Abstract
Over the past years, CRISPR/Cas9 mediated genome editing has developed into a powerful tool for modifying genomes in various organisms. In high-throughput screens, CRISPR/Cas9 mediated gene perturbations can be used for the systematic functional analysis of whole genomes. Discoveries from such screens provide a wealth of knowledge about gene to phenotype relationships in various biological model systems. However, a database resource to query results efficiently has been lacking. To this end, we developed GenomeCRISPR (http://genomecrispr.org), a database for genome-scale CRISPR/Cas9 screens. Currently, GenomeCRISPR contains data on more than 550 000 single guide RNAs (sgRNA) derived from 84 different experiments performed in 48 different human cell lines, comprising all screens in human cells using CRISPR/Cas published to date. GenomeCRISPR provides data mining options and tools, such as gene or genomic region search. Phenotypic and genome track views allow users to investigate and compare the results of different screens, or the impact of different sgRNAs on the gene of interest. An Application Programming Interface (API) allows for automated data access and batch download. As more screening data will become available, we also aim at extending the database to include functional genomic data from other organisms and enable cross-species comparisons.
INTRODUCTION
High-throughput screening experiments have been an indispensable tool in functional genome research for many years. Functional screens can systematically interrogate genotype to phenotype relationships and identify key dependencies of biological systems – an essential requirement in understanding how genes function in the context of a cell or organism in health and disease (1). In the past, RNA interference has been widely used to perform such screens (2–5). Discoveries from many of these experiments have been made accessible through centralized data resources such as GenomeRNAi (6,7) and have fueled further developments in the area of systems genetics.
Recently, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats with the CRISPR associated protein Cas9) has gained substantial use in genetic screens in various organisms including human, mouse and zebrafish (8–10). In order to support the rapid development of this new field, accessibility of CRISPR/Cas9-derived functional data is crucial (11). Several resources have been established to address this need. These include CRISPRz (12), a database of sgRNAs validated in zebrafish, WGE (13), a data resource that contains information about sgRNAs that can be used to target genes of interest and CrisprGE (14), a platform that provides knowledge about sequence mutations caused by sgRNAs previously used in various experimental settings. Nevertheless, a database that allows to compare screening results, such as perturbation phenotypes or sgRNA efficiency, of many different experiments on a genome-wide scale has so far not been available. To fill this gap, we here report GenomeCRISPR (Supplementary Figure S1), a database for high-throughput screening experiments using CRISPR/Cas9. At the time of submission, GenomeCRISPR comprises data from a total of 84 different high-throughput screening experiments performed across 48 human cell lines. In such pooled screening format, similar to shRNA screens, high-complexity sgRNA collections are transduced into culture cells using lentiviruses approaches (4,15). Following CRISPR/Cas9 induced mutagenesis, a particular condition (e.g. drug selection) is applied to the pooled mutant cell library, and enrichment or depletion of sgRNAs are measured as phenotypes using next generation sequencing. GenomeCRISPR is rapidly updateable to include future active submissions and data curated from the scientific literature. GenomeCRISPR features information about reported performance of 553 122 sgRNAs used in these screens and focuses on interactive data visualization to allow intuitive comparison of the results among different experiments. Of these we report 85 564 sgRNA targeting genes identified as hits or positive controls by authors of the original publication. Here, we present GenomeCRISPR as a resource that will help users to query experimental data sets for questions, such as:
Has a gene of interest been perturbed before in CRISPR/Cas9 screens?
Which sgRNAs had the largest impact on the function of a specific gene?
Which phenotypes were observed upon perturbation of a gene under specific conditions?
DATABASE CONTENT
Currently, GenomeCRISPR contains data from 84 different high-throughput experiments reported in recently published publications using human tissue cell culture. These screens cover a variety of different experimental approaches such as applying CRISPR/Cas9k.o. to induce null alleles or using transcriptional activator or interference (‘CRISPRa/i’) (16). Further, these approaches have been applied in different screening experiments such as negative selection screens, where loss of a specific phenotype (e.g. fitness) is observed (8,17,15) and positive selection screens, where gain of phenotypes is measured (such as resistance to a drug) (18,19). The screens included in the database at the time of manuscript submission were carried out in 48 different human cancer cell lines and comprise negative selection screens (‘drop-out’) as well as positive selection screens for resistance to drug or virus perturbation. GenomeCRISPR was designed with flexibility in mind. Therefore, it can easily be expanded to add screening experiments in different organisms using newly discovered methods, the only necessary requirement being sgRNA sequences and quantitative phenotypes (e.g. sgRNA abundances before and after treatment). Further organisms in addition to human cultured cells will be included in the future as soon as a sufficient amount of data has become available.
To ensure high data quality standards, data were extracted from published experiments and imported into GenomeCRISPR via manual curation. Manual curation has several advantages over automated curation approaches. These include discovery of inconsistencies and errors in the data as well as the possibility of rejecting untrustworthy and incomplete information (20). Moreover, the heterogeneous nature of published data sets currently poses a big challenge for the development of automated curation pipelines. To this end, experimental information regarding screen design (pos. versus neg. selection), methodology (CRISPRa/i/n), cell line and experimental condition were determined. Details about the biological model system and the experimental condition (e.g. negative selection screen for cell viability) complement this information. Furthermore, score and hit information were annotated for perturbed genes as stated by the authors of the experiment. The ‘score’ is defined as a quantitative measure, which the study authors used to rank the tested genes by their phenotypic strength. A ‘hit’ is a gene, which exceeded a certain score threshold chosen by the authors of the publication to qualify for further validation. Finally, sequence, genomic location and sequencing read counts (a proxy for mutant abundance in pooled formats) were extracted for all sgRNAs using scripts deposited in https://github.com/boutroslab/ Supplementary Material as reported with E-CRISP (21). For pooled screening formats, log2 fold changes of all sgRNAs are calculated between their abundances in perturbed and unperturbed states or early and later time points from median normalized read counts. These fold changes are considered as the screen's signal. They were further summarized into 19 bins (Supplementary Figure S2), to enable comparisons between screens (Supplementary Methods).
DATA ACCESS
Website
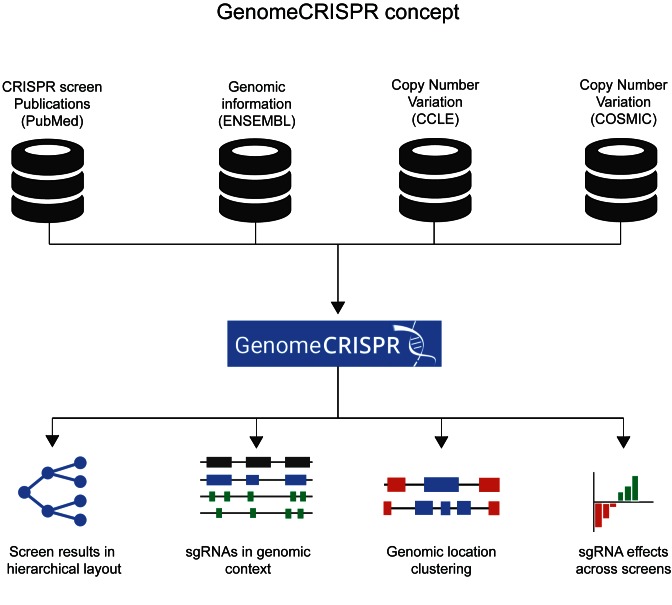
The GenomeCRISPR website is available at http://genomecrispr.org and serves as main access point to the database. A main page helps users to familiarize with the database concept by providing a short description of its contents (Figure 1). There, the database can be queried by either a gene identifier or a genomic range. It can be browsed to explore experiments reported in different publications annotated with data from ENSEMBL (genome information), CCLE (copy number variations) and COSMIC (copy number variations) (20,22,23). One use case to query GenomeCRISPR is the search for more information about a specific gene in order to investigate whether it showed a phenotype in one of the screens and, if so, which sgRNA caused the most significant functional impact. Searching for a gene using its gene symbol or ENSEMBL (22) gene ID takes the user to a result page that consists of three sections.
Figure 1.
Concept of the GenomeCRISPR database. CRISPR screening derived from publications or by directed submission is integrated with data from various sources (14,17,18). Screening data is then visualized characteristics of CRISPR reagents and screens.
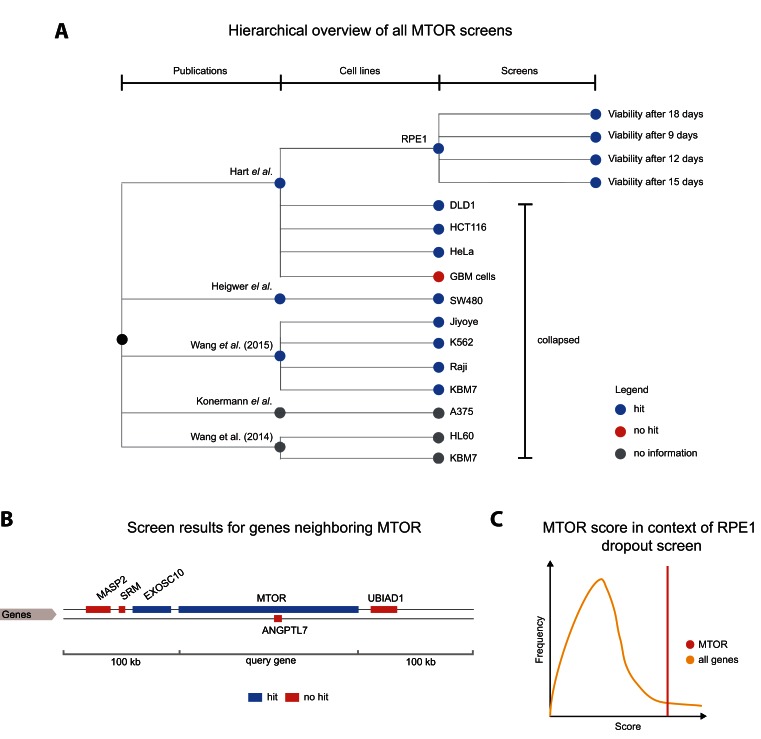
The first section displays an overview of all screens in the database that include a perturbation of the queried gene, here MTOR (Figure 2). A tabular representation of the results shows the users in which experiments MTOR has shown a significant phenotype as described by the authors. The phenotype itself can be inferred from the experiment, cell line and study title (the full title can be read in a pop-up window). A small ‘score in context’ graph will show how the gene's ‘score’ compares to all other genes tested in this experiment. Here, MTOR is scored as a ‘hit’ in several experiments, most prominently in all negative selection screens (17,15).
Figure 2.
Overview of all screens featuring a perturbation of MTOR. (A) A tree visualization shows published results across three levels of detail. An inner node represents a publication in scientific literature. Every publication node has a child node for each cell line used for experiments. The leaves of the tree illustrate different experimental conditions. Blue nodes imply that MTOR was identified as a hit in the experiment. Red nodes and gray nodes show that MTOR was not a hit, or that no hit information was provided by authors, respectively. In this example, RPE1 and SW480 cell line nodes have been expanded to show all performed experiments. (B) Results for all genes in the ±100 000 base pair neighborhood of MTOR. Blue and red colors depict hits and non-hits, respectively. (C) Hovering over one of the terminal nodes (to the right) triggers a ‘score in context’ figure. This cartoon represents the strength of the genes phenotype (score) in the context of all genes screened in this experiment. So the user can quickly judge upon the certainty of the hit identification.
A hierarchically structured screen overview is shown in the form of an interactive tree. This tree has four levels, which include (i) a root node displaying the entirety of results, (ii) publications, (iii) cell lines and a level for (iv) individual screening experiments. By default, individual screening experiments and tree nodes can be collapsed and expanded by users allowing them to filter the tree. Blue nodes indicate that a node or one of its children represents a screen in which the gene of interest was a ‘hit’. Red colour illustrates that neither the node nor any if its children correspond to a screen where the queried gene could be identified as hit. Grey nodes depict that the authors of the experiment have not provided such information. Nodes reveal information about title, abstract and authors of each publication by right-clicked. Moving the mouse cursor over an experiment node can check how well a gene scored in the context of the full screen (Figure 2C). There, a figure will be displayed showing the genes score relative to the distribution of all genes tested in the respective experiment.
Recently, Aguirre et al., Munoz et al. and others reported that already the existence of double strand breaks in endogenous DNA causes a phenotypic response to DNA-damage stress, often resulting in impaired fitness (24,25). Thus, clusters of ‘hits’ in neighbouring genes at a particular genomic location could be the consequence of a cell's response to a DNA-damage, e.g. caused by copy number variations in cancer cells (24,25). To provide users with a tool to quickly asses this source of potential false positives, experiment nodes can also be right-clicked which will display experiment results in a 100 000 base pair neighbourhood around the query gene. A help page (reachable via the ‘?’ icon) introduces new users to the screen tree. The visualization is implemented using the TnT Tree BioJavaScript module (26,27).
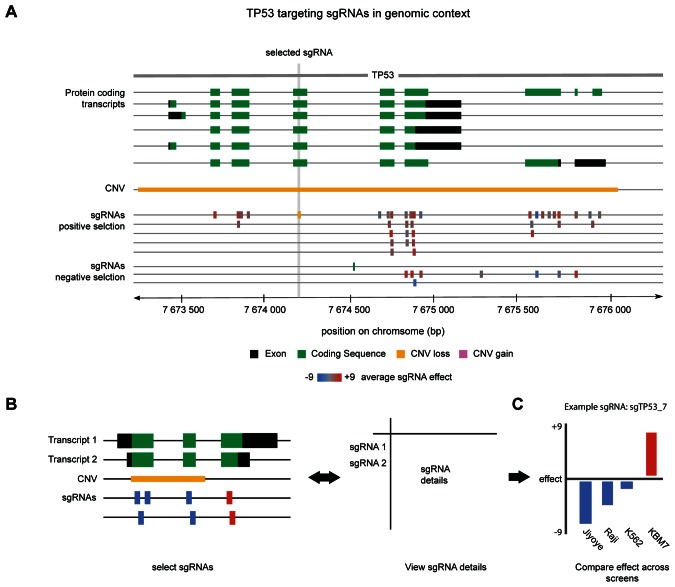
In the second section of the GenomeCRISPR results pages, users find detailed information about individual sgRNAs. Differently scoring sgRNAs can be identified and the ones that had the highest functional impact on the query gene can be singled out. In the upper half of the page an overview of all sgRNAs used in experiments is visualized in their genomic context (Figure 3A) using the neXtprot interactively zoomable feature viewer widget (28). As an example TP53's exon structure is shown in black and its coding sequence is displayed in green (Figure 3A). An sgRNA track is added for each screen type in which at least one reported experiment features a perturbation of the queried gene. In this example, 34 sgRNAs are illustrated as color-coded rectangles mapped to their genomic locus. The colour code represents their average functional effects across all experiments this sgRNA has been used in. Additional copy number variation tracks are shown for different cell lines (Supplementary Methods, Supplementary File 1) to help users evaluate observed sgRNA effects. By repeatedly clicking on sgRNA track-labels (indicated by the screen type), users can zap through the sgRNA contents. An exportable table positioned in the lower half of the page holds details about sgRNAs, such as location, sequence and direction. A click on an sgRNA feature will select it and focus the corresponding row in the sgRNA table (Figure 3B). Vice versa selection of an sgRNA in the table directs the genome browser to its location. These also include specifics about sgRNA Protospacer Adjacent Motif, targeted gene plus the least and the highest score effect size reached in any experiment. This information is complemented by an interactive bar plot (reachable at ‘screen details’), showing the measured signal (here log2 fold change between sgRNA abundances) in different conditions. The plot holds one bar for each experimental context the sgRNA was used in. Bar heights represent the binned measurement values (Supplementary Figure S2). On mouse-over, the maximum and minimum fold changes observed in the experiment can be inspected. This helps users get an idea about the effect size of the sgRNA-induced perturbation in the context of the entire experiment. Positive value bars are shown in blue and negative value bars are coloured red to provide a quick impression of effect type and size without having to examine axis labels in detail. The chart can be sorted by effect value yielding a waterfall plot, or experimental condition, which will result in grouping of bars by cell lines and conditions. For example, looking at the observed effect of the TP53 targeting sgRNA sgTP53_7 across multiple screens show clear differences between KBM7, Jiyoye, Raji and K562 cell lines, reflecting their differential dependencies on the activity of this gene (Figure 3C).
Figure 3.
sgRNAs used in screening experiments in their genomic context. (A) Feature viewer showing all sgRNAs used to screen TP53 in the selected genomic range. Black and green bars represent exons and coding sequence (CDS), respectively. As in most common genome browsers one transcript is shown per line. The yellow bar indicates a known loss of copy number at this position. sgRNA tracks are grouped according to the type of screen that was performed. Depending on the average effect size (Supplementary Figure S2) observed across all screens, sgRNA features are colored on a blue to gray to red color scale. (B) sgRNA features in the genome browser can be selected. Upon selection, details like sequence, genomic location or Protospacer Adjacent Motif (PAM) are highlighted in an sgRNA table. Furthermore, it is possible to select reagents in the sgRNA table and the genome browser will automatically zoom to their position. (C) A bar plot showing sgRNA effect (Supplementary Figure S2) observed across experiments can be displayed on demand. Positive and negative values indicate enrichment or depletion of the sgRNA, respectively. The plot is interactive and can be ordered and filtered according to user preference. Experiment details will be displayed as hover text.
Finally, a short section contains basic details about genomic location and function of the query gene and provides links to other data resources such as ENSEMBL (22) and GeneCards (29). Users can follow these links to find more detailed information about the query. And, an overview of all screening experiments in the database is provided on the about page in form of an interactive tree map, implemented using the JavaScript library D3 (30).
Computational access
GenomeCRISPR provides a RESTful API that can be used to query the database. Using this API, data of various scale can be downloaded on demand from a local user or other databases. Experiment and sgRNA data can be retrieved in JSON format according to several different selection criteria. Users can for example download all sgRNAs which ever reached a score of nine in any experiment. Likewise, one can also download just all sgRNAs and corresponding information for a two experiments and compare them to each other. Or users can download all sgRNA information for a particular gene in JSON format. A more detailed documentation service is available at http://genomecrispr.dkfz.de/api/documentation.
Example results
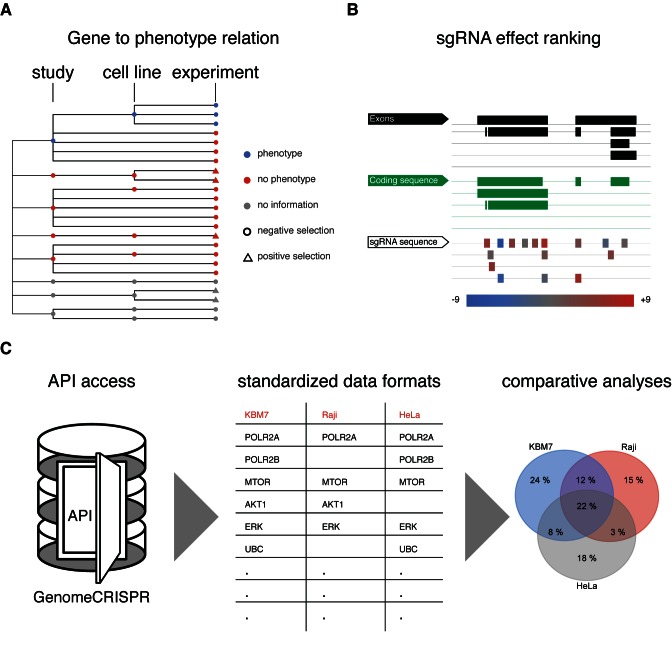
GenomeCRISPR provides functionalities to retrieve the following types of data (Figure 4). Firstly, it provides information of genotype to phenotype associations by displaying in which of the experiments a gene X has shown a significant phenotype (Figure 4A). For example, TP53 shows only in GBM cells a significant phenotype, clearly separating those from other cell lines. Secondly, GenomeCRISPR provides insight into which sgRNA constructs have been used to target gene X and what their phenotypic impact (in terms of discretized measured values) has been in different experimental setups. This allows, e.g. to identify for example the ‘best’ sgRNA to target gene X in follow-up experiments or other functional studies or allows to draw conclusion on future design of sgRNAs (Figure 4B). Our example demonstrates, that while all sgRNAs share this region of the TP53 gene model as target, they vary greatly in their functional penetrance. Thus, one could avoid sgRNAs that show no effect for further study. Finally, GenomeCRISPR provides a unique repository of the largest screens carried out using the CRISPR/Cas technology. Its data, when downloaded using the programmatic access features (API) or export functionality can also be utilized to perform large scale comparative analyses. For example, this could enable cross-screen or cross-sgRNA analysis of large sets and built new models of sgRNA efficacy (31,32) (Figure 4C).
Figure 4.
Example results of querying GenomeCRISPR. (A) Querying GenomeCRISPR for TP53 yields a tree view where it becomes apparent that only GBM cells (blue dots, label not shown) show a significant lack of viability phenotype upon loss of TP53. This allows to infer on the cell lines’ dependency on TP53 functional status and reveals its general difference to the other cell lines in the database. (B) In case a user wants to for example replicate the experiment or learn, which sgRNA has been best performing on perturbing TP53 in many instances a colour code directs to a quantitative answer of these questions. Here, most of the sgRNAs have shown the expected phenotype across screens (red bars). However, for unclear reasons two of the sgRNAs have shown the opposite phenotype (blue bars) and should be avoided. (C) All data in GenomeCRISPR can be downloaded programmatically through an API. Users can for example retrieve cell line specific hit lists using ‘R’ code (Supplementary File 2) or any other programming language. With this data, also large scale comparative analyses could be performed, e.g. searching for common hits across screens.
CONCLUSION
GenomeCRISPRs mission is to provide an easy to use resource for users to query, compare and visualize the results of high-throughput screening experiments by CRISPR/Cas9 genome-editing. It allows a wide range of user groups easy access to readily formatted data for a variety of applications. Moreover, while submission of data from the CRISPR screening community is highly encouraged a major challenge for robust curation of published data is the lack of a standardized format for the publication of CRISPR screens. Often, personal communications with authors of experiments is needed to acquire all necessary data. Also, data are frequently published after normalization, making it difficult to compare with data from other experiments. Therefore, we would like to propose a Minimal Information About CRISPR/Cas Screen (MIACCS) file as described in Supplementary Table S1. In the future, standardized analysis workflows might facilitate the submission and comparison of data sets (33). More complex phenotypes, such as high content phenotypes (34), might complement existing selection and sequencing-based phenotypic readouts. We are confident that GenomeCRISPR will be a useful resource for scientists to help them plan, design and evaluate CRISPR/Cas9 screening experiments as well as compare their results with existing data.
Acknowledgments
The authors thank Torben Brenner for support. Furthermore, the authors thank F. Port, T. Zhan, N. Rindtorff, L. Henkel, G. Ambrosi and additional members of the Boutros lab for feedback and discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
BMBF-funded Heidelberg Center for Human Bioinformatics (HD-HuB) within the German Network for Bioinformatics Infrastructure (de.NBI) [#031A537A]; Research in the lab of M.B. is supported by an ERC Advanced Grant of the European Commission. Funding for open access charge: German Cancer Research Center (DKFZ).
Conflict of interest statement. None declared.
REFERENCES
- 1.Carpenter A.E., Sabatini D.M. Systematic genome-wide screens of gene function. Nat. Rev. Genet. 2004;5:11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- 2.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Boutros M., Ahringer J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 4.Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 5.Bernards R. A missing link in genotype-directed cancer therapy. Cell. 2012;151:465–468. doi: 10.1016/j.cell.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt E., Pelz O., Buhlmann S., Kerr G., Horn T., Boutros M. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41:D1021–D1026. doi: 10.1093/nar/gks1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flockhart I.T., Booker M., Hu Y., McElvany B., Gilly Q., Mathey-Prevot B., Perrimon N., Mohr S.E. FlyRNAi.org - The database of the Drosophila RNAi screening center: 2012 update. Nucleic Acids Res. 2012;40:D715–D719. doi: 10.1093/nar/gkr953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.-R.J., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S., Sanjana N.E., Zheng K., Shalem O., Lee K., Shi X., Scott D.A., Song J., Pan J.Q., Weissleder R., et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 12.Varshney G.K., Zhang S., Pei W., Adomako-Ankomah A., Fohtung J., Schaffer K., Carrington B., Maskeri A., Slevin C., Wolfsberg T., et al. CRISPRz: a database of zebrafish validated sgRNAs. Nucleic Acids Res. 2016;44:D822–D826. doi: 10.1093/nar/gkv998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgkins A., Farne A., Perera S., Grego T., Parry-Smith D.J., Skarnes W.C., Iyer V. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015;31:3078–3080. doi: 10.1093/bioinformatics/btv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaur K., Tandon H., Gupta A.K., Kumar M. CrisprGE: a central hub of CRISPR/Cas-based genome editing. Database (Oxford) 2015;2015:bav055. doi: 10.1093/database/bav055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K.R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., Sabatini D.M. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heigwer F., Zhan T., Breinig M., Winter J., Brügemann D., Leible S., Boutros M. CRISPR library designer (CLD): software for multispecies design of single guide RNA libraries. Genome Biol. 2016;17:55. doi: 10.1186/s13059-016-0915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heigwer F., Kerr G., Boutros M. E-CRISP: fast CRISPR target site identification. Nat. Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G. V., Sonkin D., et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz D.M., Cassiani P.J., Li L., Billy E., Korn J.M., Jones M.D., Golji J., Ruddy D.A., Yu K., McAllister G., et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- 25.Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.-Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B., et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pignatelli M. TnT: a set of libraries for visualizing trees and track-based annotations for the web. Bioinformatics. 2016;32:2524–2525. doi: 10.1093/bioinformatics/btw210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corpas M., Jimenez R., Carbon S.J., García A., Garcia L., Goldberg T., Gomez J., Kalderimis A., Lewis S.E., Mulvany I., et al. BioJS: an open source standard for biological visualisation - its status in 2014. F1000Research. 2014;3:55. doi: 10.12688/f1000research.3-55.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaudet P., Michel P.-A., Zahn-Zabal M., Cusin I., Duek P.D., Evalet O., Gateau A., Gleizes A., Pereira M., Teixeira D., et al. The neXtProt knowledgebase on human proteins: current status. Nucleic Acids Res. 2015;43:D764–D770. doi: 10.1093/nar/gku1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H., et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bostock M., Ogievetsky V., Heer J. D3: Data-Driven Documents. IEEE Trans. Vis. Comput. Graph. 2011;17:2301–2309. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 31.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter J., Breinig M., Heigwer F., Bru D., Leible S., Pelz O., Zhan T., Boutros M. caRpools: An R package for exploratory data analysis and documentation of pooled CRISPR/Cas9 screens. Bioinformatics. 2015;32:632–634. doi: 10.1093/bioinformatics/btv617. [DOI] [PubMed] [Google Scholar]
- 34.Boutros M., Heigwer F., Laufer C. Microscopy-based high-content screening. Cell. 2015;163:1314–1325. doi: 10.1016/j.cell.2015.11.007. [DOI] [PubMed] [Google Scholar]