Abstract
The FlyRNAi database of the Drosophila RNAi Screening Center (DRSC) and Transgenic RNAi Project (TRiP) at Harvard Medical School and associated DRSC/TRiP Functional Genomics Resources website (http://fgr.hms.harvard.edu) serve as a reagent production tracking system, screen data repository, and portal to the community. Through this portal, we make available protocols, online tools, and other resources useful to researchers at all stages of high-throughput functional genomics screening, from assay design and reagent identification to data analysis and interpretation. In this update, we describe recent changes and additions to our website, database and suite of online tools. Recent changes reflect a shift in our focus from a single technology (RNAi) and model species (Drosophila) to the application of additional technologies (e.g. CRISPR) and support of integrated, cross-species approaches to uncovering gene function using functional genomics and other approaches.
INTRODUCTION
Functional genomics, which includes the large-scale application of techniques such as RNA interference (RNAi) and CRISPR-Cas9 approaches, opens the doors to both unbiased and focused assays of gene function. Applying functional genomics approaches at large scale requires expertise in reagent design, reagent production, high-throughput screening and large-scale data management, analysis and integration. The Drosophila RNAi Screening Center (DRSC) at Harvard Medical School (HMS) was established in 2003 to support genome-wide RNAi screening in Drosophila cultured cells. In 2008, we additionally launched large-scale production of the Transgenic RNAi Project (TRiP) fly stock collection and added support for in vivo RNAi reagent identification and data sharing. Ongoing efforts now include the large-scale application of CRISPR-Cas9 approaches in Drosophila cells and in vivo, including design and production of single guide RNA (gRNA) fly stocks. We have expanded the FlyRNAi database and website over the years to accommodate new reagent types and data sets, as well as to support a growing number of bioinformatics analyses and approaches (1).
Bioinformatics and database tracking are essential to every step in our functional genomics platform. Our needs include reagent design, reagent production tracking, data management, analysis and integration, and public availability of online tools, reagent information and data sets. For Drosophila research specifically, we have developed robust approaches for design and large-scale production of reagents (1–4), as well as made reagents and resulting data publicly available online (1,2). We also provide searchable access to information from other sources, including FlyBase (5). For functional genomics applications more generally, we have developed a portal for RNAi reagent evaluation and retrieval that automatically updates with new gene annotations releases (fly, worm, human) and when public RNAi resources are available (6); developed a data analysis and visualization tool for high-throughput data analysis that is based on protein complexes (fly, yeast, human) (7); and developed an approach to ortholog identification (8) now adopted by other resources, e.g. FlyBase (5). Altogether, this suite of tools allows us to support a number of applications with broad utility (Table 1) and those of specific interest to the Drosophila research community (Table 2).
Table 1. Navigating resources at DRSC/TRiP relevant to Drosophila and other species.
| Question | Species supported | Resource |
|---|---|---|
| Where can I view an overview of the online tools? | Overview page http://fgr.hms.harvard.edu/tools | |
| What miRNA-like seed sequence targets are enriched in my large-scale siRNA or shRNA screen data? | fly, human, mouse (and other if a reference file is provided) | Online GESS http://www.flyrnai.org/gess/ |
| What known and predicted protein complexes are enriched in my high-throughput functional genomics data set? | fly, budding yeast, human | COMPLEAT http://www.flyrnai.org/compleat/ |
| Has Gene X been conserved across species? | fly, budding yeast, fission yeast, worm, frog (X. tropicalis), zebrafish, mouse, rat, human | DIOPT http://www.flyrnai.org/diopt |
| Is the human ortholog of Gene X associated with disease? | fly, budding yeast, fission yeast, worm, frog (X. tropicalis), zebrafish, mouse, rat, human | DIOPT-DIST http://www.flyrnai.org/diopt-dist |
Table 2. Navigating Drosophila-specific resources at DRSC/TRiP.
| Question | Resource |
|---|---|
| Where can I view an overview of the online tools? | Overview page http://fgr.hms.harvard.edu/tools |
| In what cells, stages, or tissues is Gene X expressed? | DGET http://flyrnai.org/tools/dget/web/ |
| To what functional groups does Gene X belong?a | GLAD http://flyrnai.org/tools/glad/web/ |
| Where can I design dsRNAs against Gene X? | SnapDragon http://flyrnai.org/cgi-bin/RNAi_find_primers.pl |
| Where can I learn if Gene X scored in a cell-based screen? | Gene Lookup http://flyrnai.org/cgi-bin/DRSC_gene_lookup.pl |
| Where can I view all public DRSC cell-based screens and data? | Screen Summary http://flyrnai.org/screensummary |
| Where can I find all existing cell RNAi reagents targeting Gene X? | UP-TORR http://flyrnai.org/up-torr/ |
| Where can I find all existing in vivo RNAi fly stocks targeting Gene X? | UP-TORR http://flyrnai.org/up-torr/ |
| Where can I learn if existing in vivo RNAi fly stocks have phenotypes? | RSVP http://flyrnai.org/cgi-bin/RSVP_search.pl |
| Where can I find qPCR primers for Gene X? | FlyPrimerBank http://flyrnai.org/FlyPrimerBank |
| Where can I view gRNAs in the context of Gene X? | Find CRISPR http://flyrnai.org/crispr2/ |
| Where can I evaluate predicted efficiency of a given gRNA? | CRISPR efficiency http://flyrnai.org/evaluateCrispr |
| Where can I find protocols for Drosophila research? | Drosophila Protocols Portal http://flyrnai.org/tools/protocols/web/ |
| How do my hits connect with each other physically and phenotypically? | SignedPPI http://flyrnai.org/SignedPPI/ |
aAlso recommended, FlyBase Gene Groups (http://flybase.org/static_pages/FBgg/browse.html).
IMPROVEMENTS TO THE DATABASE AND WEBSITE
Updates to the database for reagent production and screen data management
RNAi reagent production and screen data management at the DRSC and TRiP has been supported by a relational database at the backend since 2003. The organization of the backend database has changed significantly since our 2012 update to accommodate new reagent types, new data types and to support our growing suite of online tools (see below and Tables 1 and 2), e.g. with new gene annotation information from model organism databases. Major additions in terms of RNAi reagents and data since 2012 include the reagents and data for new focused cell-based RNAi sub-libraries, for example, genes that encode RNA-binding proteins (complete list at http://fgr.hms.harvard.edu/drsc-focused-sub-libraries), as well as reagent and validation data for the large number of in vivo fly stocks built by our TRiP reagent production platform (3), for example, a human disease related collection (HuDis, http://www.flyrnai.org/HuDis-TRiP).
Updates to the website
We recently re-designed and reorganized our website (Figure 1), including a shift to a content managed system. The new DRSC/TRiP-Functional Genomics Resources website (http://fgr.hms.harvard.edu/) serves as a portal to the FlyRNAi database and other resources. At the site, we organized supporting information for our online tools (name, URL, version, etc.) in a standard format, with the expectation this will help researchers find and use the most appropriate online tools at our site for their applications. For each tool, we now have an overview page aimed at new users (e.g. http://fgr.hms.harvard.edu/compleat) and a more detailed documentation page (e.g. http://fgr.hms.harvard.edu/compleat-documentation). The website is hosted by a third party in the cloud. Since the FlyRNAi database and online tools themselves remain hosted by the HMS Research Computing Group, the tools remain at their original flyrnai.org URLs (e.g. http://www.flyrnai.org/compleat/). By moving to a content-managed system (OpenScholar) and using an online form for bug reports (Qualtrics), we are able to more easily receive and respond to community feedback, as well as more readily alert the community to new online tools, updates, and features. We have also shifted from an in-house built reagent request and equipment sign-up system to using Stratocore PPMS software to manage online requests for instrument training, instrument hours, and reagent libraries.
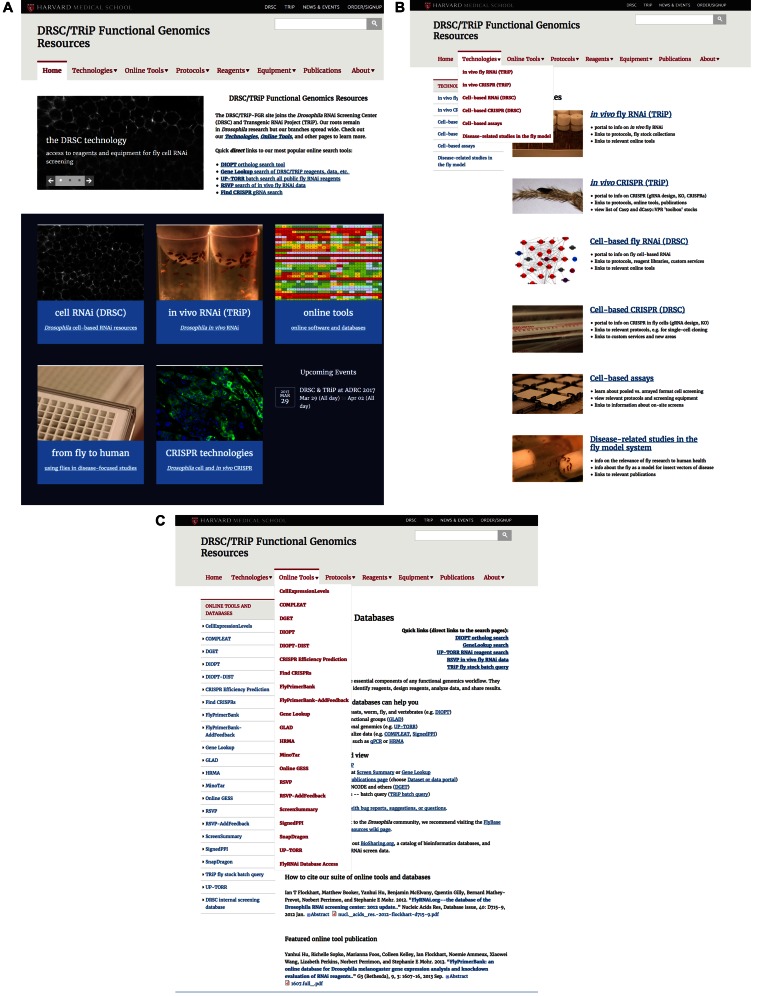
Figure 1.
DRSC/TRiP Functional Genomics Resources website: a new portal to the FlyRNAi database and online tools. We performed a complete re-design and reorganization of our website (http://fgr.hms.harvard.edu), which provides information about our RNAi and CRISPR platforms for Drosophila cell-based and in vivo systems, as well as providing information about and access to our database and online tools. (A) Home page. (B) Technologies overview page. (C) Online tools overview page, including the tools menu bar.
RECENT DEVELOPMENTS
Improvements to the DIOPT ortholog search tool
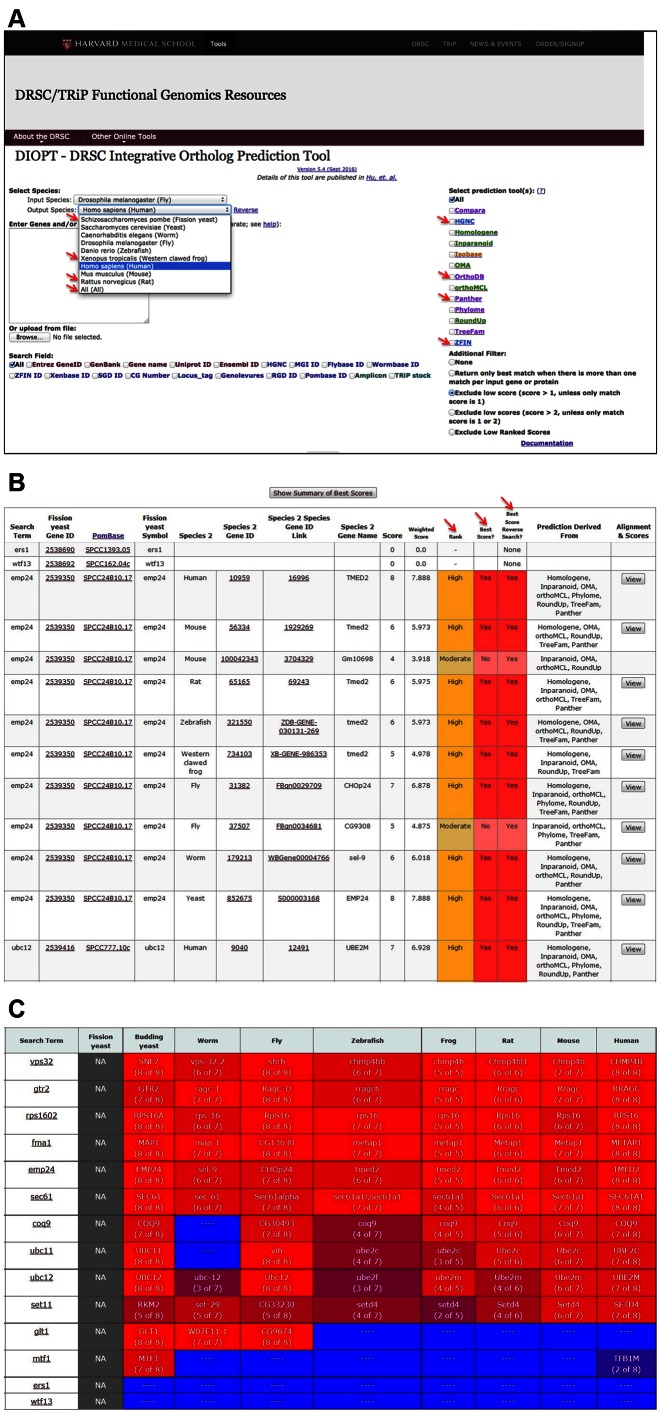
Identification of orthologs is a common step in functional genomics pipelines, for example during construction of a gene list for a targeted functional approach. Our DRSC Integrative Ortholog Prediction Tool, DIOPT (8) (Figure 2, Table 1), provides a convenient, downloadable mechanism for high-confidence or ‘wide net’ ortholog mapping and is our most-used online tool. The underlying data at DIOPT were updated to include three more species, the frog X. tropicalis, fission yeast, and rat, adding to existing coverage of human, mouse, fish, fly, worm and budding yeast. We also expanded DIOPT to include ortholog predictions from four additional algorithms or sources, i.e. orthoDB (9), Panther (10) and manually assembled ortholog relationships from HGNC (11) and Zfin (12). Notably, DIOPT was recently integrated into FlyBase. Thus, researchers can initiate a DIOPT-based ortholog search either at our site (http://www.flyrnai.org/diopt) or from the FlyBase home page (http://flybase.org/). DIOPT originally supported pairwise searches (species A vs. species B). We recently added to functionality and display at our DIOPT user interface (Figure 2). For example, users can now opt to view results for all species (i.e. species A versus all species). Moreover, in results tables, users now see a color-coded indication of which predicted ortholog pair has the best score, as well as whether or not that pair is the top-scoring match for the reverse search (species B versus species A). When a user selects ‘all’ as the output, the results page additionally includes a link (‘Show Summary of Best Scores’) to a heat map view of the best-scoring ortholog pair for each comparison species. As shown in Figure 2, this can give a quick look at conservation across major groups. The related tool DIOPT-Diseases and Traits, DIOPT-DIST, facilitates identification of orthologs of model species in humans, and further, connects those human genes with diseases; users can start a search with a model organism gene(s) or with a disease term.
Figure 2.
Improved user interface and additions to the DIOPT ortholog search tool. Since the initial release of DIOPT in 2013 (vs1), there have been four additional major releases. (A) DIOPT search page. As indicated (red arrows), more model organisms and prediction algorithms have been added. (B) Example results page. At the most recent release (vs5), we added annotation that indicates, for a given ortholog pair, if that pair is the best match based on the DIOPT score. We also indicate if it is the best match if the reverse search was done. If the pair is the best score with both forward and reverse searches, and the DIOPT score is 2 or up, we annotate this pair as ‘high’ in rank. Detailed information about the ranking system can be found http://fgr.hms.harvard.edu/diopt-documentation. Red arrows highlight these three added features in the results page. We also added an option for an ‘all species’ search that allows the user to retrieve the orthologs from all other model organisms. As shown in panel (C), this includes an option to view a summary of the best matches in a heat map format.
Defining gene and reagent lists for functional genomics and other projects
We have a number of tools that help researchers define the optimal set of genes to be included in a high- or low-throughput study (Tables 1 and 2, Figure 3). DIOPT and DIOPT-DIST can help people to select genes based on data or annotations from different species, or on human disease associations. Our Gene List Annotation for Drosophila, GLAD (13), allows researchers to view genes in curated functional groups, such as kinases or components of a signaling pathway. At the Drosophila Gene Expression Tool, DGET (http://biorxiv.org/content/early/2016/09/15/075358), a user can retrieve expression profile data from RNAseq studies by the modENCODE consortium and specific laboratories (14–16) for Drosophila cell lines, tissues, developmental stages and treatments. Using DGET, researchers can refine target gene lists or design experiments based on gene expression (e.g. include only those genes for which the existing public data suggests that the gene is expressed in the tissue of interest).
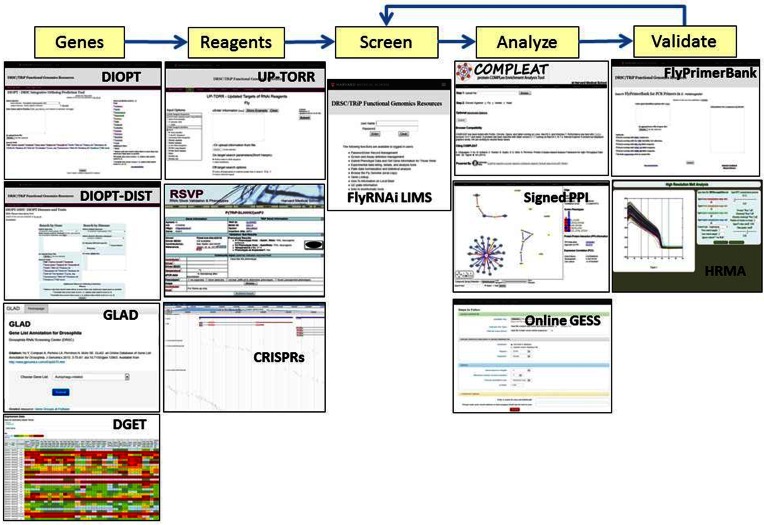
Figure 3.
Using online tools in a functional genomics workflow. Various online tools supported by our database can be used for specific steps in a functional genomics workflow, including defining a gene list, defining a reagent list, data analysis and data integration and reagent or experimental validation.
We also have online tools that help researchers to identify optimal reagents (Figure 3). We established the RNAi Stock Validation and Phenotype tool, RSVP, to store qPCR and phenotype data for RNAi transgenic fly stocks. Data in RSVP were collected from our own publications and unpublished phenotyping data, and from curated information at FlyBase. There is also an option for users to input their own results with specific combinations of in vivo RNAi fly stocks and Gal4 drivers. Updated Targets Of RNAi Reagents, UP-TORR, allows researchers to retrieve in batch mode all RNAi reagents for cell-based or in vivo studies from our own and other public resources. The reagent information is based on automatically updated gene annotations. Drosophila RNAi reagent results at UP-TORR link to corresponding validation and phenotype data at RSVP, allowing researchers to select reagents based on both bioinformatics analysis and experimental results. FlyPrimerBank, a resource of precomputed genome-wide qPCR primer designs based on transcript sequence, also combines bioinformatics prediction with experimental results, as in addition to the pre-computed qPCR primer designs, the resource also includes our own and community-provided feedback on experimental tests of primer quality are also incorporated into the database.
Many of these resources improve as more data are shared by the community. As mentioned above, RSVP provides data submission user interface that allows researchers to provide qPCR knockdown efficiency or phenotype data for RNAi fly stocks. Moreover, as stated above, FlyPrimerBank provides a feedback submission page that allows users to provide feedback as to whether or not qPCR primers meet certain quality standards. To highlight and encourage feedback submissions, our new website treats the ‘Add Feedback’ functions of these tools as separate online tools on the list of Online Tools (see http://fgr.hms.harvard.edu/tools).
Suite of tools for CRISPR-Cas9 applications
CRISPR-Cas9 is allowing for rapid advancement in gene editing and other functional genomics approaches, including in Drosophila. Bioinformatics support is important for selecting effective guide RNAs (gRNAs) and defining target sites, e.g. for in vivo CRISPR activation (CRISPRa) (17). Support is needed for both small-scale projects and design of genome-wide gRNA libraries (18). We have three resources to help users design CRISPR-based studies and detect CRISPR-edited mutations. Find CRISPRs is a pre-computed database of CRISPR gRNA designs (19). It presents gRNA designs in the context of a JBrowse genome browser (20) to facilitate selection of gRNAs in specific gene regions. We annotated all of the potential off-target sites with different stringencies, and calculated efficiency and potential frameshift scores, and make these results available to help users choose the most appropriate gRNA designs. gRNA efficiency prediction is a stand-alone tool for calculation of efficiency scores and evaluation of gRNAs designed using any algorithm (19). The High Resolution Melt Analysis or HRMA tool was developed to help users evaluate genomic DNA changes introduced by CRISPR–Cas9 genome editing (21).
Drosophila protocols portal
Access to protocols has in one sense been facilitated by the internet, e.g. via easy search and download of relevant papers and step-by-step protocols at lab websites. However, replacement of authoritative protocol books with online resources distributed in various places (journals, YouTube, etc.) can make it hard to find reliable protocols that can be put to practical use in the lab. To address a need for a centralized resource, we built a test version of a Drosophila Protocols Portal (http://www.flyrnai.org/tools/protocols/web/). Protocols from various sources were curated by our staff, organized into topic areas, and tagged with relevant keywords. At the portal, protocols can be viewed by category or searched via query of text associated with the resource (e.g. title) and curated keywords. If there is sufficient community interest, we or others (e.g. FlyBase) will expand the resource, such as to include comments or ratings of protocols and to accept new protocol submissions by the community.
Support for data sharing, analysis and visualization
We added or updated since 2012 a number of resources and tools that support data sharing, analysis, and visualization (see Tables 1 and 2, Figure 3). The COMPlex Enrichment Analysis Tool, COMPLEAT (7), allows a researcher to do enrichment analysis of large-scale datasets based on protein complex annotations. In the Signed Protein–Protein Interactions resource, SignedPPI (22), protein–protein interaction ‘edges’ have been assigned signs based on functional genomics data that indicate positive (activation) or negative (inhibition) relationships between nodes. SignedPPI helps researchers access, build, and navigate signed PPIs for development of new hypotheses regarding network connectivity and functional interactions.Online GESS (23), which is based on the Genome-wide Enrichment of Seed Sequence matches (GESS) algorithm developed by Sigoillot et al. (24), helps researchers analyze microRNA-type off-targets in siRNA or shRNA screen datasets for human, mouse, Drosophila, or any other species for which a reference sequence is provided.
Interaction with other resources
We interact with other databases via import of external data and sharing of our reagent and screen data with meta-databases. We rely on FlyBase (5) for gene mapping and other curated data. In addition, we use data from other groups as sources of information for specific online tools; for example, DGET (http://biorxiv.org/content/early/2016/09/15/075358), includes data from the modENCODE consortium and individual labs (14–16). We have deposited DRSC reagent information into NCBI PubChem Probe and Sequence (25), and keep these data updated when we add new libraries. TRiP fly stock information is deposited with FlyBase and the Bloomington Drosophila Stock Center. We also deposit experimental data from screens done on-site at our facility or off-site using our libraries into GenomeRNAi (26) and PubChem BioAssay (25).
FUTURE DIRECTIONS
In our 2012 update, we noted two main challenges, collection of full data for screens and providing online view of screen image datasets. Availability of screen data allows researchers interested in one or a few genes to check if they were ‘hits’ in any screens, and facilitates meta-analysis, as exemplified by SignedPPI (22). Our efforts to collect and display screen data has been helped by reviewers, who are increasingly asking that authors share data in public repositories such as FlyRNAi and/or PubChem. Regarding image data, we found a practical and free way to provide access to images for a small-scale screen, using Flickr as a repository for the images and a look-up table to help researchers view images for specific reagents or genes of interest (27) (see http://www.flyrnai.org/DRSC-RBP_data.php). This is not an ideal solution, however, and online availability of screen images remains a barrier overall. OMERO (28) provides a possible mechanism for addressing this in the future and we are pursuing this in collaboration with image data management experts.
Continued overall challenges include keeping up-to-date with new gene annotations and data releases from a large number of external sources with different update schedules. Use of web analytics and our bug report form to learn about community interest will help us prioritize frequently-accessed resources for updates. We expect that support for CRISPR applications will continue to be an area of focus for us in the future. For example, we plan to modify RSVP to accommodate data from tissue-specific in vivo Drosophila CRISPR approaches. Altogether, we remain focused on responding to community needs and welcome input on how we can continue to expand and improve our database, website, and suite of online tools. Moreover, we expect to continue our efforts to build more integrated and cross-species resources, facilitating the use of information from Drosophila and other organisms to uncover conserved gene function.
Acknowledgments
The authors would like to thank the many researchers who have performed screens at the DRSC, used DRSC and TRiP reagents, and provided data and informal feedback on reagents, protocols, online tools, and so on. Their critical feedback and open sharing have helped us to improve the resources and data we provide to the community. The authors also extend our thanks to the members of the Perrimon lab and to the FlyBase consortium for helpful discussions regarding our online tools and website. We also thank Gabriel Caro and the team at Harvard Web Publishing for help with the website; Jay Copeland of the Harvard Medical School (HMS) Image Data Management Core for consultation on public availability of images; and the HMS Research Computing Group for consultation and support of our database and online tools.
FUNDING
NIH NIGMS [R01 GM067761, NIGMS R01 GM084947]; NIH [R24 RR032668, R24 OD021997 to N.P., P.I.]; Dana Farber/Harvard Cancer Center [NCI Cancer Center Support Grant # NIH 5 P30 CA06516 to S.E.M.]; Howard Hughes Medical Institute (to N.P.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Flockhart I.T., Booker M., Hu Y., McElvany B., Gilly Q., Mathey-Prevot B., Perrimon N., Mohr S.E. FlyRNAi.org–the database of the drosophila RNAi screening center: 2012 update. Nucleic Acids Res. 2012;40:D715–D719. doi: 10.1093/nar/gkr953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flockhart I., Booker M., Kiger A., Boutros M., Armknecht S., Ramadan N., Richardson K., Xu A., Perrimon N., Mathey-Prevot B. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 2006;34:D489–D494. doi: 10.1093/nar/gkj114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.S., et al. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echeverri C.J., Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat. Rev. Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 5.dos Santos G., Schroeder A.J., Goodman J.L., Strelets V.B., Crosby M.A., Thurmond J., Emmert D.B., Gelbart W.M., FlyBase C. FlyBase: introduction of the drosophila melanogaster release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015;43:D690–D697. doi: 10.1093/nar/gku1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y., Roesel C., Flockhart I., Perkins L., Perrimon N., Mohr S.E. UP-TORR: online tool for accurate and Up-to-Date annotation of RNAi Reagents. Genetics. 2013;195:37–45. doi: 10.1534/genetics.113.151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinayagam A., Hu Y., Kulkarni M., Roesel C., Sopko R., Mohr S.E., Perrimon N. Protein complex-based analysis framework for high-throughput data sets. Sci. Signal. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriventseva E.V., Tegenfeldt F., Petty T.J., Waterhouse R.M., Simao F.A., Pozdnyakov I.A., Ioannidis P., Zdobnov E.M. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015;43:D250–D256. doi: 10.1093/nar/gku1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mi H., Dong Q., Muruganujan A., Gaudet P., Lewis S., Thomas P.D. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruzicka L., Bradford Y.M., Frazer K., Howe D.G., Paddock H., Ramachandran S., Singer A., Toro S., Van Slyke C.E., Eagle A.E., et al. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015;53:498–509. doi: 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y., Comjean A., Perkins L.A., Perrimon N., Mohr S.E. GLAD: an online database of gene list annotation for drosophila. J. Genomics. 2015;3:75–81. doi: 10.7150/jgen.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marianes A., Spradling A.C. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife. 2013;2:e00886. doi: 10.7554/eLife.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta D., Dobson A.J., Houtz P.L., Glasser C., Revah J., Korzelius J., Patel P.H., Edgar B.A., Buchon N. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 2015;12:346–358. doi: 10.1016/j.celrep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 16.modEncode-Consortium. Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L., et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S., Ewen-Campen B., Ni X., Housden B.E., Perrimon N. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics. 2015;201:433–442. doi: 10.1534/genetics.115.181065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohr S.E., Hu Y., Ewen-Campen B., Housden B.E., Viswanatha R., Perrimon N. CRISPR guide RNA design for research applications. FEBS J. 2016;283:3232–3238. doi: 10.1111/febs.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Housden B.E., Valvezan A.J., Kelley C., Sopko R., Hu Y., Roesel C., Lin S., Buckner M., Tao R., Yilmazel B., et al. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci. Signal. 2015;8:rs9. doi: 10.1126/scisignal.aab3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L., et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Housden B.E., Lin S., Perrimon N. Cas9-based genome editing in Drosophila. Methods Enzymol. 2014;546:415–439. doi: 10.1016/B978-0-12-801185-0.00019-2. [DOI] [PubMed] [Google Scholar]
- 22.Vinayagam A., Zirin J., Roesel C., Hu Y., Yilmazel B., Samsonova A.A., Neumuller R.A., Mohr S.E., Perrimon N. Integrating protein-protein interaction networks with phenotypes reveals signs of interactions. Nat. Methods. 2014;11:94–99. doi: 10.1038/nmeth.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yilmazel B., Hu Y., Sigoillot F., Smith J.A., Shamu C.E., Perrimon N., Mohr S.E. Online GESS: prediction of miRNA-like off-target effects in large-scale RNAi screen data by seed region analysis. BMC Bioinformatics. 2014;15:192. doi: 10.1186/1471-2105-15-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigoillot F.D., Lyman S., Huckins J.F., Adamson B., Chung E., Quattrochi B., King R.W. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Suzek T., Zhang J., Wang J., He S., Cheng T., Shoemaker B.A., Gindulyte A., Bryant S.H. PubChem BioAssay: 2014 update. Nucleic Acids Res. 2014;42:D1075–D1082. doi: 10.1093/nar/gkt978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt E.E., Pelz O., Buhlmann S., Kerr G., Horn T., Boutros M. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41:D1021–D1026. doi: 10.1093/nar/gks1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohr S.E., Hu Y., Rudd K., Buckner M., Gilly Q., Foster B., Sierzputowska K., Comjean A., Ye B., Perrimon N. Reagent and data resources for investigation of RNA binding protein functions in Drosophila melanogaster cultured Cells. G3 (Bethesda) 2015;5:1919–1924. doi: 10.1534/g3.115.019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allan C., Burel J.M., Moore J., Blackburn C., Linkert M., Loynton S., Macdonald D., Moore W.J., Neves C., Patterson A., et al. OMERO: flexible, model-driven data management for experimental biology. Natt. Methods. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]