Abstract
Exposome-Explorer (http://exposome-explorer.iarc.fr) is the first database dedicated to biomarkers of exposure to environmental risk factors. It contains detailed information on the nature of biomarkers, their concentrations in various human biospecimens, the study population where measured and the analytical techniques used for measurement. It also contains correlations with external exposure measurements and data on biological reproducibility over time. The data in Exposome-Explorer was manually collected from peer-reviewed publications and organized to make it easily accessible through a web interface for in-depth analyses. The database and the web interface were developed using the Ruby on Rails framework. A total of 480 publications were analyzed and 10 510 concentration values in blood, urine and other biospecimens for 692 dietary and pollutant biomarkers were collected. Over 8000 correlation values between dietary biomarker levels and food intake as well as 536 values of biological reproducibility over time were also compiled. Exposome-Explorer makes it easy to compare the performance between biomarkers and their fields of application. It should be particularly useful for epidemiologists and clinicians wishing to select panels of biomarkers that can be used in biomonitoring studies or in exposome-wide association studies, thereby allowing them to better understand the etiology of chronic diseases.
INTRODUCTION
Environmental factors play a major role in the etiology of cancers, cardiovascular disease, and other chronic diseases. These factors are diverse in nature and include diet, drugs, cosmetics, household chemicals, pollutants, or infectious agents. Exposures to these factors vary widely between populations, and between individuals within the same population. Therefore, their measurement is essential to: (i) study associations in epidemiological studies with disease outcomes and assess their contribution to disease risk, (ii) monitor exposures to disease risk factors in population studies and (iii) assess subject compliance in clinical trials or large intervention studies (1–3).
Exposure measurements have traditionally relied on the use of questionnaires and self-reporting. However, these methods are known to be error-prone and biased. Molecular biomarkers, on the other hand, are more direct and objective indicators of exposure. Indeed, biomarkers of exposure have been increasingly used since the early 1980s, thanks to rapid progress in analytical techniques and the establishment of large cohorts with extensive biospecimen collections. Biomarkers of exposure can be compounds present in the environment and absorbed in the gut after ingestion, inhaled in lungs, or absorbed through the skin. They can also be metabolic end-products derived from environmental compounds that were metabolized by the liver and other tissues, and the microbiota. They may also be macromolecular indicators of environmental effects (e.g. enzymes, proteins or RNA transcripts related to the status of a nutrient or toxic agent).
Over the past 30 years several hundred biomarkers of exposure have been measured and reported in blood, urine and other biospecimens in various populations. However, this information is scattered over hundreds of publications under many diverse titles and subject headings. This makes the identification of these biomarkers along with their comparative performance, their field of application and their concentration ranges in different populations difficult. Historically, most biomarkers of exposure have been measured individually using compound-specific assays. However, with the emergence of various omics technologies there is an increasing tendency to characterize exposures more comprehensively. Indeed, modern mass spectrometry techniques now allow the measurement of thousands of compounds in blood, urine or other biospecimens in a single analytical run. These developments are leading, increasingly, to the reporting of data from multiple markers of exposure. Modern omics technologies should also allow the translation into practice of the concept of the exposome (the totality of exposures of a particular individual over lifetime (4,5)) and the development of exposome-wide association studies (EWAS) (2,6–8). These newly emerging trends in exposure science, combined with the growing volume of comprehensive exposure data being published, make the establishment of a centralized, online database on biomarkers of exposure particularly critical.
To date, relatively little effort has been directed to collecting and organizing data on biomarkers of exposure. The ExpoCastDB database contains information on exposure to environmental chemicals such as PAHs, PCBs, nonylphenols, or pesticides (http://actor.epa.gov/actor/faces/ExpoCastDB/Home.jsp). ExpoCastDB contains information on compound concentrations in various environmental matrices but very limited data in biospecimens. The Comparative Toxicogenomics Database (CTD) is the only online database containing a large number of concentration values in blood, urine and other biospecimens extracted from the scientific literature (9). The CTD contains about 35 000 concentration values in various biospecimens for ∼250 organic and inorganic compounds. While ExpoCastDB and CTD are very useful and important databases, they contain only limited information on biomarkers of exposure.
Here we describe Exposome-Explorer, the first database dedicated to exposure biomarkers. Exposome-Explorer consolidates the diffuse exposure biomarker data scattered throughout the literature. It contains comprehensive information on almost 500 biomarkers of exposure with concentrations, correlations with exposure estimates and temporal reproducibility, as well as other details on study population and analytical methods. All data in Exposome-Explorer was acquired from a careful review and analysis of nearly 500 peer-reviewed publications, with a particular focus on dietary and pollution exposures. Exposome-Explorer is freely accessible at http://exposome-explorer.iarc.fr.
DATA COLLECTION
All data in Exposome-Explorer was compiled through extensive literature analysis along with manual curation and computer-assisted validation. Literature searches were conducted using the Web of Science (WOS). Only peer-reviewed publications describing original work with biomarker measurements in observational studies conducted in human populations were considered. Publications on intervention studies, analytical method development, associations of biomarker with biological status or disease, biomarkers of biological status (e.g. inflammation, oxidative stress, disease), or animal studies were not considered for this initial stage of development. Articles not available online were omitted. For pollutants, general rather than occupational exposures were prioritized.
Every search included keywords commonly associated with biomarkers (‘biomarker’, ‘metabolite’, ‘concentration’, ‘level’, ‘excretion’, ‘indice’, ‘indicator’, ‘exposure’, ‘biological monitoring’) and biospecimens (‘blood’, ‘serum’, ‘plasma’, ‘urine’, ‘adipose tissue’, ‘hair’, ‘adduct’). For dietary biomarkers, data on the correlations between food or food compound intake and biomarker concentrations were also collected. Citations were searched for according to several intake synonyms (‘intake’, ‘consumption’, ‘diet’, ‘recall’, ‘questionnaire’), associations (‘association’, ‘comparison’, ‘correlation’, ‘relation’) and a variety of validation synonyms (‘validation’, ‘validity’, ‘reliability’, ‘evaluation’). For pollutant biomarkers, data on biospecimen concentrations were collected. Citations were searched for by common pollutant chemical group, such as polycyclic aromatic hydrocarbons (PAH), polychlorinated biphenyls (PCB), polybrominated diphenyl ethers (PBDE), polybrominated biphenyls (PBB), polychlorinated dibenzodioxins/furans (PCDD/F), heterocyclic amines (HCA), phthalates and disinfection byproducts (DBP). To search for biomarker reproducibility values, the following keywords were used: ‘variability’, ‘reliability’, ‘reproducibility’, ‘repeatability’, ‘intra-subject’, ‘inter-subject’, ‘within-subject’, ‘between-subject’.
Full-record citations from the WOS were downloaded in the BibTeX format (http://www.bibtex.org/) and handled with BibDesk, an open-source bibliography manager for BibTeX libraries (http://bibdesk.sourceforge.net/). Citations were initially screened by title and abstract to assess relevance. Those rated as relevant were then manually described using a series of attributes (‘Tags’) related to exposures, study design, type of numerical data, populations, biospecimens, biomarkers and confounding variables to facilitate the selection of references for annotators. The BibDesk ‘Smart Groups’ functionality and a combination of criteria based on the tags were used to dynamically generate a priority list of publications for further annotation. Full-texts from these articles were then retrieved and submitted to annotators for detailed analysis and upload of the data to Exposome-Explorer using the annotation interface.
DATA COMPILATION
A password protected annotation interface was used for the manual uploading of all data from scientific publications to Exposome-Explorer. This interface permits an efficient and consistent data annotation process. Through this annotation interface, the annotator is guided through successive steps to ensure comprehensive capture of the following information:
Publication with its bibliographic details (title, authors, year, journal, PubMed ID).
Subject groups studied in the publication, and the populations to which they belong. Each population is named with a short informative summary sentence (e.g. ‘Cases and controls in a case-control study on breast cancer’). Populations are subdivided into one (‘All’) or several (e.g. ‘Soy consumers’ and ‘Soy non-consumers’) subject groups according to different criteria (e.g. ‘by gender’, ‘by ethnicity’, ‘by smoking status’). A reference to a cohort (e.g. ‘EPIC’) can also be indicated in this data field. Age, height, weight, BMI, group size, gender, health condition, exclusion of supplement users, smoker proportion, country of origin and ethnicity are also specified when available.
Samples defined for each subject group. These describe number and time of collections (e.g. ‘baseline’, ‘1 year’) of different biospecimens. Biospecimens include urine, whole blood, serum, plasma, hair, nails, adipose tissue, breast milk, or fractions such as plasma phospholipids or red blood cell membranes.
Biomarkers. Each biomarker is described with name, synonyms, chemical group and subgroup. Chemical information (e.g. structure, chemical formula, average mass, monoisotopic mass) is automatically generated by the structure server. A ‘classification level’ allows to distinguish ‘single’ biomarkers corresponding to a single chemical entity from ‘combined’ biomarkers described in publications as the sum of single chemical entities. Details on individual compounds considered to calculate this sum are collected. Identifiers and links to external databases such as CAS, PubChem (10), ChEBI (11), HMDB (12) and FooDB (www.foodb.ca) are provided.
Biomarker measurements. For each sample or biospecimen type, biomarker concentrations are documented. Also included is the analytical method, the original measurement units as described in the publication, and specimen-specific concentration value adjustments (e.g. normalizing to ‘creatinine’ for urine, ‘lipid’ for blood, or a list of regression variables). Arithmetic or geometric mean, as well as median concentration values are compiled along with the minimum and maximum values, standard deviation, percentiles, confidence intervals, measurement size (number of subjects from which the concentration value was calculated), as well as the proportion of subjects for which the biomarker was detected. Information on the inclusion or exclusion of zero values in the calculated concentration is also given. This was mostly done for pollutant biomonitoring studies where a chemical is often detected only in a small proportion of the studied population. Some authors used a threshold (e.g. ‘detected in at least 30% samples’). Below this threshold, the geometric mean was not calculated and the authors considered the compound to be not detected in the studied population. In certain cases, the compound used to express the concentration is given if it is different from the compound or compound class being measured (e.g. ‘polyphenols in urine expressed as gallic acid’).
Correlations. To compile data on the correlations between specific biomarker measurements and food or dietary compound intake, information was collected on the intake of different foods, food groups or dietary compounds together with the method used for dietary assessment (record or questionnaire, food composition database, food coverage and time period). Food items in Exposome-Explorer are linked to the FooDB database (www.foodb.ca). For food groups, the list of individual foods considered to calculate the intake of that particular food group is indicated if described in the publication. For dietary compounds, the inclusion of dietary supplements in the calculation of their intake is indicated when mentioned in the publication. Correlation coefficients (Pearson's product moment or Spearman's rank-order) are collated together with p-value, confidence intervals, statistical significance (‘yes’/‘no’), and correlation size (number of measurements from which the correlation coefficient was calculated). Adjustment of intake or biomarker measurements prior to calculation of the correlation coefficient (e.g. ‘energy intake by residual method’ or ‘creatinine’) as well as a list of regression variables included in the calculation (e.g. ‘age, smoking status, BMI, gender’), and the use of measurement error de-attenuation are also indicated.
Temporal reproducibility of biomarker measurements in an individual is an important characteristic of the biomarker. A high reproducibility is required when only one sample per subject is available for biomarker measurement, as in most large cohort studies. Reproducibility is usually measured on repeated samples collected in a small group of individuals over a given time interval (generally weeks or months). Reproducibility is principally described by the intraclass correlation coefficient (ICC), defined as the ratio of between-subject variance to the sum of within- and between-subject variance. ICC values range from 0 to 1. The higher the ICC value, the higher the reliability of the measurement. In some cases, within- and between-subject coefficient of variation (CV), as well as within- and between-subject variance (VAR) are also detailed in the publications. These data are also captured by Exposome-Explorer. Reproducibility size (number of measurements from which the reproducibility value was calculated) and the confidence interval are also indicated in the database.
Exposome-Explorer's annotation interface uses a number of features to help complex, heterogeneous, literature-derived data to be easily and systematically translated into organized electronic data. Controlled vocabularies for compounds, foods, experimental methods, specimens, cohorts and units can be created and are fully documented in both the annotation interface and the database. This helps to avoid the inclusion of duplicate vocabulary items, such as different spellings or synonyms for a same item. Hierarchical associations of populations, samples and measurements can be represented through the creation of parent-children relations. For instance, populations stratified into different sub-groups can be easily described, or the association of several samples taken at different collection times. All data uploaded to the database via the Exposome-Explorer annotation interface is automatically validated, thereby preventing the uploading of erroneous records. Error checks include the usual database integrity verifications such as the presence of mandatory fields, checks on input text size, checks on allowable values or the uniqueness of specific values. The error checking utilities also include application-specific consistency verifications such as ‘correlations cannot be created between measurements of different subject groups’ or ‘measurement size cannot exceed subject group size’. These checks ensure the highest consistency for data collected by different annotators. Newly uploaded data is also manually and systematically checked by the database's chief curator. Repetitive insertion of similar data is facilitated via the support of record duplication and the ability to edit several records at once. Error messages are displayed to guide annotators toward making correct data entries or fixing erroneous entries.
QUERYING THE DATABASE
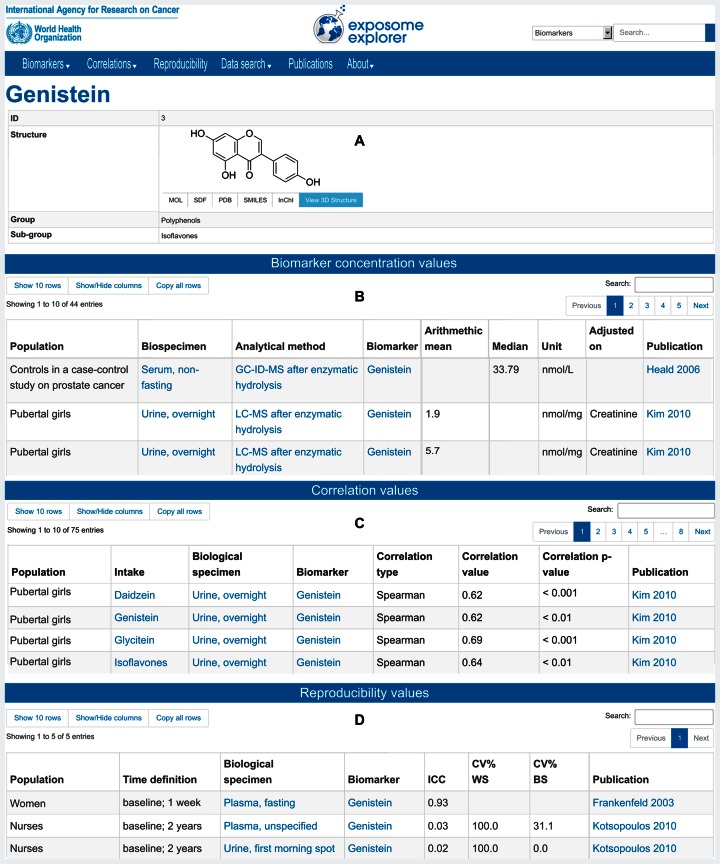
Exposome-Explorer's public user interface allows intuitive browsing and searching of almost any data type in the database (Figure 1). Data can be retrieved through the quick search functionality or through specific searches offered on the website's different menu pages.
Figure 1.
Screenshot montage of a biomarker view. Distinct areas are shown: structure and chemical information (A), concentrations (B), correlations (C) and reproducibility (D) collated from several publications. Details on populations, biospecimens, analytical methods and corresponding citations are displayed. More columns can be shown with the ‘Show/Hide columns’ button.
The ‘Biomarkers’ menu lists the biomarkers, biospecimens, analytical methods and cohorts documented in the database. Each item has its own summary page with both general details (name and classification) and all data related to this item in the database. This includes biomarker concentration values, correlation values, reproducibility values and publications. The ‘Correlations’ menu lists the correlation values between different food or dietary compound intakes and the corresponding biomarker measurements. These correlations can be searched according to the food type, dietary compound, or biomarker. The data can be filtered according to a variety of parameters including the type of biospecimen or the method used to assess the dietary exposure. The ‘Reproducibility’ menu lists the biomarker reproducibility values, which can also be filtered according to biomarker classification (as well as other parameters) and ranked in decreasing or increasing order. The ‘Data search’ menu lists all biomarker concentration values and allows searches over all data fields. Searches by chemical structure are also possible, as well as browsing biomarker classes and subclasses. The ‘Publications’ menu lists all the publications in the database. The list can be filtered by title, authors, year or PubMed ID. Full-text for these publications can be accessed via PubMed links or direct publisher URLs. For every annotated publication, the totality of collected data including the bibliographic information, detailed subject descriptions, and biomarker data (concentration values, dietary intake values, correlation values and reproducibility values) is displayed in a single page.
Tables on Exposome-Explorer's different webpages can be easily adjusted to suit user-specific needs. They can be filtered and sorted on any column. Similarly, the number of displayed rows can be personalized. Several hidden columns are available and can be shown in order to provide more details for the default display. Every original value is listed with its corresponding bibliographic citation, making it possible to link each value back to its metadata. Default conversions from highly diverse to standardized units are possible while original values are preserved. Tables can be exported in different formats (e.g. CSV, TAB) and reused in other programs (e.g. Excel, R) in order to conduct more specific analyses.
DATABASE IMPLEMENTATION
Exposome-Explorer was developed using Ruby on Rails (http://rubyonrails.org/). Ruby on Rails is a web framework which employs the Model-View-Controller (MVC) design pattern. It allows the construction of robust, responsive and reliable web applications. The Exposome-Explorer data is stored in a MySQL relational database (http://www.mysql.com/). Hierarchical associations of records (‘trees’) are implemented as a nested set model inside a single database column of the tables using the Rails gem Ancestry (https://github.com/stefankroes/ancestry). Highly diverse units and their conversions are transparently handled with the Phys-Units library (https://github.com/masa16/phys-units/), which is a Ruby implementation of the GNU Units software (http://www.gnu.org/software/units/). Chemical structures are hosted on the Wishart's MolDB structure server. Website global search is implemented using the Wishart's Unearth gem, which uses Elasticsearch indexing (https://www.elastic.co/). The web interface is built with the Bootstrap front-end framework (http://getbootstrap.com/). Tables on the different web pages are formatted using jQuery DataTables (http://www.datatables.net/).
DATA STATISTICS
To date, data from a total of 480 publications have been entered into this first release of Exposome-Explorer. This includes 10 510 concentration values for 692 biomarker entries, among which 8,861 concentrations correspond to 488 ‘single’ dietary and pollutant biomarkers. Approximately one third of these biomarkers are dietary biomarkers (Table 1). For dietary biomarkers, almost half of the concentrations are related to fatty acids, followed by carotenoids and polyphenols. With regard to pollutant biomarkers, about two thirds of the concentration data in Exposome-Explorer are related to PCBs and PBDEs, followed by PAHs, PCDD/Fs and phthalates. Most reported concentration measurements are from blood and urine (Table 1). Some concentration data are also available for other biospecimens such as hair, nails, adipose tissue or breast milk.
Table 1. Content of the Exposome-Explorer database.
| Data type | Records (n) | Description | Publications (n) |
|---|---|---|---|
| Single biomarkers | 488 | Dietary biomarkers (142), pollutants (346) | 430 |
| Concentrations | 10 510 | Blood (6461), urine (2073), other (1976) | 458 |
| Correlations | 8034 | On dietary biomarkers for 50 foods and 78 food compounds | 196 |
| Reproducibility | 536 | Diet (303), pollution (233) | 52 |
CONCLUSIONS AND FUTURE ENHANCEMENTS
Exposome-Explorer currently contains the most complete and comprehensive information on exposure biomarkers ever compiled from peer-reviewed literature. It is also the first publicly available, web-enabled database specifically dedicated to exposure biomarkers. We believe it provides a good starting point for selecting markers of interest or for defining panels of biomarkers that can be used in exposome-wide association studies. Through Exposome-Explorer, biomarker concentrations can be compared in different cohorts or population groups at different levels of exposure (e.g. consumers and non-consumers of a particular food), or between different geographical areas. All of the database's manually curated information is fully linked with other online databases and with the original publications. These features make it unique among all biomarker databases that we are aware of. The high granularity of the data in the database should allow users to conduct very diverse and advanced analyses or comparisons across publications.
Looking both at the range of studied compounds and the number of corresponding studies, Exposome-Explorer also allows users to quickly identify poorly studied exposures or biomarkers that may require further work for validation. Plans for future enhancements to Exposome-Explorer include its extension to other categories of exposures (nutritional status, pesticides, occupational exposures), the addition of other types of biomarkers (DNA and protein adducts), and the inclusion of more information characterizing biomarkers such as their half-life and other pharmacokinetic parameters. Plans are also being made to add putative or non-validated biomarkers identified in (pre)clinical studies, but never measured in populations.
Overall, we believe Exposome-Explorer will help in the generation of hypotheses for discovery of new biomarkers to be tested in the laboratory. It should also help in evaluating the performance of existing biomarkers and integrating exposure data based on biomarkers with data collected with other technologies. Exposome-Explorer should contribute to the translation of the exposome into practice in epidemiological research.
FUNDING
European Commission: EXPOsOMICS FP7-KBBE-2012 [308610]; NutriTech FP7-KBBE-2011-5 [289511]; Joint Programming Initiative FOODBALL 2014–17. Funding for open access charge: EXPOsOMICS FP7-KBBE-2012 [308610].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wild C.P., Vineis P., Garte S. Molecular Epidemiology of Chronic Diseases. 1st edn. Hoboken, NJ: Wiley; 2008. [Google Scholar]
- 2.Scalbert A., Brennan L., Manach C., Andres-Lacueva C., Dragsted L.O., Draper J., Rappaport S.M., van der Hooft J.J., Wishart D.S. The food metabolome: a window over dietary exposure. Am. J. Clin. Nutr. 2014;99:1286–1308. doi: 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- 3.Alves A., Kucharska A., Erratico C., Xu F., Den Hond E., Koppen G., Vanermen G., Covaci A., Voorspoels S. Human biomonitoring of emerging pollutants through non-invasive matrices: state of the art and future potential. Anal. Bioanal. Chem. 2014;406:4063–4088. doi: 10.1007/s00216-014-7748-1. [DOI] [PubMed] [Google Scholar]
- 4.Wild C.P. Complementing the genome with an ‘exposome’: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 5.Wild C.P., Scalbert A., Herceg Z. Measuring the exposome: a powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen. 2013;54:480–499. doi: 10.1002/em.21777. [DOI] [PubMed] [Google Scholar]
- 6.Rappaport S.M. Biomarkers intersect with the exposome. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2012;17:483–489. doi: 10.3109/1354750X.2012.691553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel C.J., Bhattacharya J., Butte A.J. An environment-wide association study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzoulaki I., Patel C.J., Okamura T., Chan Q., Brown I.J., Miura K., Ueshima H., Zhao L., Van Horn L., Daviglus M.L., et al. A nutrient-wide association study on blood pressure. Circulation. 2012;126:2456–2464. doi: 10.1161/CIRCULATIONAHA.112.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis A.P., Grondin C.J., Lennon-Hopkins K., Saraceni-Richards C., Sciaky D., King B.L., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database's 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastings J., de Matos P., Dekker A., Ennis M., Harsha B., Kale N., Muthukrishnan V., Owen G., Turner S., Williams M., et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–D463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., et al. HMDB 3.0–The human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]