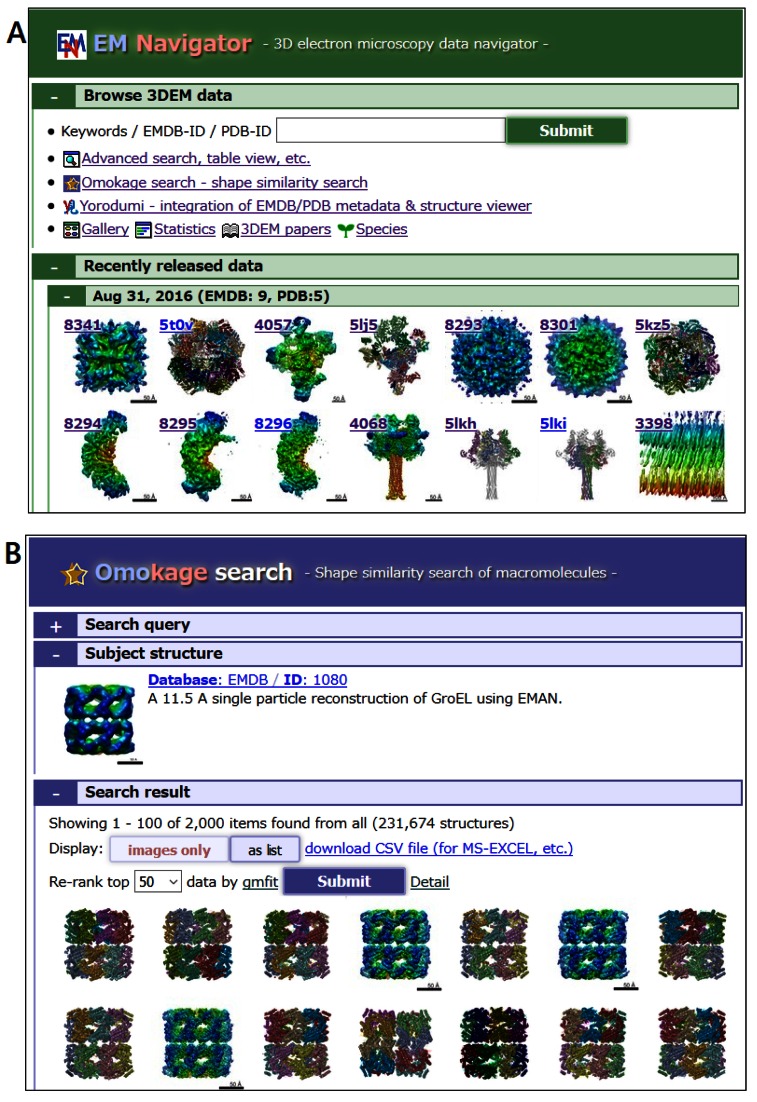
One of the exciting trends in structural biology in the recent years is the elucidation of very large complex structures by hybrid methods that combine electron microscopy (EM) and other methods. Since the information of EM structures is of very different characters than those of traditional PDB structures, it requires special treatment in visualization and analysis. We at the PDBj have been providing the EM Navigator (http://pdbj.org/emnavi/) for aiding the animated visualization of EM structures. The EM Navigator is now integrated with the Yorodumi browser (http://pdbj.org/emnavi/viewtop.php) so that entries in the PDB and EMDB can be browsed in a single integrated interface (Figure 3A). As the number of EM structures increases, there will be a demand for comparing (and classifying) those structures. The Omokage search (23) provides structural similarity search over macromolecular assemblies in both the PDB and EMDB based on electron density, independent of sequence order and number of subunits. For a given query structure (in the form of either atomic coordinates or an electron density map), Omokage first searches globally similar structures in the PDB and EMDB that have a similar one-dimensional profile (a vector of global shape features), and then generates the ‘alignment’ of structures by fitting Gaussian mixture models (Figure 3B) (24).
Figure 3.
Analysis tools for the PDB and EMDB. (A) The EM Navigator/Yorodumi (http://pdbj.org/emnavi) provides an integrated interface to the PDB and EMDB combined. (B) The Omokage search (http://pdbj.org/emnavi/omo-search.php) performs shape similarity searches through both the PDB and EMDB.
Another recent trend in structural biology focuses on complex structures between proteins or between proteins and chemical compounds (25). To support analyses and modeling of complex structures, we have added HOMCOS (26), a tool for searching, analyzing and modeling of complex structures based on structural similarities. The HOMCOS server supports several functionalities. First, possible dimeric complexes of proteins are searched for a given pair of protein sequences (based on sequence similarities using BLAST (27)), which serve as templates for modeling the complex structures. Second, possible protein–compound complexes are searched for a pair of a protein sequence and the structure of a chemical compound (in either 3D or 2D). While the protein similarities are detected by BLAST, the similarities between chemical compounds are detected by the KCOMBU program (28,29) and superposed by the FKCOMBU program (30). Third, for a given protein sequence, residue-wise annotations of possible ligand binding sites and effects of mutations are listed. These annotations are imported from the UniProt (18) database in addition to complex structures in the PDB.
Understanding protein functions often requires analysis of protein dynamics. However, running molecular dynamics simulations and analyzing the trajectories are not always easy for the casual user of the PDB. We therefore provide ProMode (31–33) which is a database of protein dynamics computed from normal mode analysis (NMA). The original ProMode database was derived from the NMA based on a physicochemical potential function (31). It was, however, computationally very demanding and could not scale to large structures. We have therefore developed ProMode Elastic (32) which is based on the (all-atom) elastic network model (34) of native protein structures and thus alleviates energy minimization. We are currently in the process of extending the ProMode Elastic to cover many of the (moderately) large complexes in the PDB.
CONCLUSION
PDBj accepts and processes the deposited macromolecular structural data through the OneDep system as one of the members of the wwPDB. In addition to keeping and providing the common structural archive, PDBj develops our own services. In particular, semantic web tools based on RDF could be useful to link PDB data with other external biological data resources. Other tools to analyze large and complex macromolecular structures following the recent trends in structural biology should also support researchers in the wide range of the life science field.
Acknowledgments
The authors thank the PDBj annotators, Reiko Igarashi, Yumiko Kengaku, Hasumi Cho, Junko Sato for their constant effort that makes the wwPDB possible at all, and Reiko Yamashita and Takahiro Kudou for technical support. A. R. K. thanks Shuichi Kawashima (DBCLS) and Katsuhiko Okubo (NBDC) for their help in improving the design of the wwPDB/RDF, and Michelle Ragsac for assisting with SQL queries in Figure 2. Y. T. thanks Shigeru Endo for helping with the ProMode Elastic service.
FUNDING
Database Integration Coordination Program from the National Bioscience Database Center (NBDC); Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from Japan Agency for Medical Research and Development (AMED) (in part); JSPS KAKENHI [26440078] (in part). Funding for open access charge: Database Integration Coordination Program from the National Bioscience Database Center (NBDC), JST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kinjo A.R., Suzuki H., Yamashita R., Ikegawa Y., Kudo T., Igarashi R., Kengaku Y., Cho H., Standley D.M., Nakagawa A., et al. Protein Data Bank Japan (PDBj): Maintaining a structural data archive and Resource Description Framework format. Nucleic Acids Res. 2012;40:D453–D460. doi: 10.1093/nar/gkr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H., Henrick K., Nakamura H., Markley J.L. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H.M., Burley S.K., Kleywegt G.J., Markley J.L., Nakamura H., Velankar S. The archiving and dissemination of biological structure data. Curr. Opin. Struct. Biol. 2016;40:17–22. doi: 10.1016/j.sbi.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velankar S., van Ginkel G., Alhroub Y., Battle G.M., Berrisford J.M., Conroy M.J., Dana J.M., Gore S.P., Gutmanas A., Haslam P., et al. PDBe: improved accessibility of macromolecular structure data from PDB and EMDB. Nucleic Acids Res. 2016;44:D385–D395. doi: 10.1093/nar/gkv1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J., et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., et al. BioMagResBank. Nuleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinoshita K., Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–1330. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- 8.Bekker G.-J., Nakamura H., Kinjo A.R. Molmil: a molecular viewer for the PDB and beyond. J. Cheminform. 2016;8:42. doi: 10.1186/s13321-016-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita M., Igarashi Y., Ito M., Chen Y.-A., Nagao C., Sakaguchi Y., Sakate R., Masui T., Mizuguchi K. Sagace: a web-based search engine for biomedical databases in Japan. BMC Res. Notes. 2012;5:1–5. doi: 10.1186/1756-0500-5-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cereto-Massagué A., Ojeda M.J., Valls C., Mulero M., Garcia-Vallvé S., Pujadas G. Molecular fingerprint similarity search in virtual screening. Methods. 2015;71:58–63. doi: 10.1016/j.ymeth.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 11.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminform. 2011;3:1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinjo A.R., Yamashita R., Nakamura H. PDBj mine: design and implementation of relational database interface for Protein Data Bank Japan. Database. 2010;2010:baq021. doi: 10.1093/database/baq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westbrook J., Ito N., Nakamura H., Henrick K., Berman H.M. PDBML: the representation of archival macromolecular structure data in XML. Bioinformatics. 2005;21:988–992. doi: 10.1093/bioinformatics/bti082. [DOI] [PubMed] [Google Scholar]
- 14.Westbrook J.D., Bourne P.E. STAR/mmCIF: an ontology for macromolecular structure. Bioinformatics. 2000;16:159–168. doi: 10.1093/bioinformatics/16.2.159. [DOI] [PubMed] [Google Scholar]
- 15.Velankar S., Dana J.M., Jacobsen J., van Ginkel G., Gane P.J., Luo J., Oldfield T.J., O'Donovan C., Martin M.J., Kleywegt G.J. SIFTS: structure integration with function, taxonomy and sequences resource. Nucleic Acids Res. 2013;41:D483–D489. doi: 10.1093/nar/gks1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. The Pfam protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama T., Wilkinson M.D., Micklem G., Kawashima S., Yamaguchi A., Nakao M., Yamamoto Y., Okamoto S., Oouchida K., Chun H.-W., et al. The 3rd DBCLS BioHackathon: improving life science data integration with Semantic Web technologies. J. Biomed. Semantics. 2013;4:1–17. doi: 10.1186/2041-1480-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokochi M., Kobayashi N., Ulrich E.L., Kinjo A.R., Iwata T., Ioannidis Y.E., Livny M., Markley J.L., Nakamura H., Kojima C., et al. Publication of nuclear magnetic resonance experimental data with semantic web technology and the application thereof to biomedical research of proteins. J. Biomed. Semantics. 2016;7:16. doi: 10.1186/s13326-016-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wimalaratne S.M., Bolleman J., Juty N., Katayama T., Dumontier M., Redaschi N., Le Novère N., Hermjakob H., Laibe C. SPARQL-enabled identifier conversion with Identifiers.org. Bioinformatics. 2015;31:1875–1877. doi: 10.1093/bioinformatics/btv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolleman J.T., Mungall C.J., Strozzi F., Baran J., Dumontier M., Bonnal R. P.J., Buels R., Hoehndorf R., Fujisawa T., Katayama T., et al. FALDO: a semantic standard for describing the location of nucleotide and protein feature annotation. J. Biomed. Semantics. 2016;7:1–12. doi: 10.1186/s13326-016-0067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki H., Kawabata T., Nakamura H. Omokage search: shape similarity search service for biomolecular structures in both the PDB and EMDB. Bioinformatics. 2016;32:619–620. doi: 10.1093/bioinformatics/btv614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawabata T. Multiple subunit fitting into a low-resolution density map of a macromolecular complex using a gaussian mixture model. Biophys. J. 2008;95:4643–4658. doi: 10.1529/biophysj.108.137125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiebel J., Krimmer S.G., Röwer K., Knörlein A., Wang X., Park A.Y., Stieler M., Ehrmann F.R., Fu K., Radeva N., et al. High-throughput crystallography: reliable and efficient identification of fragment hits. Structure. 2016;24:1398–1409. doi: 10.1016/j.str.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Kawabata T. HOMCOS: an updated server to search and model complex 3D structures. J. Struct. Funct. Genomics. 2016 doi: 10.1007/s10969-016-9208-y. doi:10.1007/s10969-016-9208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.L. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawabata T. Build-up algorithm for atomic correspondence between chemical structures. J. Chem. Info. Model. 2011;51:1775–1787. doi: 10.1021/ci2001023. [DOI] [PubMed] [Google Scholar]
- 29.Kawabata T., Sugihara Y., Fukunishi Y., Nakamura H. LigandBox: a database for 3D structures of chemical compounds. Biophysics. 2013;9:113–121. doi: 10.2142/biophysics.9.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawabata T., Nakamura H. 3D flexible alignment using 2D maximum common substructure: dependence of prediction accuracy on target-reference chemical similarity. J. Chem. Info. Model. 2014;54:1850–1863. doi: 10.1021/ci500006d. [DOI] [PubMed] [Google Scholar]
- 31.Wako H., Kato M., Endo S. ProMode: a database of normal mode analyses on protein molecules with a full-atom model. Bioinformatics. 2004;20:2035–2043. doi: 10.1093/bioinformatics/bth197. [DOI] [PubMed] [Google Scholar]
- 32.Wako H., Endo S. Normal mode analysis based on an elastic network model for biomolecules in the Protein Data Bank, which uses dihedral angles as independent variables. Comput. Biol. Chem. 2013;44:22–30. doi: 10.1016/j.compbiolchem.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Tsuchiya Y., Kinoshita K., Endo S., Wako H. Dynamic features of homodimer interfaces calculated by normal-mode analysis. Protein Sci. 2012;21:1503–1513. doi: 10.1002/pro.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tirion M.M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 35.Standley D.M., Toh H., Nakamura H. ASH structure alignment package: sensitivity and selectivity in domain classification. BMC Bioinformatics. 2007;8:116. doi: 10.1186/1471-2105-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Standley D.M., Yamashita R., Kinjo A.R., Toh H., Nakamura H. SeSAW: balancing sequence and structural information in protein functional mapping. Bioinformatics. 2010;26:1258–1259. doi: 10.1093/bioinformatics/btq116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinjo A.R., Nakamura H. Similarity search for local protein structures at atomic resolution by exploiting a database management system. Biophysics. 2007;3:75–84. doi: 10.2142/biophysics.3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Standley D.M., Toh H., Nakamura H. GASH: an improved algorithm for maximizing the number of equivalent residues between two protein structures. BMC Bioinformatics. 2005;6:221. doi: 10.1186/1471-2105-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 40.Kinjo A.R., Nishikawa K. Predicting secondary structures, contact numbers, and residue-wise contact orders of native protein structure from amino acid sequence using critical random networks. Biophysics. 2005;1:67–74. doi: 10.2142/biophysics.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinjo A.R., Nishikawa K. CRNPRED: highly accurate prediction of one-dimensional protein structures by large-scale critical random networks. BMC Bioinformatics. 2006;7:401. doi: 10.1186/1471-2105-7-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lis M., Kim T., Sarmiento J., Kuroda D., Dinh H., Kinjo A.R., Amada K., Devadas S., Nakamura H., Standley D.M. Bridging the gap between single-template and fragment based protein structure modeling using Spanner. Immunome Res. 2011;7:1–8. [Google Scholar]
- 43.Kinoshita K., Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami Y., Kinoshita K., Kinjo A.R., Nakamura H. Exhaustive comparison and classification of ligand-binding surfaces in proteins. Protein Sci. 2013;22:1379–1391. doi: 10.1002/pro.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodsell D.S., Dutta S., Zardecki C., Voigt M., Berman H.M., Burley S.K. The RCSB PDB ‘Molecule of the Month’: Inspiring a Molecular View of Biology. PLoS Biol. 2015;13:e1002140. doi: 10.1371/journal.pbio.1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Standley D.M., Toh H., Nakamura H. Detecting local structural similarity in proteins by maximizing number of equivalent residues. Proteins. 2004;57:381–391. doi: 10.1002/prot.20211. [DOI] [PubMed] [Google Scholar]
- 47.Kinjo A.R., Nakamura H. Comprehensive structural classification of ligand binding motifs in proteins. Structure. 2009;17:234–246. doi: 10.1016/j.str.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Kinjo A.R., Nakamura H. Geometric similarities of protein-protein interfaces at atomic resolution are only observed within homologous families: an exhaustive structural classification study. J. Mol. Biol. 2010;399:526–540. doi: 10.1016/j.jmb.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 49.Kinjo A.R., Nakamura H. GIRAF: a method for fast search and flexible alignment of ligand binding interfaces in proteins at atomic resolution. Biophysics. 2012;8:79–94. doi: 10.2142/biophysics.8.79. [DOI] [PMC free article] [PubMed] [Google Scholar]