Abstract
Interactions between nitrogen-fixing (i.e. diazotrophic) cyanobacteria and their viruses, cyanophages, can have large-scale ecosystem effects. These effects are mediated by temporal alterations in nutrient availability in aquatic systems owing to the release of nitrogen and carbon sources from cells lysed by phages, as well as by ecologically important changes in the diversity and fitness of cyanobacterial populations that evolve in the presence of phages. However, ecological and evolutionary feedbacks between phages and nitrogen-fixing cyanobacteria are still relative poorly understood. Here, we used an experimental evolution approach to test the effect of interactions between a common filamentous, nitrogen-fixing cyanobacterium (Nodularia sp.) and its phage on cellular nitrogen release and host properties. Ecological, community-level effects of phage-mediated nitrogen release were tested with a phytoplankton bioassay. We found that cyanobacterial nitrogen release increased significantly as a result of viral lysis, which was associated with enhanced growth of phytoplankton species in cell-free filtrates compared with phage-resistant host controls in which lysis and subsequent nutrient release did not occur after phage exposure. We also observed an ecologically important change among phage-evolved cyanobacteria with phage-resistant phenotypes, a short-filamentous morphotype with reduced buoyancy compared with the ancestral long-filamentous morphotype. Reduced buoyancy might decrease the ability of these morphotypes to compete for light compared with longer, more buoyant filaments. Together, these findings demonstrate the potential of cyanobacteria–phage interactions to affect ecosystem biogeochemical cycles and planktonic community dynamics.
Keywords: cyanobacteria, eco-evolutionary dynamics, Nodularia sp., cyanophages, host–parasite interaction, marine nitrogen cycle
1. Introduction
Cyanobacteria are prominent components of phytoplankton in marine ecosystems, accounting for a significant proportion of global primary production and nitrogen fixation [1–8]. Buoyancy conferred by gas vesicles and the ability to fix atmospheric nitrogen (i.e. diazotrophy) are important factors promoting the competitive dominance of bloom-forming filamentous species in genera such as Nodularia, Aphanizomenon and Anabaena [9,10]. Once blooms have formed, they may be disrupted by physical factors such as intensification of wind [11,12], increase in salinity [13] and decrease in temperature [14], light radiation [15] and the availability of phosphorus [16,17].
Lytic viruses occur in high titres in the upper illuminated layers of the ocean and have been associated with cyanobacterial bloom decay in marine ecosystems across the globe [1,18–22]. However, the more specific role of viruses in bloom termination is relatively poorly understood. In addition to performing photosynthesis, certain cyanobacteria can fulfil their nitrogen demand by nitrogen fixation. Thus, interactions between cyanobacteria and their viruses, cyanophages may play an important role in global biogeochemical cycles [1,23,24]. However, not many studies exist on the eco-evolutionary feedbacks between nitrogen-fixing, or diazotrophic, cyanobacteria and their phages [25]. Fixed nitrogen is released from cyanobacterial blooms, and may be routed to higher trophic levels, through associated heterotrophic bacteria and phytoplankton species [6,7,26,27]. These include members of the nitrogen-limited unicellular picocyanobacterial genera Synechococcus and Prochlorococcus, which are key primary producers in oceans and display mass occurrence in association with nitrogen-fixing cyanobacteria [2,28,29]. Nitrogenous compounds not only leak out of living cells, primarily as dissolved organic nitrogen and ammonium, but are also released in high concentrations as a result of cellular lysis, such as during bloom decay [26,30]. Because lytic viruses can cause rapid cellular lysis and bloom decay, they can be hypothesized to affect the release of nitrogen from cyanobacterial hosts, altering the flow of nitrogen in planktonic food webs.
Instead of being stable, however, interactions between cyanobacteria and cyanophages can be altered through antagonistic coevolution. Resistance to co-occurring phages has been commonly observed in natural cyanobacterial communities [18,31], suggesting reduction in ecosystem-level effects that result from host lysis. Phage resistance, in turn, has been associated with reduced fitness [32], commonly interpreted as reduced growth rate [33]. However, in a number of studies on picocyanobacteria and their phages, a growth ability cost has not been observed in all resistance mutants [34–37]. Further, it has been proposed that reduced fitness may result from trade-offs between various fitness-related host properties, not only growth ability [33]. Besides affecting host fitness, phages have been found to maintain host diversity at all levels from genomes to populations and communities [38,39]. In addition to the immediate ecological effects resulting from the invasion of a host population by a lytic phage, alterations in host fitness and diversity caused by antagonistic coevolution can thus have wide-ranging ecological and evolutionary consequences.
Here, we used an experimental evolution approach to study the effect of evolving interactions between the filamentous, nitrogen-fixing cyanobacterium Nodularia sp. strain AV2 and its lytic phage vB_NpeS-2AV2 (Siphoviridae) [25] on cellular nitrogen release and host fitness and diversity. Nodularia spumigena is among the most common planktonic cyanobacteria and the most important nitrogen-fixers in marine ecosystems such as the Baltic Sea [5,40–42]. We found that rapid evolution in host resistance altered the flow of nitrogen from the cyanobacterium to the environment, with strong community-level effects among other phytoplankton species. Further, we discovered a novel cost of phage resistance, decreased light-competing ability, in association with increased phenotypic diversity in filament morphology and growth ability. These observations shed light on eco-evolutionary dynamics in key species interaction, in our case between nitrogen-fixing cyanobacteria and their phages, which ultimately can have large-scale ecosystems-level effects via altered biogeochemical cycles.
2. Material and methods
2.1. Test species, culture media and culture conditions
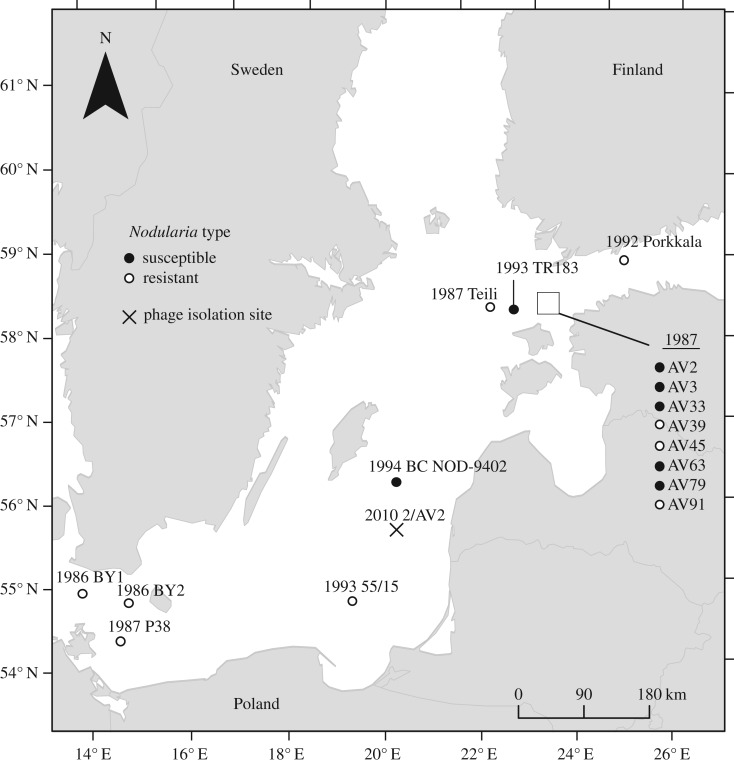
The cyanophage vB_NpeS-2AV2, identified as a member of Siphoviridae, was isolated from the surface water in the Baltic proper in June of 2010 after the disappearance of a Nodularia bloom [25]. Cyanobacterial host strains of the filamentous, nitrogen-fixing species Nodularia sp., isolated from the Baltic Sea, were obtained from the University of Helsinki Culture Collection (HAMBI). The host range of the cyanophage vB_NpeS-2AV2 was tested against 45 Nodularia genera isolates from a wide geographical area in the Baltic Sea (figure 1; electronic supplementary material, table S2; for methods, see resistance assay below), isolated between the years 1987 and 1994. When estimating the ecological effects of release of intracellular nitrogen on phytoplankton community, we used 11 phytoplankton strains representing different taxonomic groups, including green algae, diatoms and picocyanobacteria. Details of these strains are provided in the electronic supplementary material, table S1.
Figure 1.
Spatial and temporal occurrence of the Nodularia host in the Baltic Sea. (Complete information for strains is provided in the electronic supplementary material, table S2.)
In all experiments, we used the cyanobacterial culture medium Z8 with salt and without nitrogen [15,43]. Medium was prepared in glass bottles in type 2 analytical grade water (ELIX® water purification system, Millipak® 40 0.22 µm filter, Merck Millipore, Billerica, MA) and sterilized by autoclaving. All cultures were kept in static conditions in plastic cell culture vials (Sarstedt or VWR) at 25 ± 1°C and a continuous light intensity of 5–8 µmol m−2 s−1.
2.2. Obtaining phenotypes with different evolutionary history
The populations of Nodularia sp. strain AV2 studied here were the result of a 22-week-long microcosm experiment consisting of two treatments, host alone (naive population) and host with the phage vB_NpeS-2AV2 (evolved population), with three replicates in both treatments (electronic supplementary material, figure S1; experimental details in [25]). Samples used in this study were collected at the end of the experiment (week 22). From these population-level samples, we isolated approximately 20 clonal strains from each of the three replicates in both treatments (n = 118). Strains were isolated and purified by culturing samples in 0.55% agarose plates containing modified Z8 medium. Upon plating, individual filaments were allowed to grow into colonies, and transferred to 20 ml of liquid medium. All isolated clones were tested for phage resistance as described above, and because all clones were cultured in liquid medium for more than 20 generations (more than three months) without the phage, we can safely assume that the resistance trait is heritable.
2.3. Identifying potential hosts strains for vB_NpeS-2AV2 phage among the Baltic Sea isolates and testing host resistance
We determined potential host strains for phage vB_NpeS-2AV2 and the phage resistance of experimental isolates with a previously used optical density (OD)-based method [44]. Phage susceptible phenotypes (host strains) and phage-resistant phenotypes were determined as quantitative traits by comparing OD values between cultures with active and inactivated phage, with no difference indicating phage resistance and a significant difference indicating phage susceptibility. In the case of determining host strains among natural isolates from the culture collection (strain details in the electronic supplementary material, table S2), 100 µl of phage stock (approx. 3.6 × 107 pfu ml−1) or the same stock inactivated by autoclaving was added to 8 ml containing 2 ml of a dense culture of a Nodularia strain and 6 ml of fresh liquid medium in a six-well plate (n = 4). After culturing for 7 days, OD at 750 nm was measured (Tecan Infinite M200). Based on the analysis of 45 culture collection isolates, the strain AV2 was chosen as a model for a susceptible Nodularia strain in the preceding experimental evolution study [25]. When assessing the resistance of clonal Nodularia sp. AV2 isolates obtained from the end of the evolutionary experiment, a volume of 20 µl (containing approx. 3.6 × 107 pfu ml−1) of ancestral phage or the same stock inactivated by autoclaving was added to 200 µl of clonal host cultures in liquid medium in a 96-well plate (n = 4). OD was measured after culturing for 7 and 14–16 days. To test for the possibility of coevolution, we repeated the same assay for all Nodularia isolates with 22 week coexistent phage obtained from the same experimental population as the isolate tested.
2.4. Measuring evolved changes in Nodularia filament length morphology, adhesion and buoyancy
Mean filament length was determined for 34 randomly selected isolates from the naive population and all isolates from the evolved population (n = 58). For each isolate, a length-representative sample of 20 filaments was measured using light microscopy. A resistant, short-filamentous type that was detected is hereafter referred as R-short and the ancestral strain-like long type as R-long.
The ability of strains to remain in suspended form in liquid medium was tested for randomly selected naive, R-long and R-short isolates (n = 9 for all treatments). A volume of 2 ml from cultures with similar cell densities was added to a 24-well tissue culture plate (Sarstedt), and cultured for 7 days without perturbation. The culture medium and suspended filaments were subsequently removed by a pipette, with special care taken to avoid stirring medium, and the amount of filaments per area in the bottom of the well was counted by light microscopy.
We tested difference in filament buoyancy by taking 1 ml samples of randomly selected long-filamentous and short-filamentous isolates (n = 4 for both treatments). The sample was taken from the surface layer of two month old 20 ml cultures with similar overall cell density after the culture had settled for 7 days, as well as after mixing cultures. Difference in sample cell density was then determined by light microscopy.
2.5. Measuring growth ability of naive and evolved clones
To examine the effect of phage-mediated evolution on host growth ability, a growth experiment was performed in liquid medium without nitrogen and with limiting concentrations of iron (Fe) or phosphorus (P). Limitation by P and Fe were chosen, because these two are among the key limiting nutrients for cyanobacteria [45,46]. Based on previous studies, concentrations of 2 µM P and 4 µM Fe were selected [17,47]. Chosen concentrations were achieved by adjusting the amount of FeCl3 × 6H2O in Fe-limiting medium and replacing a part of K2HPO4 × 3H2O with KCl in P-limiting medium. The experiment was performed under both nutrient limitations with the same 27 clones as with the suspension test described above. Prior to beginning the experiment, isolates were washed by centrifugation (7 min at 7000g/4°C; repeated once) and nutrient-starved by culturing isolates in low Fe or P conditions for 7 days in order to avoid the transference of excess nutrients into experimental vials. During the experiment, cells were enumerated weekly by light microscopy, and growth rate was calculated as r = ln(Nt + 1/Nt)/t, where Nt is population size at time t.
2.6. Release of cellular nitrogen after phage exposure
To test the effect of bacteria–phage interactions on the release of cellular nitrogen and phytoplankton growth ability, populations of Nodularia with different evolutionary histories (naive and evolved) were precultured for three weeks in liquid culture medium. Culturing was performed in a volume of 500 ml in a 850 ml tissue culture flask (VWR). After reaching a high cell density (approx. 3.2 × 106 cells ml−1), both naive and evolved populations were exposed to the ancestral phage (approx. 4 × 107 pfu ml−1) at a high multiplicity of infection of approximately 13. We monitored the following community dynamics by performing host and phage counts daily for 4 days. Host counts were performed by light microscopy, and phage counts were performed with a plaque assay on agarose plates. To acquire cell-free filtrates, cultures at day 4 were centrifuged (7 min at 7000g/4°C) and filtered through a 70 µm nylon cell strainer (Falcon®, NY, USA), followed by filtration through 0.22 µm. Total nitrogen in the filtrate was quantified from samples taken at the beginning and end of the experiment by an accredited laboratory (MetropoliLab, Helsinki, Finland) using the standard SFS-EN ISO 11095-1 that follows a previously described method [48].
2.7. Effect of Nodularia lysate on phytoplankton growth ability
To examine the large-scale effect of bacteria–phage interactions on other members of the phytoplankton community, strains representing green algae, diatoms and picocyanobacteria were cultured in filtrates from naive and evolved population cultures following phage exposure and assumed to contain released cellular nitrogen. The bacteria–phage lysate studies were conducted in nitrogen-free medium (see §2a) with other nutrients at abundant concentrations (i.e. with nitrogen as the only limiting nutrient and nitrogenous compounds from Nodularia serving as the only nitrogen source). To allow diatom growth and equalize culturing conditions for all phytoplankton strains, Na2SiO3 × 9H2O was added to a final concentration of 106 mM. In total, 11 phytoplankton strains were studied (electronic supplementary material, table S1). Prior to beginning the experiment, phytoplankton strains were centrifuged and nitrogen-starved for 3 days. We started the growth experiment by adding phytoplankton strains (final cell density of 104 cells ml−1) in 40 ml of filtrate in a 75 ml tissue culture flask (Sarstedt). We enumerated cells weekly by using a compound microscope (Zeiss Axioskop 2 plus, Oberkochen, Germany) with a 40× objective and a haemocytometer counting chamber (Improved Neubauer, Marienfeld, Germany). Eukaryotic phytoplankton strains were determined by light microscopy and picocyanobacterial strains (Synechococcus and Synechocystis) by epifluorescence microscopy. We converted cell numbers to biovolumes using species-specific standardized geometric formulae and size-classes [49].
2.8. Statistical analyses
Repeated-measures ANOVA (RMANOVA) was used to analyse host and phage densities and nitrogen concentrations in the nitrogen release test, phytoplankton densities in the bioassay and coevolution of re-infectivity in the phage. One-way ANOVA was used to analyse differences between clonal isolates in filament length, heterocyst density, growth ability, ability to remain in suspended form in medium and formation of oxygenic bubbles. A t-test was used to determine the phage resistance of clonal isolates, as well as buoyancy. RMANOVA, one-way ANOVA and post hoc analyses were performed with SPSS Statistics v. 22 (IBM SPSS Statistics, Chicago, IL).
3. Results
3.1. Host resistance evolution and phage coevolution
All isolates from the naive host population (0% resistant, n = 60) were susceptible to phage infection, and all isolates from the evolved host population were resistant (100% resistant, n = 58). However, within the course of the experiment, re-infectivity detectable with the method used had not evolved in the phage against phage-resistant isolates.
3.2. Evolution of new host morphotype
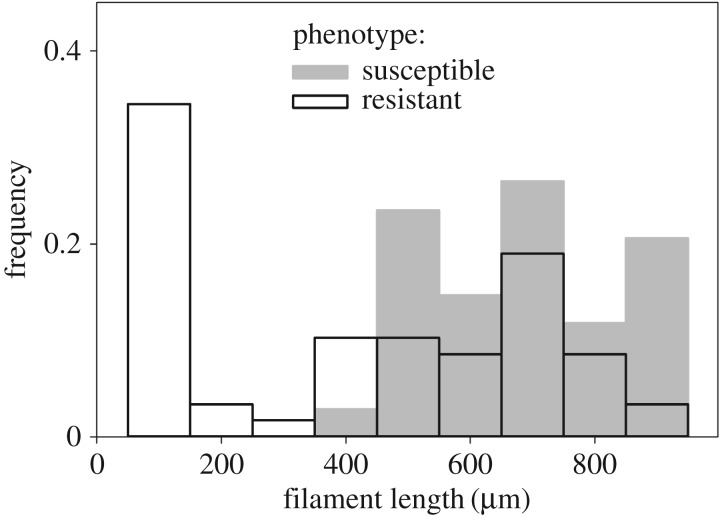
Based on visual and light-microscopic observations, two morphologically distinct phenotypes were observed among phage-resistant isolates (figures 2 and 3). A novel short-filamentous-resistant phenotype was present in 20.0–57.9% (means: 40.0 ± 19.0%) of isolates from evolved population microcosm replicates, and was not detected among the susceptible phenotype. The short-filamentous phenotype had a highly significantly lower mean filament length compared with both the susceptible phenotype (Tukey's HSD: p < 0.001) and the resistant long-filamentous phenotype (Tukey's HSD: p < 0.001; ANOVA: F89,2 = 130, p < 0.001). The long-filamentous-resistant phenotype did not differ from the susceptible phenotype (Tukey's HSD: p = 0.339).
Figure 2.
Filament length frequency distribution among susceptible (grey bars, n = 34) and resistant phenotype isolates (black bars, n = 58). Among resistant isolates, there is a distinct short-filamentous morphotype in addition to an ancestor-like long-filamentous morphotype.
Figure 3.
Light micrographs of ancestral and phage-resistant Nodularia sp. AV2 isolates. (a) Ancestral isolates, (b) phage-resistant long-filamentous isolates, and (c) phage-resistant short-filamentous isolates.
3.3. Differences in growth rates and buoyancy between phenotypes
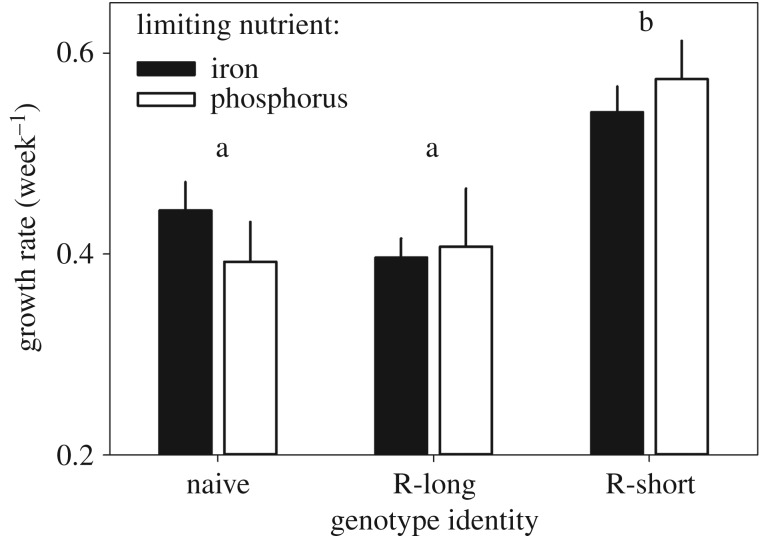
In both iron- and phosphorus-limited conditions, the short-filamentous-resistant phenotype had a higher maximum growth rate compared with both the susceptible phenotype (Tukey's HSD: Fe-lim p = 0.026, P-lim p = 0.026) and the long-filamentous-resistant phenotype (Tukey's HSD: Fe-lim p = 0.002; ANOVA: F21,2 = 8.75, p = 0.002, P-lim p = 0.046; ANOVA: F21,2 = 4.81, p = 0.019; figure 4). No difference was observed between the susceptible and resistant long-filamentous phenotypes (Tukey's HSD: Fe-lim p = 0.430, P-lim p = 0.971).
Figure 4.
Maximum weekly growth rates (rmax ± s.e.) of Nodularia sp. phenotypes cultured in iron and phosphorus-limited Z8 medium. Different letters indicate significant differences between phenotypes (Tukey's HSD: p < 0.05, all differences within phenotypes are non-significant.)
The ability of the R-short phenotype to stay in suspension in liquid medium was lower (adherence to well plate surface: 1.2 × 105 ± 4.9 × 104 µm mm−2) compared with both the susceptible phenotype (2.4 × 104 ± 3.3 × 104 µm mm−2; Tukey's HSD: p < 0.001) and the R-long phenotype (1.6 × 104 ± 1.5 × 104 µm mm−2; Tukey's HSD: p < 0.001; ANOVA: F24,2 = 20.7, p < 0.001). The long-filamentous-resistant phenotype did not differ from the susceptible phenotype (Tukey's HSD: p = 0.903). Visible oxygen bubble formation in the filament matrix was observed in 97% of susceptible phenotype cultures, 100% of long-filamentous-resistant phenotype cultures and in none of short-filamentous-resistant phenotype cultures, differing from the others (Tukey's HSD: p < 0.001; ANOVA: F114,2 = 904, p < 0.001). The long-filamentous-resistant phenotype did not differ from the susceptible phenotype (Tukey's HSD: p = 0.651). The density of long-filamentous phenotypes was significantly higher in the surface of the culture compared with the short-filamentous-resistant phenotype (t-test: t3.00 = 4.41, p = 0.02), and the densities did not differ in samples collected after mixing cultures (t-test: t3.39 = 1.22, p = 0.30).
3.4. Release of cellular nitrogen and bioassay with phytoplankton strains
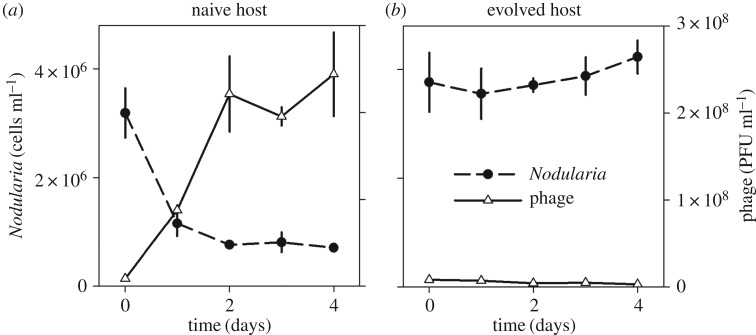
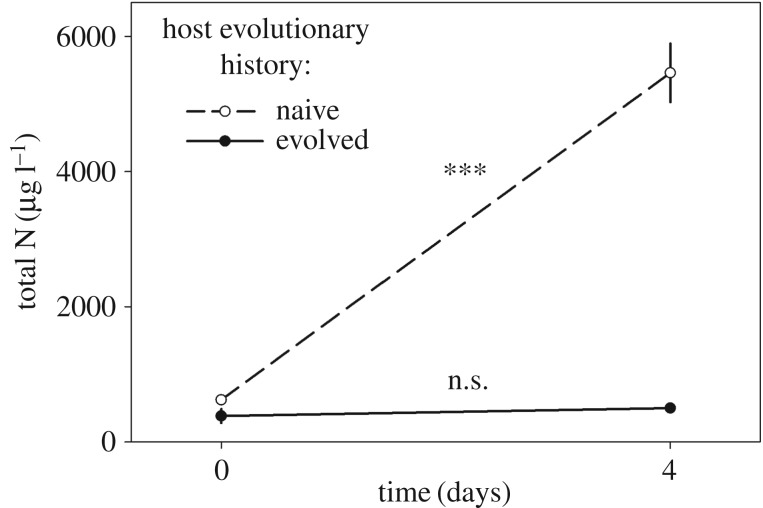
In the naive population, phage addition resulted in a mass mortality event among the host and substantial increase in phage particle number (figure 5). In the evolved population, phage addition did not result in a change in either host or phage numbers. The treatments differed significantly for both host (F1,20 = 151, p < 0.001) and phage (F1,20 = 123, p < 0.001). Total nitrogen increased in the cell-free filtrate from the naive population, differing significantly from the evolved population in which no increase was detected (F1,4 = 90.1, p < 0.001; figure 6).
Figure 5.
Host (dashed line) and phage (solid line) population dynamics in experimental treatments following phage addition (mean ± s.e.). (a) A mass mortality event took place among the naive host population, whereas the evolved host population remained at a high density. (b) The phage with naive host increased rapidly, whereas the phage with evolved host remained at a low particle number.
Figure 6.
Total nitrogen concentrations in filtrates from experimental treatments at the beginning and end of 4 day-long phage exposure (mean ± s.e.). Total nitrogen increased significantly in the cell-free filtrate from the naive host population (dashed line), whereas remaining at the same level in the filtrate from the evolved host population (solid line).
All 11 studied phytoplankton strains grew significantly better in the filtrate from the naive population compared with the evolved population (RMANOVA: p < 0.001; figure 7).
Figure 7.
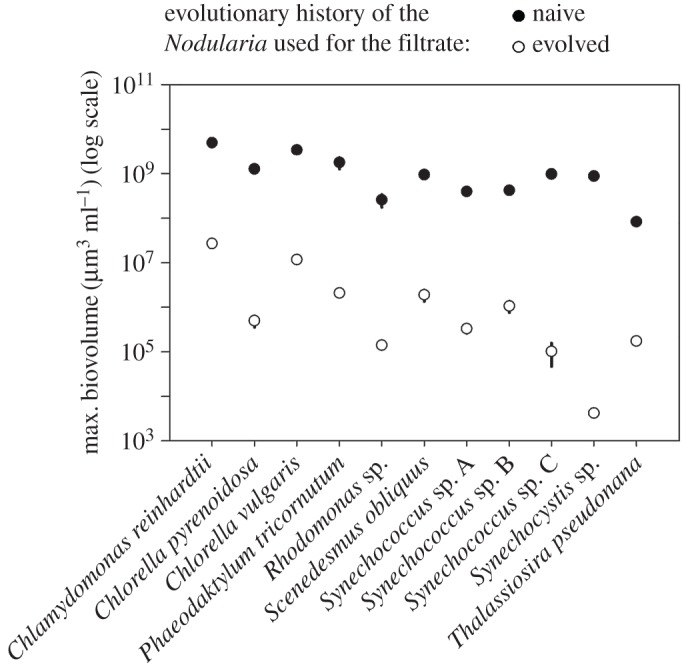
Maximum phytoplankton biovolume sustained by experimental filtrates (mean ± s.e.). The biovolumes of phytoplankton strains cultured in the filtrate from the naive population (black dots) are 1.9 × 102–2.1 × 105 higher (Chlamydomonas reinhardtii UTEX 89 and Synechocystis sp. UHCC 0318, respectively) compared with the evolved population (white dots). (Details of the phytoplankton species used can be found in the electronic supplementary material, table S1.)
4. Discussion
We found that high levels of cellular nitrogen were released from the nitrogen-fixing cyanobacterium Nodularia sp. strain AV2 upon viral lysis. This process can have ecosystem scale effects, because nitrogen-fixing cyanobacteria can reach a high biomass and thus be an important source of nitrogen in aquatic environments. In the case of Nodularia and phage vB_NpeS-2/AV2, it also seems that susceptible host types have a rather wide spatio-temporal distribution (figure 1) in the Baltic Sea, highlighting its potential importance concerning the nitrogen cycle. In general, in the Baltic Sea, the filamentous cyanobacteria Nodularia spumigena, Aphanizomenon spp. and Anabaena spp. are considered to be the principal nitrogen-fixing organisms [2,5]. Nodularia spumigena, in particular, can play a substantial role in the total carbon flux even at low abundance [5]. By causing host lysis and, subsequently, release of cellular nitrogen from the host, cyanophages infecting nitrogen-fixing cyanobacteria can therefore be key players in planktonic communities, even though viral biomass is very small compared with other microbes. In this sense, phages that lyse nitrogen-fixing cyanobacteria can be considered to be keystone species in aquatic ecosystems [50]. This reasoning is in line with previous estimates concerning the significance of the viral loop in rerouting organic matter in marine ecosystems [1,23,24,51].
However, the importance of the link between cyanobacteria–phage interactions and the nitrogen cycle depends on the evolutionary background of the host. Evolution of host resistance, a common occurrence in cyanobacterial populations in nature and the laboratory alike [18,20,21,23,25,31,33,34,36,37,52] can reduce the importance of this link. This is evidenced by our observation that cellular nitrogen release did not occur with phage resistance phenotypes of Nodularia sp. Furthermore, in the Baltic Sea, Nodularia sp. AV strains that were isolated from the same bloom within a 8 day time window, showed variability in phage susceptibility, suggesting that both susceptible and resistant phenotypes can coexist in a single bloom (figure 1). This indicates the possibility for temporal phenotype dynamics in natural blooms. Antagonistic coevolution between nitrogen-fixing cyanobacteria and their phages may therefore cause spatio-temporal changes in the availability of dissolved organic nitrogen, contributing to a dynamic nutrient landscape in planktonic ecosystems. In addition to causing nitrogen release via lysis of sensitive host cells, cyanophages might also alter the nitrogen metabolism of live cyanobacteria by expression of specific proteins, as has been shown for carbon metabolism [53].
Evolution in key species interaction can have large, community-level effects. In experimental evolution studies, dynamics have been traditionally studied with simple one resource one consumer communities, such as bacteria–phage [54–60], bacteria–protozoan [61–63] or algae–rotifer [64–66] systems. However, experimental evidence on eco-evolutionary community dynamics is also needed in more complex food webs. Nevertheless, to date, a relatively low number of studies have addressed the effects of evolution on ecological dynamics in multispecies communities [67]. Here, we present indirect evidence of the effect of evolution in one key interaction on biologically relevant community dynamics. We found that strains representing common phytoplankton groups, green algae, diatoms and picocyanobacteria, grew considerably better in cell-free filtrates from phage-exposed susceptible host cultures compared with resistant host cultures. These results indicate that eco-evolutionary dynamics in a focal host–parasite interaction can resonate into wider community dynamics via causing fluctuations in resource release. More specifically, these observations also support the hypothesis that passing of fixed nitrogen from cyanobacterial blooms to nitrogen-limited picocyanobacteria may explain their mass occurrence in association with nitrogen-fixing cyanobacteria [2,28]. This specific interaction is especially significant since picocyanobacteria are among the most important photosynthetic organisms in marine ecosystems.
We also found that evolution of phage resistance resulted in a novel short-filamentous Nodularia sp. morphotype with decreased buoyancy and increased growth ability compared with other resistant and susceptible isolates. This observation has several potential ecological and evolutionary consequences. First, this lends support from a novel filamentous cyanobacteria–phage system to the hypothesis that phages act to maintain bacterial diversity [38,39]. Second, we propose that decreased buoyancy is a new type of fitness cost caused by phage resistance, because buoyancy can be considered to be critical to the survival of cyanobacteria. Because photosynthesis is their primary mode of energy metabolism, decreased buoyancy may significantly lower cyanobacterial light-competing ability. By allowing upward movement in the vertical light gradient of the water column, bloom-formation and overshadowing of competing phototrophs [9,68], the light-competing ability conferred by buoyancy can be considered to be an essential component in both the interspecies and intraspecies competitive ability of cyanobacteria. In this context, a fitness trade-off may be considered to occur between phage resistance and competition for nutrients and light, with reduced overall fitness in a subset of the resistant population. Development of large phenotypic differences may also lead to expansion into new ecological niches, such as from planktonic to benthic zones as is imaginable in the case of reduced buoyancy, facilitating genetic divergence between morphotypes through adaptive radiation [69]. This can be important since the genus Nodularia includes not only planktonic, but also benthic species, N. sphaerocarpa and N. harveyana [70–72], viruses can be speculated to play a role in niche diversification processes. A low number of Nodularia species supported by molecular studies is contrasted by a much higher number of traditional taxonomical species assignments based on morphological diversity [73,74]. Our observations give rise to the question of whether the high observed morphological diversity can, in part, be explained by phage-mediated evolution, potentially promoting intraspecies adaptive radiation. These findings may have implications concerning the evolutionary history and diversification of filamentous nitrogen-fixing cyanobacteria.
The observations in this study demonstrate the potential of interactions between diazotrophic cyanobacteria and their phages to have large-scale ecosystem effects. Future directions include focusing on more specific aspects of the system, such as the molecular basis of phage resistance and differential filament morphology, and the potential fitness trade-offs associated with each identified phenotype, to provide a more explicit understanding of the mechanistic basis of these effects.
Supplementary Material
Acknowledgements
We thank Ana Dienstbier and Elina Roine for the phage isolate, Ruhui Wen for help with data collection, and Lotta-Riina Sundberg for valuable comments.
Data accessibility
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.nd58h [75].
Authors' contributions
Conceived and designed experiment: J.C. and T.H. Performed the experiment: J.C. and S.C. Analysed the data: J.C. and S.C. Wrote the paper: J.C. and T.H. T.H. and K.S. provided the laboratory materials for the study. All authors contributed on and accept the final version of the manuscript.
Competing interests
The authors declare no competing financial interest.
Funding
The study was supported by the Academy of Finland 106993 to T.H. This work was partially supported by the BLUEPRINT project, funded by the European Community's Seventh Framework Programme (FP/2007–2013) under implementation agreement no. R&I/I3/2012/BONUS made with BONUS, the joint Baltic Sea Research and Development Programme, and from the Academy of Finland 1273652 to K.S. The support provided by the Finnish Cultural Foundation grant 160149 to J.C. is also gratefully acknowledged.
References
- 1.Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. BioScience 49, 781–788. (doi:10.2307/1313569) [Google Scholar]
- 2.Stal LJ, Albertano P, Bergman B, von Bröckel K, Gallon JR, Hayes PK, Sivonen K, Walsby AE. 2003. BASIC: Baltic Sea cyanobacteria. An investigation of the structure and dynamics of water blooms of cyanobacteria in the Baltic Sea: responses to a changing environment. Cont. Shelf. Res. 23, 1695–1714 (doi:10.1016/j.csr.2003.06.001)
- 3.Berman-Frank I, Quigg A, Finkel ZV, Irwin AJ, Haramaty L.. 2007. Nitrogen-fixation strategies and Fe requirements in cyanobacteria. Limnol. Oceanogr.52, 2260–2269 (doi:10.4319/lo.2007.52.5.2260)
- 4.Gruber N. 2008. The marine nitrogen cycle: overview and challenges. In Nitrogen in the marine environment (eds Capone DG, Bronk DA, Mulholland MR, Carpenter EJ), pp. 1–50, 2nd edn Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 5.Plough H, Adam B, Musat N, Kalvelage T, Lavik G, Wolf-Gladrow D, Kuypers MMM. 2011. Carbon, nitrogen and O2 fluxes associated with the cyanobacterium Nodularia spumigena in the Baltic Sea. ISME J. 5, 1549–1558. (doi:10.1038/ismej.2011.20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez-Espinosa RM, Cole JA, Richardson DJ, Watmough NJ. 2011. Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 39, 175–178 (doi:10.1042/BST0390175) [DOI] [PubMed]
- 7.IPCC. 2013. Climate change 2013: the physical science basis. In Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (eds Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, Nauels A, Xia Y, Bex V, Midgley PM), 1535 p Cambridge, UK: Cambridge University Press. [Google Scholar]
- 8.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. 2015. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594 (doi:10.1126/science.1257594) [DOI] [PubMed] [Google Scholar]
- 9.Walsby AE, Hayes PK, Boje R.. 1995. The gas vesicles, buoyancy and vertical distribution of cyanobacteria in the Baltic Sea. Eur. J Phycol 30, 87–94. (doi:10.1080/09670269500650851) [Google Scholar]
- 10.Beversdorf LJ, Miller TR, McMahon KD. 2013. The role of nitrogen fixation in cyanobacterial bloom toxicity in a temperate, eutrophic lake. PLoS ONE 8, e56103 (doi:10.1371/journal.pone.0056103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galat DL, Verdin JP. 1989. Patchiness, collapse and succession of a cyanobacterial bloom evaluated by synoptic sampling and remote sensing. J. Plankton Res. 11, 925–948 (doi:10.1093/plankt/11.5.925)
- 12.Kahru M, Leppänen J-M, Rud O.. 1993. Cyanobacterial blooms cause heating of the sea surface. Mar. Ecol. Prog. Ser. 101, 1–7 (doi:10.3354/meps101001)
- 13.Kahru M, Leppänen J-M, Rud O, Savchuk OP. 2000. Cyanobacteria blooms in the Gulf of Finland triggered by saltwater inflow into the Baltic Sea. Mar. Ecol. Prog. Ser. 207, 13–18 (doi:10.3354/meps207013)
- 14.Kanoshina I, Lips U, Leppänen J-M. 2003. The influence of weather conditions (temperature and wind) on cyanobacterial bloom development in the Gulf of Finland (Baltic Sea). Harmful Algae 2, 29–41. (doi:10.1016/S1568-9883(02)00085-9) [Google Scholar]
- 15.Lehtimäki J, Sivonen K, Luukkainen R, Niemelä SI. 1994. The effects of incubation time, temperature, light, salinity, and phosphorus on growth and hepatotoxin production by Nodularia strains. Arch. Hydrobiol 130, 269–282. [Google Scholar]
- 16.Kononen K, Kuparinen J, Mäkelä K, Laanemets J, Pavelson J, Nõmmann S.. 1996. Initiation of cyanobacterial blooms in a frontal region at the entrance to the Gulf of Finland, Baltic Sea. Limnol. Oceanogr. 41, 98–112. (doi:10.4319/lo.1996.41.1.0098)
- 17.Lehtimäki J, Moisander P, Sivonen K, Kononen K.. 1997. Growth, nitrogen fixation, and nodularin production by two Baltic Sea cyanobacteria. Appl. Environ. Microbiol. 63, 1647–1656. [DOI] [PMC free article] [PubMed]
- 18.Waterbury JB, Valois FW. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59, 3393–3399. [DOI] [PMC free article] [PubMed]
- 19.Suttle CA, Chan AM. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60, 3167–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann NH. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev 27, 17–34. (doi:10.1016/S0168-6445(03)00016-0) [DOI] [PubMed] [Google Scholar]
- 21.Marston MF, Sallee JL. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ Microbiol 69, 4639–4647. (doi:10.1128/AEM.69.8.4639-4647.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mühling M, et al. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol 7, 499–508. (doi:10.1111/j.1462-2920.2005.00713.x) [DOI] [PubMed] [Google Scholar]
- 23.Stoddard LI, Martiny JBH, Marston MF. 2007. Selection and characterization of cyanophage resistance in marine Synechococcus strains. Appl. Environ Microbiol 73, 5516–5522. (doi:10.1128/AEM.00356-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martiny JBH, Riemann L, Marston MF, Middleboe M.. 2014. Antagonistic coevolution of marine planktonic viruses and their hosts. Annu. Rev. Mar. Sci. 6, 393–414 (doi:10.1146/annurev-marine-010213-135108)
- 25.Coloma SE, Dienstbier A, Bamford DH, Sivonen K, Roine E, Hiltunen T.. In press Newly isolated Nodularia phage influences cyanobacterial community dynamics. Environ. Microbiol (doi:10.1111/1462-2920.13601) [DOI] [PubMed]
- 26.Ploug H. 2008. Cyanobacterial surface blooms formed by Aphanizomenon sp. and Nodularia spumigena in the Baltic Sea: small-scale fluxes, pH, and oxygen microenvironments. Limnol. Oceanogr. 53, 914–921. (doi:10.4319/lo.2008.53.3.0914) [Google Scholar]
- 27.Woodland RJ, Holland DP, Beardall J, Smith J, Scicluna T, Cook PLM. 2013. Assimilation of diazotrophic nitrogen into pelagic food webs. PLoS ONE 8, e67588 (doi:10.1371/journal.pone.0067588) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stal LJ, Staal M, Villbrandt M.. 1999. Nutrient control of cyanobacterial blooms in the Baltic Sea. Aquat. Microb. Ecol. 18, 165–173 (doi:10.3354/ame018165)
- 29.Zwirglmaier K, et al. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161. (doi:10.1111/j.1462-2920.2007.01440.x) [DOI] [PubMed] [Google Scholar]
- 30.Engström-Öst J, Koski M, Schmidt K, Viitasalo M, Jónasdóttir SH, Kokkonen M, Repka S, Sivonen K. 2002. Effects of toxic cyanobacteria on a plankton assemblage: community development during decay of Nodularia spumigena. Mar. Ecol Prog Ser 232, 1–14. (doi:10.3354/meps232001) [Google Scholar]
- 31.McDaniel LD, delaRosa M, Paul JH. 2006. Temperate and lytic cyanophages from the Gulf of Mexico. J. Mar. Biol. Assoc. UK86, 517–527 (doi:10.1017/S0025315406013427)
- 32.Våge S, Storesund JE, Thingstad TF. 2013. Adding a cost of resistance description extends the ability of virus–host model to explain observed patterns in structure and function of pelagic microbial communities. Environ. Microbiol. 15, 1842–1852 (doi:10.1111/1462-2920.12077)
- 33.Avrani S, Schwartz DA, Lindell D.. 2012. Virus-host swinging party in the oceans: incorporating biological complexity into paradigms of antagonistic coexistence. Mob. Genet Elements 2, 88–95. (doi:10.4161/mge.20031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lennon JT, Khatana SAM, Marston MF, Martiny JBH. 2007. Is there a cost of virus resistance in marine cyanobacteria? ISME J. 1, 300–312. (doi:10.1038/ismej.2007.37) [DOI] [PubMed] [Google Scholar]
- 35.Meyer JR, Agrawal AA, Quick RT, Dobias DT, Schneider D, Lenski RE. 2010. Parallel changes in host resistance to viral infection during 45,000 generations of relaxed selection. Evolution 64, 3024–3034. (doi:10.1111/j.1558-5646.2010.01049.x) [DOI] [PubMed] [Google Scholar]
- 36.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. 2011. Genomic island variability facilitates Prochlorococcus–virus coexistence. Nature 474, 604–608. (doi:10.1038/nature10172) [DOI] [PubMed] [Google Scholar]
- 37.Avrani S, Lindell D. 2015. Convergent evolution toward an improved growth rate and a reduced resistance range in Prochlorococcus strains resistant to phage. Proc. Natl Acad Sci USA 112, E2191–E2200. (doi:10.1073/pnas.1420347112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinbauer MG, Rassoulzadegan F.. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6, 1–11 (doi:10.1046/j.1462-2920.2003.00539.x)
- 39.Koskella B, Brockhurst MA. 2014. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev 38, 916–931. (doi:10.1111/1574-6976.12072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivonen K, Kononen K, Esala A-L, Niemelä SI. 1989. Toxicity and isolation of the cyanobacterium Nodularia spumigena from the southern Baltic Sea in 1986. Hydrobiologia185, 3–8 (doi:10.1007/BF00006062)
- 41.Sivonen K, Kononen K, Carmichael WW, Dahlem AM, Rinehart KL, Kiviranta J, Niemelä SI. 1989. Occurrence of the hepatotoxic cyanobacterium Nodularia spumigena in the Baltic Sea and structure of the toxin. Appl. Environ. Microbiol. 55, 1990–1995. [DOI] [PMC free article] [PubMed]
- 42.Bolch CJS, Orr PT, Jones GJ, Blackburn SI. 1999. Genetic, morphological, and toxicological variation among globally distributed strains of Nodularia (Cyanobacteria). J. Phycol. 35, 339–355 (doi:10.1046/j.1529-8817.1999.3520339.x)
- 43.Kótai J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae. Norwegian Institute for Water Research Publication B-11/69, pp. 1–5. Oslo, Bildern.
- 44.Torres-Barceló C, Arias-Sánchez FI, Vasse M, Ramsayer J, Kaltz O, Hochberg ME. 2014. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PLoS ONE 9, e106628 (doi:10.1371/journal.pone.0106628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly L, Ding H, Huang KH, Osburne MS, Chisholm SW. 2013. Genetic diversity in cultured and wild marine cyanomyoviruses reveals phosphorus stress as a strong selective agent. ISME J. 7, 1827–1841. (doi:10.1038/ismej.2013.58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biller SJ, Berube PM, Lindell D, Chisholm SW. 2015. Prochlorococcus: the structure and function of collective diversity. Nat. Rev. Microbiol 13, 13–27. (doi:10.1038/nrmicro3378) [DOI] [PubMed] [Google Scholar]
- 47.Paczuska L, Kosakowska A. 2003. Is iron a limiting factor of Nodularia spumigena blooms? Oceanologia 45, 679–692. [Google Scholar]
- 48.Grasshoff K. 1976. Methods of seawater analysis, 317 p Weinheim, Germany: Chemie. [Google Scholar]
- 49.Olenina I, et al. 2006. Biovolumes and size-classes of phytoplankton in the Baltic Sea. HELCOM Balt. Sea Environ. Proc. No. 106, 144 p. Helsinki, Baltic Marine Environment Protection Commission.
- 50.Paine RT. 1995. A conversation on refining the concept of keystone species. Conserv. Biol. 9, 962–964 (doi:10.1046/j.1523-1739.1995.09040962.x)
- 51.Bratbak G, Heldal M, Norland S, Thingstad TF. 1990. Viruses as partners in spring bloom microbial tropho-dynamics. Appl. Environ. Microbiol. 56, 1400–1405. [DOI] [PMC free article] [PubMed]
- 52.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4, e234 (doi:10.1371/journal.pbio.0040234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW. 2011. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl Acad Sci USA 108, E757–E764. (doi:10.1073/pnas.1102164108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buckling A, Rainey PB. 2002. The role of parasites in sympatric and allopatric host diversification. Nature 420, 496–499. (doi:10.1038/nature01164) [DOI] [PubMed] [Google Scholar]
- 55.Buckling A, Rainey PB. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc. R. Soc. Lond. B 269, 931–936. (doi:10.1098/rspb.2001.1945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buckling A, Maclean RC, Brockhurst MA, Colegrave N. 2009. The beagle in a bottle. Nature 457, 824–829. (doi:10.1038/nature07892) [DOI] [PubMed] [Google Scholar]
- 57.Brockhurst MA, Rainey PB, Buckling A. 2004. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. R. Soc. Lond. B 271, 107–111. (doi:10.1098/rspb.2003.2556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brockhurst MA, Morgan AD, Fenton A, Buckling A. 2007. Experimental coevolution with bacteria and phage: the Pseudomonas fluorescens–φ2 model system. Infect. Genet Evol 7, 547–552. (doi:10.1016/j.meegid.2007.01.005) [DOI] [PubMed] [Google Scholar]
- 59.Koskella B, Lin DM, Buckling A, Thompson JN. 2011. The costs of evolving resistance in heterogeneous parasite environments. Proc. R. Soc. B 279, 1896–1903. (doi:10.1098/rspb.2011.2259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823. (doi:10.3390/v5030806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friman VP, Hiltunen T, Laakso J, Kaitala V. 2008. Availability of prey resources drives evolution of predator-prey interaction. Proc. R. Soc. B 275, 1625–1633. (doi:10.1098/rspb.2008.0174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiltunen T, Becks L.. 2014. Consumer co-evolution as an important component of the eco-evolutionary feedback. Nat. Commun. 5, 5226 (doi:10.1038/ncomms6226)
- 63.Hiltunen T, Ayan GB, Becks L. 2015. Environmental fluctuations restrict eco-evolutionary dynamics in predator-prey system. Proc. R. Soc. B 282, 20150013 (doi:10.1098/rspb.2015.0013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG Jr. 2003. Rapid evolution drives ecological dynamics in a predator-prey system. Nature 424, 303–306. (doi:10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 65.Hiltunen T, Ellner SP, Hooker G, Jones LE, Hairston NG Jr. 2014. Eco-evolutionary dynamics in a three-species food web with intraguild predation: intriguingly complex. Adv. Ecol. Res. 50, 41–73 (doi:10.1016/B978-0-12-801374-8.00002-5) [DOI] [PubMed]
- 66.Becks L, Ellner SP, Jones LE, Hairston NG Jr. 2010. Reduction of adaptive genetic diversity radically alters eco-evolutionary community dynamics. Ecol. Lett. 13, 989–997 (doi:10.1111/j.1461-0248.2010.01490.x)
- 67.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, Barraclough TG. 2012. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 10, e1001330 (doi:10.1371/journal.pbio.1001330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huisman J, Sharples J, Stroom JM, Visser PM, Kardinaal WEA, Verspagen JMH, Sommeijer B. 2004. Changes in turbulent mixing shift competition for light between phytoplankton species. Ecology 85, 2960–2970. (doi:10.1890/03-0763) [Google Scholar]
- 69.Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394, 69–72. (doi:10.1038/27900) [DOI] [PubMed] [Google Scholar]
- 70.Hayes PK, Barker GLA. 1997. Genetic diversity within Baltic Sea populations of Nodularia (cyanobacteria). J. Phycol. 33, 919–923 (doi:10.1111/j.0022-3646.1997.00919.x)
- 71.Lehtimäki J, Lyra C, Suomalainen S, Sundman P, Rouhiainen L, Paulin L, Salkinoja-Salonen M, Sivonen K.. 2000. Characterization of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int. J. Syst. Evol. Microbiol. 50, 1043–1053 (doi:10.1099/00207713-50-3-1043) [DOI] [PubMed]
- 72.Lyra C, Laamanen M, Lehtimäki JM, Surakka A, Sivonen K.. 2005. Benthic cyanobacteria of the genus Nodularia are non-toxic, without gas vacuoles, able to glide and genetically more diverse than planktonic Nodularia. Int. J. Syst. Evol. Microbiol. 55, 555–568 (doi:10.1099/ijs.0.63288-0) [DOI] [PubMed]
- 73.Komárek J, Hübel M, Hübel H, Šmarda J.. 1993. The Nodularia studies 2. Taxonomy. Algol. Stud. 68, 1–25.
- 74.Baker PD. 1991. Identification of common noxious cyanobacteria. Part I. Nostocales. Urban Water Research Association of Australia Research Report no. 29, 204 p.
- 75.Cains J, Coloma S, Sivonen K, Hiltunen T. 2016. Evolving interactions between diazotrophic cyanobacterium and phage mediate nitrogen release and host comptetitive ability. Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.nd58h)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.nd58h [75].