Abstract
The increasing number of experimentally detected interactions between proteins makes it difficult for researchers to extract the interactions relevant for specific biological processes or diseases. This makes it necessary to accompany the large-scale detection of protein–protein interactions (PPIs) with strategies and tools to generate meaningful PPI subnetworks. To this end, we generated the Human Integrated Protein–Protein Interaction rEference or HIPPIE (http://cbdm.uni-mainz.de/hippie/). HIPPIE is a one-stop resource for the generation and interpretation of PPI networks relevant to a specific research question. We provide means to generate highly reliable, context-specific PPI networks and to make sense out of them. We just released the second major update of HIPPIE, implementing various new features. HIPPIE grew substantially over the last years and now contains more than 270 000 confidence scored and annotated PPIs. We integrated different types of experimental information for the confidence scoring and the construction of context-specific networks. We implemented basic graph algorithms that highlight important proteins and interactions. HIPPIE's graphical interface implements several ways for wet lab and computational scientists alike to access the PPI data.
INTRODUCTION
Protein–protein interactions (PPIs) are experimentally detected at a large scale. Several databases exist that collect the knowledge about human and model organism PPIs by manual curation (1,2) and provide the community with invaluable resources on the functional organization of the cell. Many studies use these PPI networks to identify commonalities among a group of proteins (3), to infer novel functions for a gene of interest (4) or for gene-disease association prediction (5,6).
Despite these successful applications, there are several fundamental problems with the way in which PPIs are experimentally determined: (i) PPI detection methods are associated with high error rates (both false positives and false negative rates). Some genome-wide screens might be associated with false positive rates exceeding 50% (7–10). (ii) PPI detection methods typically do not reveal information associated with the context in which PPIs are realized. Even more, many of these methods measure PPIs under unphysiological conditions (e.g. human proteins expressed in yeast) and even when they do use physiological conditions, it is uncertain whether the same PPI also occurs in different tissues, cellular compartments or time points (e.g. the adaptor protein GRB2 changes its interaction partners dramatically in response to different cellular stimuli (11)). (iii) PPI detection methods do not reveal important qualities of PPIs such as their directionality and effect (which exist for example in a signaling context for kinase–substrate and phosphatase–substrate interactions). (iv) PPI detection methods are associated with technical biases and the choice of proteins tested for interaction partners introduces a study bias (10,12–13). The noisy and biased nature of the PPI networks can severely impact the biological hypotheses generated from these data (14–16).
Standards and protocols have been developed that allow the exchange and aggregation of these data (17), facilitating the development of a number of databases that collect the information from different curated sources (2). However, the growing knowledge of interaction partners for most human proteins makes it difficult for researchers to extract the relevant information when proteins are studied under specific cellular conditions. In this scenario, the objective becomes to retrieve a small set of high-confidence interaction partners that are, for example, involved in disease progression, expressed in a certain body part or signaling substrates of the protein of interest. This makes it necessary to accompany the experimental detection of PPIs with bioinformatics strategies to annotate PPIs with context information, to select meaningful subsets from the entire network and to provide means to interpret them.
To this end, our web resource, the Human Integrated Protein-Protein Interaction rEference or HIPPIE [http://cbdm.uni-mainz.de/hippie/], aims to provide highly reliable, context-specific PPI networks and guide their interpretation. This is achieved by stringent confidence scoring of PPIs, integrating different types of experimental information and offering basic graph algorithms that highlight important proteins. We implemented a versatile interface that allows wet lab and computational scientists alike to access the data. Visualization of the output facilitates the inspection of relevant parts of the human interactome. Integrating and interfacing external tools allow to interpret the results of HIPPIE queries and to pass them on for subsequent analyses.
Since its first release in early 2011 (18), HIPPIE has grown significantly both in size and functionality. It has been maintained by a team of biologists and developers and has established itself as a one-stop resource for querying and analyzing the human interactome. In its second version, we have added a large amount of PPIs and implemented several new features to facilitate the study of PPI networks.
MATERIALS AND METHODS
PPI retrieval
We use the PSICQUIC interface (19) to regularly update our PPI repository. HIPPIE contains experimentally detected PPIs from IntAct (20), MINT (21), BioGRID (1), HPRD (22), DIP (23), BIND (24) and MIPS (25). In annual cycles, we branch out major releases of HIPPIE, which we provide for download in the HIPPIE tab-separated or MITAB-format files (26).
We map all source database entries to gene names, Entrez gene ids and UniProt ids or accessions, which are all valid options for querying HIPPIE.
PPI annotation
PPIs in HIPPIE are context annotated using gene expression information, gene ontology (GO) terms and MeSH disease headings. For the generation of tissue-specific networks, we retrieve gene expression data from 53 healthy human tissues from GTEx (27). GTEx provides gene-level expression quantifications (for the technical details see their documentation: http://www.gtexportal.org/home/documentationPage). We consider that a gene is expressed, if its median expression over samples in a tissue exceeds a stringent RPKM threshold of 1 (28). We apply a node removal approach to generate tissue-specific PPI networks, which means that nodes representing non-expressed genes are excluded from the network.
The annotation of PPIs in HIPPIE with respect to gene function and disease is achieved using GO biological process and cellular compartment terms and MeSH disease headings. This is done by taking into account the hierarchical structure of the GO and MeSH ontologies: We associate each PPI with the lowest common ancestor of each pair of terms/headings annotating the two interaction partners. Details of the annotation procedure are described elsewhere (29).
We predict directionality using the shortest-path approach described in (29). We infer whether a PPI is activating or repressing using phenotypic image data of a genome-wide cellular RNAi knockdown screen (30). We add expert-curated PPI directionality and effect information from KEGG (31).
Enrichment analysis
Disease and functional enrichment analyses for the members of the subnetwork resulting from a protein or network query are carried out via the third-party tools Gene Set to Diseases (32) and PANTHER (33), respectively. The former associates genes with diseases based on biomedical literature, the latter considers GO terms. In both cases, the statistically over-represented diseases or terms associated with sets of proteins are determined. In more detail, Gene Set to Diseases uses associations of diseases to all human genes based on the over-representation of disease MeSH terms attached to the PubMed records related to each human gene; each association disease:gene is evaluated by a P-value (from a one-tailed Fisher's exact test), a false discovery rate (FDR; derived from the P-value using the Benjamini and Hochberg method) and by the number of PubMed records that associate the gene to the disease. In its current implementation, HIPPIE uses Gene Set to Diseases with a threshold of at least three PubMed records and a FDR < 0.05, on the disease:gene associations.
HIPPIE REST web services
HIPPIE v2.0 can be queried via its recently-implemented REST web service. This means that the partners of a protein of interest or the way in which a group of proteins interact can be easily determined and integrated into bioinformatics pipelines.
Detailed information about this service can be consulted at http://cbdm-01.zdv.uni-mainz.de/∼mschaefer/hippie/information.php#api.
RESULTS
Selecting reliable and meaningful PPIs in HIPPIE v2.0
HIPPIE provides access to the manually curated set of human PPIs from several source databases (see Materials and Methods). As our aim is to integrate only the most reliable interactions, we do not include any predicted associations between proteins but only experimentally determined ones. In order to compute a confidence score, we use information on the experiments performed to detect a PPI. This score is half manual, half computationally optimized (18) and weights the amount and quality of the experimental evidence supporting each PPI. With this, users can select the most reliable PPIs and dramatically reduce the amount of false positives within the notoriously error-prone human PPI network.
Our own work (29,34) and that of other groups (35–37) demonstrated that the addition of functional and tissue-expression data, in combination with basic graph algorithms, enables the construction of PPI networks that are highly relevant to a specific research question. In particular, we showed that when cellular signaling is studied in an infectious or genetic disease context, combining both gene expression information from the tissue affected by the disease and predicted network information flow, serves to highlight important mediators of disease from the PPI hairball surrounding the studied proteins (29). Indeed, disease-causing proteins tend to form an increased number of tissue-specific PPIs in the tissues that they affect (38).
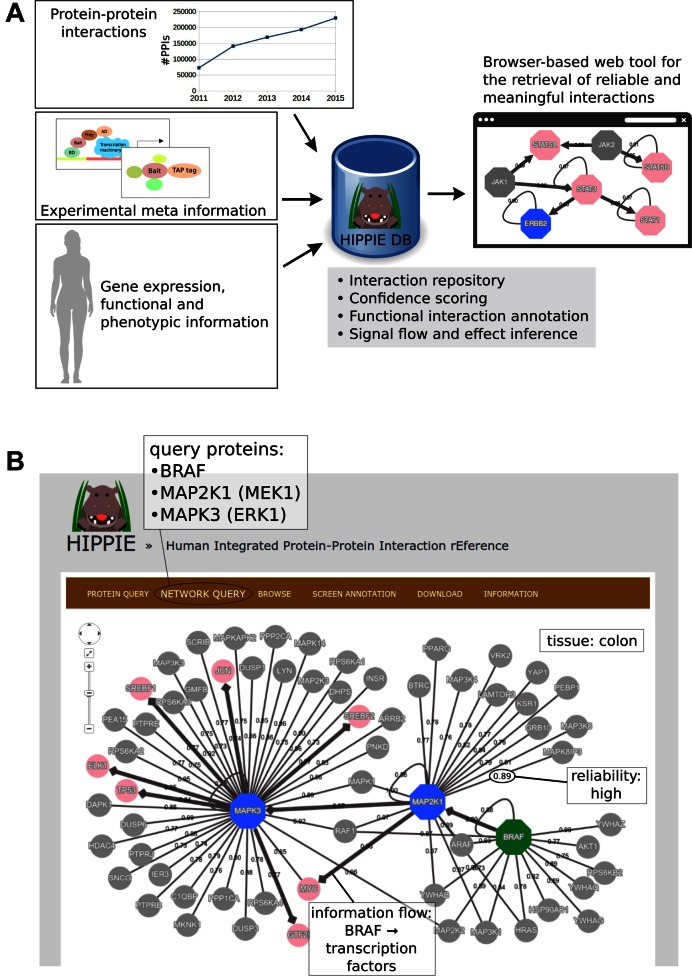
We therefore annotate PPIs in HIPPIE using a controlled vocabulary derived from annotations of the interacting proteins. PPIs are annotated with respect to protein function, disease-relevance and tissue expression (29). To this end, HIPPIE integrates various types of experimental and protein annotation data (Figure 1A).
Figure 1.
Data in Human Integrated Protein–Protein Interaction rEference (HIPPIE) and how it can be used to reconstruct signaling events. (A) HIPPIE integrates heterogeneous data types: PPIs from expert-curated source databases are constantly updated and integrated into HIPPIE. Experimental meta-information (e.g. on the experimental methods employed to detect PPIs and on their reproducibility) is extracted and used to compute a confidence score for each interaction in HIPPIE. Gene expression, gene function and phenotypic data are aggregated and used to annotate the PPIs stored in HIPPIE and to infer edge effect and directionality. (B) The reconstruction of central components of the MAPK pathway downstream of BRAF is shown as a HIPPIE query example. The kinases BRAF, MEK1 (MAP2K1) and ERK1 (MAPK3) are members of the Mitogen-activated protein kinase (MAPK) signaling cascade and activate each other in the stated order. BRAF is frequently mutated in several cancers, colon cancer among them, where BRAF mutations are found in approximately 9% of all patients (51). Querying HIPPIE with the kinases BRAF, MEK1 and ERK1 and filtering for high confidence PPIs and colon expression results in the depicted network. Displaying shortest paths between BRAF (‘source’, in green) and transcription factors (‘sinks’) correctly reproduces the chain of signaling events (BRAF activates MEK1, which activates ERK1 in turn). All terminal nodes in pink (ELK1, MYC, JUN, TP53, SREBF1/2) are known substrates of ERK1 (52).
Additionally, we take advantage of network topological properties and other complementary data to offer options like adding edge directionality or activation/repression properties to the networks; these predictions are done by means of the shortest-path approach described in (29) and using an algorithm based on phenotypic image data for unannotated PPIs described in (30), respectively. In both cases, we complement the predicted data with expert curated information retrieved from KEGG (31).
The PPI annotations are implemented as filters in HIPPIE: either single PPI annotations or combinations of them can be selected by the user. Subsequently, a network is constructed around the query proteins consisting only of PPIs annotated with the selected categories. Interaction directionality and effect can be visualized in the resulting network. In this way, informative subnetworks of the entire human PPI network can be generated. As an example, Figure 1B illustrates the usefulness of HIPPIE's unique capability of combining different types of information to reconstruct signaling flow.
Novel features in HIPPIE v2.0
HIPPIE has been growing since its first release in 2011, from ∼73 000 to now more than 273 900 experimentally determined interactions among 17 000 human proteins. We classify 42 600 of the PPIs as high confidence interactions (18). For more than 15 000 of the proteins in HIPPIE, we have expression or functional information, allowing us to annotate the interactions between them. We recently updated HIPPIE's expression data to GTEx RNA-Seq gene expression quantifications in 53 healthy human tissues from post-mortem samples (27).
A bottleneck of querying HIPPIE has been the data-intensive loading of the filters and the subsequent computation of subnetworks from heterogeneous data, which forced us to previously limit queries to 100 proteins. In HIPPIE v2.0 we were able to remarkably improve performance, allowing us to relax previous upload limits and providing a much faster subnetwork generation from input lists of hundreds of proteins. At the same time, we enhanced the visual appeal of the HIPPIE output by porting the network visualization to Cytoscape.js (39) and added new visual control features for a more precise network exploration.
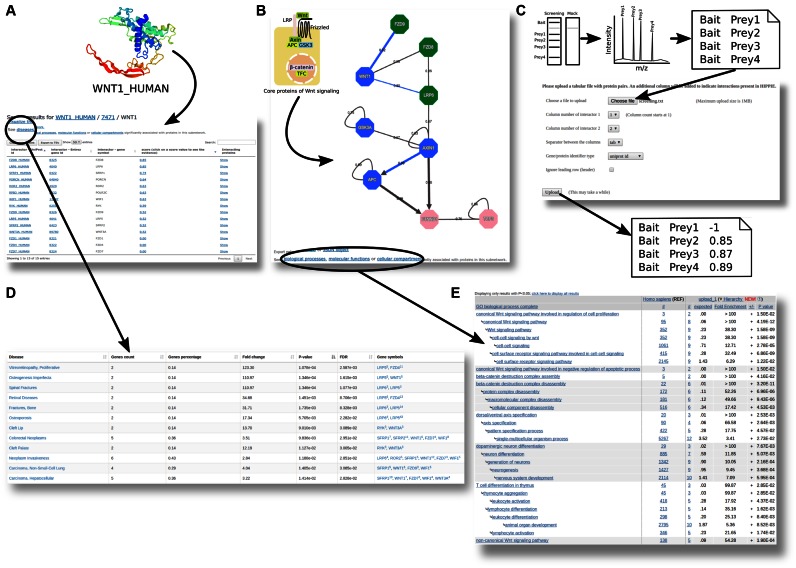
We also updated the input interface so that the data stored in HIPPIE can be accessed in several ways: (i) We recently implemented a browsing option, which provides a comprehensive view of the proteins for which PPI data are available, together with network summary statistics like number of interacting partners or protein ID mappings. (ii) HIPPIE can be queried by either single proteins, groups of proteins or even entire networks (Figure 2A–C visualizes the different input options). (iii) It is now possible to test whether the subnetworks resulting from such queries are significantly associated to certain human disorders, according to the literature (Figure 2D) (32), or gene ontology terms (Figure 2E) (33). (iv) The output of experimental PPI screens can be directly uploaded in order to highlight already known PPIs and their HIPPIE confidence scores (Figure 2C). (v) It is now much easier to integrate HIPPIE into bioinformatics pipelines, thanks to its newly implemented REST web service (see the Materials and Methods for more details).
Figure 2.
HIPPIE query types and result network interpretation. (A) When a single protein is used as an input in HIPPIE's Protein Query Tab, it produces a table with the interacting partners of that protein. The confidence score for each interaction is also listed and the interactors of each partner can be easily queried with a single click. (B) If a list of proteins or interactions is used as an input in HIPPIE's Network Query Tab, it produces a network of interactions between these proteins. In this query type, it is possible to implement filters that put the list of interactions in a functional and cellular context and the user can choose between different output types. In the example, HIPPIE was queried for high-confidence interactions between the core members of the Wnt signaling pathway, showing predicted information flow (arrow direction) and interaction effects (activating interactions are indicated by triangle arrowheads). (C) HIPPIE's Screen Annotation Tab allows to check whether a list of measured interactions is present in the database and how reliable each link is. In the example, a tab-separated file is uploaded to HIPPIE and it outputs a new file with the confidence score of each interaction or −1 if it is not present. (D and E) The results of the Protein and Network Queries allow one to perform a disease or functional enrichment analysis for the members of the resulting protein subnetwork. These analyses are carried out via the tools Gene Set to Diseases (32) and PANTHER (33), respectively.
Comparison with other PPI resources
HIPPIE differs from other PPI resources in that it enables the simultaneous generation of highly reliable and meaningful networks. There are other resources integrating PPIs and gene functional interactions (40,41), and some of them make it possible to generate tissue-specific (42,43) or more general context-specific networks (44). For example, the GIANT web server (45) predicts functional protein maps in 144 tissues by integrating thousands of expression and PPI experiments. HIPPIE's sources intendedly incorporate only experimentally determined data (both for interactions and expression) without inferred gene–gene or gene–tissue associations. In contrast, many other resources consider inferred associations (which in some cases can be excluded by the user (e.g. IID, STRING and MyProteinNet)). Moreover, HIPPIE provides means to score the amount and quality of experimental PPI evidence, providing a comprehensive, computationally optimized score, which is, to the best of our knowledge, the only PPI confidence score combining expert knowledge on experimental reliability with computationally optimized parameters of the scoring formula (18). In fact, HIPPIE's scoring system has recently has been shown to perform best among confidence scores incorporating only PPI experimental information (46). Our approach integrates experimental information to score all known experimentally detected PPIs, without creating or deleting any of them, but allowing the selection of well-grounded PPIs under the solid assumption that coherent experimental information increases the reliability of PPI data [e.g. (10)]. Additionally, users can restrict the resulting networks to PPIs from experiments reporting only direct physical binding events (such as Y2H) or include indirect associations (e.g. co-purified members of the same protein complex).
HIPPIE's scope stands out from other resources in that it integrates heterogeneous data types with basic network algorithms to not only highlight meaningful PPIs but also set them in their appropriate functional-, tissue-, effect- or even disease-specific context. This is achieved by filtering networks with respect to PPI annotations and highlighting important edges with inferred information flow. We previously demonstrated that our approach enriches the resulting networks in canonical signaling events and more reliable PPIs (29).
DISCUSSION
The need for accurate PPI networks to generate biological hypotheses has been widely recognized (47). Still, a problem that few databases simultaneously address is: which interactions from the huge human PPI hairball network are meaningful for a specific research question and which PPIs can we trust. Here, we described the development and extension of HIPPIE, which aims at filling this gap. Over the last years, HIPPIE has become a popular human PPI resource in the research community and many users have taken advantage of its unique features to generate highly reliable and meaningful PPI networks, centering around specific groups of proteins and research questions.
HIPPIE can be used at different levels. First, it is a resource integrating experimental PPIs from different manually curated sources. Its confidence scoring allows to easily distinguish high-confidence interactions from possible false positive measurements. On the other hand, we implemented PPI annotation and subnetwork generation algorithms that allow to perform more sophisticated queries, addressing particular research questions under more physiological conditions. When implementing HIPPIE, we put an emphasis on usability and, over the last years, reacted to numerous user requests to improve its documentation and interface. For example, the HIPPIE webpage now contains a step-by-step manual to reproduce the biological pathway reconstruction depicted in Figure 1B and to guide new users through the process of performing more complex analyses in the resource. Also, the possibility to upload experimental data, which then get scanned for known PPIs and annotated with basic summary information, was implemented upon request from wet lab scientists performing the experimental screening of protein interactions and is one of HIPPIE's most commonly used features.
Thanks to the implementation of the annotation of PPIs with disease terms, HIPPIE is particularly useful in a biomedical context. But even without explicitly considering known gene–disease associations in HIPPIE queries, we demonstrated here and in previous studies (29,30) the power of HIPPIE to reveal disease mechanisms by integrating heterogeneous types of experimental data.
Gene expression changes over tissues are an important modulator of cell type-specific signaling (48,49) and disease (50). Therefore, projecting accurate estimates of gene abundance onto PPI networks will provide more physiological insights into the functional organization of the cell under healthy and disease conditions. With GTEx, we incorporated one of the most extensive and accurate catalogs of gene expression in human tissues.
Future developments of HIPPIE will include, for example, developing ways to deal with the inherent study bias in PPI data sets to generate bias-reduced networks (14), and improving our method to annotate PPIs based on GO terms (see Materials and Methods) by taking into account frequencies of GO terms.
In summary, HIPPIE makes an important step in the direction of extracting meaningful and reliable PPI subnetworks from the growing amount of human PPI data and of guiding the development of biological hypotheses from network data.
AVAILABILITY
HIPPIE is freely available to academic users via http://cbdm.uni-mainz.de/hippie/.
FUNDING
German Research Foundation [SCHA 1933/1-1]; Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013–2017’ [SEV-2012-0208]; European Union Seventh Framework Programme (FP7/2007-2013) [n° HEALTH-F4-2011-278568 (PRIMES)]; Spanish Ministerio de Economía y Competitividad [Plan Nacional BIO2012-39754]; European Fund for Regional Development (EFRD). Funding for open access charge: University of Mainz.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chatr-Aryamontri A., Breitkreutz B.-J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N., et al. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel C.L., Owens C.C., Murali T.M., Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., Bono de B., Jassal B., et al. Network-based functional enrichment. BMC Bioinformatics. 2011;12:S14. doi: 10.1186/1471-2105-12-S13-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radivojac P., Clark W.T., Oron T.R., Schnoes A.M., Wittkop T., Sokolov A., Graim K., Funk C., Verspoor K., Ben-Hur A., et al. A large-scale evaluation of computational protein function prediction. Nat. Methods. 2013;10:221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piñero J., Queralt-Rosinach N., Bravo À., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun K., Gonçalves J.P., Larminie C., Przulj N. Predicting disease associations via biological network analysis. BMC Bioinformatics. 2014;15:304. doi: 10.1186/1471-2105-15-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart G.T., Ramani A.K., Marcotte E.M. How complete are current yeast and human protein-interaction networks. Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesan K., Rual J.F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T., Hao T., Zenkner M., Xin X., Goh K.I., et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mrowka R., Patzak A., Herzel H. Is there a bias in proteome research. Genome Res. 2001;11:1971–1973. doi: 10.1101/gr.206701. [DOI] [PubMed] [Google Scholar]
- 10.Von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 11.Bisson N., James D.A., Ivosev G., Tate S.A., Bonner R., Taylor L., Pawson T. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat. Biotechnol. 2011;29:653–658. doi: 10.1038/nbt.1905. [DOI] [PubMed] [Google Scholar]
- 12.Futschik M.E., Chaurasia G., Herzel H. Comparison of human protein–protein interaction maps. Bioinformatics. 2007;23:605–611. doi: 10.1093/bioinformatics/btl683. [DOI] [PubMed] [Google Scholar]
- 13.Gillis J., Ballouz S., Pavlidis P. Bias tradeoffs in the creation and analysis of protein-protein interaction networks. J. Proteomics. 2014;100:44–54. doi: 10.1016/j.jprot.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer M.H., Serrano L., Andrade-Navarro M.A. Correcting for the study bias associated with protein–protein interaction measurements reveals differences between protein degree distributions from different cancer types. Front. Genet. 2015;6:260. doi: 10.3389/fgene.2015.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brito G.C., Andrews D.W., Tan S., Tan H., Chung M., Engelenburg Van S., Palmer A., Moffat J., Sabatini D., Costanzo M., et al. Removing bias against membrane proteins in interaction networks. BMC Syst. Biol. 2011;5:169. doi: 10.1186/1752-0509-5-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Silva E., Thorne T., Ingram P., Agrafioti I., Swire J., Wiuf C., Stumpf M.P., de Silva E., Stumpf M., Stelzl U., et al. The effects of incomplete protein interaction data on structural and evolutionary inferences. BMC Biol. 2006;4:39. doi: 10.1186/1741-7007-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranda B., Blankenburg H., Kerrien S., Brinkman F.S., Ceol A., Chautard E., Dana J.M., De Las Rivas J., Dumousseau M., Galeota E., et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer M.H., Fontaine J.-F., Vinayagam A., Porras P., Wanker E.E., Andrade-Navarro M.A. HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLoS One. 2012;7:e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del-Toro N., Dumousseau M., Orchard S., Jimenez R.C., Galeota E., Launay G., Goll J., Breuer K., Ono K., Salwinski L., et al. A new reference implementation of the PSICQUIC web service. Nucleic Acids Res. 2013;41:W601–W606. doi: 10.1093/nar/gkt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerrien S., Aranda B., Breuza L., Bridge A., Broackes-Carter F., Chen C., Duesbury M., Dumousseau M., Feuermann M., Hinz U., et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E., Sacco F., Palma A., Nardozza A.P., Santonico E., et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A., et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isserlin R., El-Badrawi R.A., Bader G.D. The biomolecular interaction network database in PSI-MI 2.5. Database (Oxford) 2011;2011:baq037. doi: 10.1093/database/baq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagel P., Kovac S., Oesterheld M., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Mark P., Stumpflen V., Mewes H.W., et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 26.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 27.Ardlie K.G., Deluca D.S., Segre A. V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., Lek M., et al. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 29.Schaefer M.H., Lopes T.J.S., Mah N., Shoemaker J.E., Matsuoka Y., Fontaine J.-F., Louis-Jeune C., Eisfeld A.J., Neumann G., Perez-Iratxeta C., et al. Adding protein context to the human protein-protein interaction network to reveal meaningful interactions. PLoS Comput. Biol. 2013;9:e1002860. doi: 10.1371/journal.pcbi.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suratanee A., Schaefer M.H., Betts M.J., Soons Z., Mannsperger H., Harder N., Oswald M., Gipp M., Ramminger E., Marcus G., et al. Characterizing protein interactions employing a genome-wide siRNA cellular phenotyping screen. PLoS Comput. Biol. 2014;10:e1003814. doi: 10.1371/journal.pcbi.1003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrade-Navarro M.A., Fontaine J.F. Gene set to Diseases (GS2D): disease enrichment analysis on human gene sets with literature data. Genomics and Computational Biology. 2016 In press. [Google Scholar]
- 33.Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–D342. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes T.J.S., Schaefer M., Shoemaker J., Matsuoka Y., Fontaine J.-F., Neumann G., Andrade-Navarro M.A., Kawaoka Y., Kitano H. Tissue-specific subnetworks and characteristics of publicly available human protein interaction databases. Bioinformatics. 2011;27:2414–2421. doi: 10.1093/bioinformatics/btr414. [DOI] [PubMed] [Google Scholar]
- 35.Lan A., Ziv-Ukelson M., Yeger-Lotem E. A context-sensitive framework for the analysis of human signalling pathways in molecular interaction networks. Bioinformatics. 2013;29:i210–i216. doi: 10.1093/bioinformatics/btt240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magger O., Waldman Y.Y., Ruppin E., Sharan R., Schunkert H., König I., Kathiresan S., Reilly M., Assimes T., Birnbaum S., et al. Enhancing the Prioritization of Disease-Causing Genes through Tissue Specific Protein Interaction Networks. PLoS Comput. Biol. 2012;8:e1002690. doi: 10.1371/journal.pcbi.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuncbag N., McCallum S., Huang S.-S.C., Fraenkel E. SteinerNet: a web server for integrating ‘omic’ data to discover hidden components of response pathways. Nucleic Acids Res. 2012;40:W505–W509. doi: 10.1093/nar/gks445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barshir R., Shwartz O., Smoly I.Y., Yeger-Lotem E. Comparative analysis of human tissue interactomes reveals factors leading to tissue-specific manifestation of hereditary diseases. PLoS Comput. Biol. 2014;10:e1003632. doi: 10.1371/journal.pcbi.1003632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franz M., Lopes C.T., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics. 2016;32:309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner B., Razick S., Turinsky A.L., Vlasblom J., Crowdy E.K., Cho E., Morrison K., Donaldson I.M., Wodak S.J. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database. 2010;2010:baq023–baq023. doi: 10.1093/database/baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kotlyar M., Pastrello C., Sheahan N., Jurisica I. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016;44:D536–D541. doi: 10.1093/nar/gkv1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barshir R., Basha O., Eluk A., Smoly I.Y., Lan A., Yeger-Lotem E. The TissueNet database of human tissue protein-protein interactions. Nucleic Acids Res. 2013;41:D841–D844. doi: 10.1093/nar/gks1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basha O., Flom D., Barshir R., Smoly I., Tirman S., Yeger-Lotem E. MyProteinNet: build up-to-date protein interaction networks for organisms, tissues and user-defined contexts. Nucleic Acids Res. 2015;43:W258–W263. doi: 10.1093/nar/gkv515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C., et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karagoz K., Sevimoglu T., Arga K.Y. Integration of multiple biological features yields high confidence human protein interactome. J. Theor. Biol. 2016;403:85–96. doi: 10.1016/j.jtbi.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Vidal M., Cusick M.E., Barabási A.-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaefer M.H., Yang J.-S., Serrano L., Kiel C. Protein Conservation and Variation Suggest Mechanisms of Cell Type-Specific Modulation of Signaling Pathways. PLoS Comput. Biol. 2014;10:e1003659. doi: 10.1371/journal.pcbi.1003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiel C., Verschueren E., Yang J.-S., Serrano L. Integration of protein abundance and structure data reveals competition in the ErbB signaling network. Sci. Signal. 2013;6:ra109. doi: 10.1126/scisignal.2004560. [DOI] [PubMed] [Google Scholar]
- 50.Lage K., Hansen N.T., Karlberg E.O., Eklund A.C., Roque F.S., Donahoe P.K., Szallasi Z., Jensen T.S., Brunak S. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20870–20875. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon S., Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]