Abstract
Rhea (http://www.rhea-db.org) is a comprehensive and non-redundant resource of expert-curated biochemical reactions designed for the functional annotation of enzymes and the description of metabolic networks. Rhea describes enzyme-catalyzed reactions covering the IUBMB Enzyme Nomenclature list as well as additional reactions, including spontaneously occurring reactions, using entities from the ChEBI (Chemical Entities of Biological Interest) ontology of small molecules. Here we describe developments in Rhea since our last report in the database issue of Nucleic Acids Research. These include the first implementation of a simple hierarchical classification of reactions, improved coverage of the IUBMB Enzyme Nomenclature list and additional reactions through continuing expert curation, and the development of a new website to serve this improved dataset.
INTRODUCTION
Rhea (http://www.rhea-db.org) is a comprehensive and non-redundant resource of expert-curated biochemical reactions designed for the functional annotation of enzymes and the description of metabolic networks (1). Rhea covers reactions of the hierarchical enzyme classification of the Enzyme Nomenclature committee of the IUBMB (hereafter referred to as the ‘EC’) (2,3) as represented by the ENZYME (4) and IntEnz (5) resources as well as additional enzymatic and transport reactions and spontaneously occurring reactions described in the literature.
Rhea reactions are defined by their participants and a specific reaction direction. Rhea represents small molecules and the functional groups of large macromolecules such as proteins using chemical entities from the ChEBI ontology (6), selecting the major microspecies (protonation state) for each ChEBI entity at an arbitrary pH of 7.3 and balancing all reactions for mass and charge accordingly. The curation of small molecule data is an integral part of the Rhea curation workflow, and Rhea curators have submitted thousands of new compounds to ChEBI during its development. Each set of reaction participants is associated to four potential directions: left to right (LR, =>), right to left (RL, <= ), bidirectional (BI, <=>) and undefined (UN, <?>), each with its own unique reaction identifier. Rhea reactions can be used to annotate the preferred direction ( =>, <=, <=>) of experimentally characterized enzymatic reactions in knowledgebases, to describe metabolic networks and derived models where reaction fluxes are not defined a priori (<?>), and to link knowledgebases and models. Knowledgebases that use Rhea for the annotation of enzyme and metabolite data include the SwissLipids knowledgebase for lipid biology (7), the EBI Enzyme portal (8), the MetaboLights repository of metabolomics data (9) and IntEnz (5). Resources that use Rhea for the annotation of metabolic networks and models include MetaNetX (10,11) and Microscope (12). Rhea also provides links to other metabolite and pathway databases such as KEGG (13), MetaCyc (14) and Reactome (15). More information about Rhea reactions and their use can be found in our previous publications (1,16).
In the following sections we summarize recent developments in Rhea since our last publication (1). These include the first implementation of a simple hierarchical classification of reactions, improved coverage of known enzymatic activities and additional reactions through continuing expert curation and the development of a new website to serve this enhanced dataset.
CURRENT DEVELOPMENTS IN RHEA
Rhea reaction classification
The EC uses a hierarchy of exactly four levels to classify enzyme activities according to a single representative reaction (referred to hereafter as the ‘representative reaction of the IUBMB’). Rhea (like other reaction databases) aims to provide complete coverage of all representative reactions of the IUBMB as well as other additional reactions described in the literature (whether or not these are related to an EC number).
To improve the annotation and classification of enzyme functions using Rhea we recently introduced a simple hierarchical reaction classification that covers representative reactions of the IUBMB as well as these additional reactions. This reaction classification uses ‘is a’ relationships to link ‘child’ reactions to their (more generic) ‘parent’ reactions, and allows any number of levels in order to facilitate the classification of extant and ancestral enzyme functions in ways that are meaningful to biologists. This may include the introduction of reaction classes that lie between existing levels of the EC and a finer grained classification of related reactions that are currently ‘compressed’ into the fourth level of the EC.
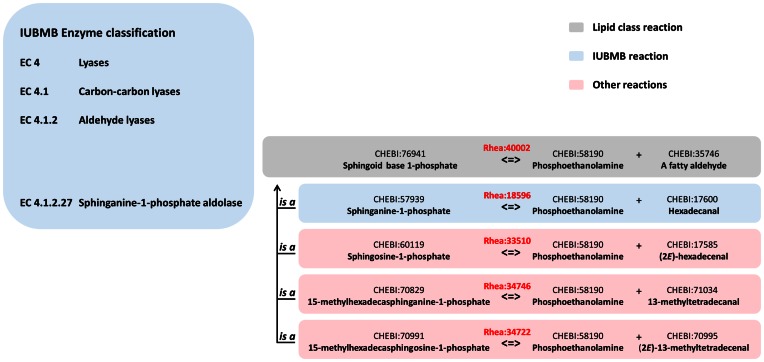
Figure 1 illustrates the use of the Rhea reaction hierarchy to classify the reactions associated with the enzyme activity sphinganine-1-phosphate aldolase (EC 4.1.2.27). The representative reaction of the IUBMB for this enzyme is ‘sphinganine 1-phosphate = phosphoethanolamine + palmitaldehyde’ (RHEA:18596), which is catalyzed by the enzyme SGPL1 of Homo sapiens as well as orthologous enzymes in other species (17). Sphinganine 1-phosphate is one member of a class of related phosphorylated sphingoid bases which vary in chain length, degree of saturation and branching. Additional related reactions (not mentioned by the EC reference list (3)) feature other members of this class such as sphingosine-1-phosphate (18) (RHEA:33510), the main phosphorylated sphingoid base in H. sapiens, as well as 15-methylhexadecasphinganine-1-phosphate (RHEA:34746) and 15-methylhexadecasphingosine-1-phosphate (RHEA:34722) (19), two phosphorylated sphingoid bases of Caenorhabditis elegans and the presumed substrates for the C. elegans SPGL1 homolog spl-1. The common ancestral function of H. sapiens SPGL1 and C. elegans spl-1 could be summarized as ‘sphingoid-1-phosphate lyase’, which is a specialization of the ‘aldehyde-lyases’ (the third level of the EC classification, EC 4.1.2) and a generalization of ‘sphinganine-1-phosphate aldolase’ (the fourth and final level, EC 4.1.2.27). This common ancestral function therefore lies between levels of the EC. In Rhea a grouping reaction ‘a sphingoid 1-phosphate = a fatty aldehyde + phosphoethanolamine’ (RHEA:40002) was created using the newly defined grouping class of metabolite ‘a sphingoid 1-phosphate’ (CHEBI:76941) and existing metabolite classes. This grouping reaction links the representative reaction of the IUBMB to these additional related reactions. It can be used to annotate extant members of this orthologous group as well as ancestral functions in phylogenetic trees (20) at a greater level of precision than the generic ‘aldehyde lyase’ annotation EC 4.1.2.
Figure 1.
The Rhea reaction classification. The IUBMB enzyme classification (left) describes the enzyme activity sphinganine-1-phosphate aldolase (EC 4.1.2.27), which catalyzes the reaction Rhea:18596 (blue). This reaction, and other reactions including Rhea:33510, Rhea:34720 and Rhea:34746 (pink), are specific forms of the more generic reaction Rhea:40002 (grey), which lies between EC 4.1.2 and EC 4.1.2.27. Enzymes are omitted for the sake of clarity.
The Rhea reaction classification was introduced in July 2016. The current release (release 75 of 30 July 30 2016) features over 600 expert-curated reaction relations. We are currently developing methods to calculate reaction relations for legacy data using curated relations in the ChEBI ontology (6) and computed measures of chemical structure and reaction similarity (21–23). We expect the number of reaction relations to increase significantly in the near future as we continue to check and validate calculated relations.
Rhea content
Rhea has steadily grown since our last report through the expert curation of new chemical entities in ChEBI and reactions from peer-reviewed literature (see http://www.rhea-db.org/ statistics for details). At the time of writing, Rhea (release 75 of 30 July 30 2016) includes 9273 unique reactions (not considering directions) involving 8124 unique reaction participants, an increase of 2152 unique reactions and 2094 unique reaction participants since our last publication (release 53 of July 2014). Rhea covers over 94% of EC numbers with a defined reaction (4794 of 5124 EC numbers), and provides 4479 additional reactions. Many of these additional reactions were curated to support the generation of lipid libraries in the SwissLipids resource (7).
Rhea cites 8905 unique PubMed identifiers, an increase of 6142 since our last publication. This large increase in the amount of curated literature is the result of a concerted effort to map all enzymatic activities described in UniProtKB/Swiss-Prot to Rhea (including the representative reactions of the IUBMB and additional reactions) (see ‘Future directions’ section). During this process the existing literature from UniProtKB/Swiss-Prot was reviewed by Rhea curators and curated into Rhea where necessary.
Rhea website
Since our last publication we have developed and deployed a new website at http://www.rhea-db.org. This website provides the same options for interactive and programmatic access as the previous version (1). Users can search for reaction and compound identifiers and names, EC numbers, UniProtKB/Swiss-Prot accession numbers, bibliographic citations and identifiers from external cross-referenced resources at http://www.rhea-db.org/advancedsearch. Reaction data can be downloaded in BioPax2 (24), RXN and RD (25) formats at http://www.rhea-db.org/download, which also provides access to the newly introduced reaction relations (described above) in tab-delimited form. Individual reactions can be bookmarked by adding the required identifier to the URL template http://www.rhea-db.org/reaction?id=, as in this example: http://www.rhea-db.org/reaction?id=10499. Reaction data in BioPax2 (24), RXN (25) and CMLReact (26) formats can also be obtained using RESTful web services at http://www.rhea-db.org/webservice.
DISCUSSION
The UniProt consortium will use Rhea as a vocabulary for the annotation of enzymatic activities in UniProtKB from late 2017/early 2018 onward. To this end we continue to increase the coverage of Rhea through expert curation of new reactions, including representative reactions of the IUBMB and additional reactions described in peer reviewed literature. We also plan to develop an automated pipeline that assists Rhea reaction curation by identifying and prioritizing candidate reactions from the MetaNetX resource of genome scale metabolic models (10,11). More immediate developments are focused on a new RDF representation of Rhea data. This will be made available at a dedicated SPARQL endpoint to be hosted by the Vital-IT infrastructure (https://www.vital-it.ch/), which currently maintains a number of SPARQL endpoints such as http://sparql.uniprot.org/ and http://snorql.nextprot.org.
Acknowledgments
The authors would like to thank Jerven Bolleman and Sebastien Gehant of the Swiss-Prot group of SIB for stimulating discussions on many areas of development including RDF and database design and Simone Badoer of the Web Production team of EMBL-EBI for invaluable technical assistance. We would also like to thank the Cheminformatics and Metabolism Team at the European Bioinformatics Institute for their invaluable work in maintaining and developing ChEBI. We gratefully acknowledge the software contributions of ChemAxon [https://www.chemaxon.com/products/marvin/].
FUNDING
Swiss Federal Government through the State Secretariat for Education, Research and Innovation (SERI); SwissLipids project of the SystemsX.ch, the Swiss Initiative in Systems Biology (in part); EMBL. Funding for open access charge: SERI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Morgat A., Axelsen K.B., Lombardot T., Alcantara R., Aimo L., Zerara M., Niknejad A., Belda E., Hyka-Nouspikel N., Coudert E., et al. Updates in Rhea-a manually curated resource of biochemical reactions. Nucleic Acids Res. 2015;43:D459–D464. doi: 10.1093/nar/gku961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDonald A.G., Tipton K.F. Fifty-five years of enzyme classification: advances and difficulties. FEBS J. 2014;281:583–592. doi: 10.1111/febs.12530. [DOI] [PubMed] [Google Scholar]
- 3.McDonald A.G., Boyce S., Tipton K.F. ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009;37:D593–D597. doi: 10.1093/nar/gkn582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fleischmann A., Darsow M., Degtyarenko K., Fleischmann W., Boyce S., Axelsen K.B., Bairoch A., Schomburg D., Tipton K.F., Apweiler R. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aimo L., Liechti R., Hyka-Nouspikel N., Niknejad A., Gleizes A., Gotz L., Kuznetsov D., David F.P., van der Goot F.G., Riezman H., et al. The SwissLipids knowledgebase for lipid biology. Bioinformatics. 2015;31:2860–2866. doi: 10.1093/bioinformatics/btv285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcantara R., Onwubiko J., Cao H., Matos P., Cham J.A., Jacobsen J., Holliday G.L., Fischer J.D., Rahman S.A., Jassal B., et al. The EBI enzyme portal. Nucleic Acids Res. 2013;41:D773–D780. doi: 10.1093/nar/gks1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haug K., Salek R.M., Conesa P., Hastings J., de Matos P., Rijnbeek M., Mahendraker T., Williams M., Neumann S., Rocca-Serra P., et al. MetaboLights - an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013;41:D781–D786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moretti S., Martin O., Van Du Tran T., Bridge A., Morgat A., Pagni M. MetaNetX/MNXref—reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 2016;44:D523–D526. doi: 10.1093/nar/gkv1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernard T., Bridge A., Morgat A., Moretti S., Xenarios I., Pagni M. Reconciliation of metabolites and biochemical reactions for metabolic networks. Brief Bioinform. 2014;15:123–135. doi: 10.1093/bib/bbs058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallenet D., Belda E., Calteau A., Cruveiller S., Engelen S., Lajus A., Le Fevre F., Longin C., Mornico D., Roche D., et al. MicroScope–an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A., et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcantara R., Axelsen K.B., Morgat A., Belda E., Coudert E., Bridge A., Cao H., de Matos P., Ennis M., Turner S., et al. Rhea–a manually curated resource of biochemical reactions. Nucleic Acids Res. 2012;40:D754–D760. doi: 10.1093/nar/gkr1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiss U., Oskouian B., Zhou J., Gupta V., Sooriyakumaran P., Kelly S., Wang E., Merrill A.H., Jr, Saba J.D. Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J. Biol. Chem. 2004;279:1281–1290. doi: 10.1074/jbc.M309646200. [DOI] [PubMed] [Google Scholar]
- 18.Brizuela L., Ader I., Mazerolles C., Bocquet M., Malavaud B., Cuvillier O. First evidence of sphingosine 1-phosphate lyase protein expression and activity downregulation in human neoplasm: implication for resistance to therapeutics in prostate cancer. Mol. Cancer Ther. 2012;11:1841–1851. doi: 10.1158/1535-7163.MCT-12-0227. [DOI] [PubMed] [Google Scholar]
- 19.Mendel J., Heinecke K., Fyrst H., Saba J.D. Sphingosine phosphate lyase expression is essential for normal development in Caenorhabditis elegans. J. Biol. Chem. 2003;278:22341–22349. doi: 10.1074/jbc.M302857200. [DOI] [PubMed] [Google Scholar]
- 20.Gaudet P., Livstone M.S., Lewis S.E., Thomas P.D. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman S.A., Torrance G., Baldacci L., Martinez Cuesta S., Fenninger F., Gopal N., Choudhary S., May J.W., Holliday G.L., Steinbeck C., et al. Reaction decoder tool (RDT): extracting features from chemical reactions. Bioinformatics. 2016;32:2065–2066. doi: 10.1093/bioinformatics/btw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giri V., Sivakumar T.V., Cho K.M., Kim T.Y., Bhaduri A. RxnSim: a tool to compare biochemical reactions. Bioinformatics. 2015;31:3712–3714. doi: 10.1093/bioinformatics/btv416. [DOI] [PubMed] [Google Scholar]
- 23.Rahman S.A., Cuesta S.M., Furnham N., Holliday G.L., Thornton J.M. EC-BLAST: a tool to automatically search and compare enzyme reactions. Nat. Methods. 2014;11:171–174. doi: 10.1038/nmeth.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demir E., Cary M.P., Paley S., Fukuda K., Lemer C., Vastrik I., Wu G., D'Eustachio P., Schaefer C., Luciano J., et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalby A., Nourse J.G., Hounshell W.D., Gushurst A.K.I., Grier D.L., Leland B.A., Laufer J. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J. Chem. Inform. Comput. Sci. 1992;32:244–255. [Google Scholar]
- 26.Holliday G.L., Murray-Rust P., Rzepa H.S. Chemical markup, XML, and the world wide web. 6. CMLReact, an XML vocabulary for chemical reactions. J. Chem. Inf. Model. 2006;46:145–157. doi: 10.1021/ci0502698. [DOI] [PubMed] [Google Scholar]