The cytoplasmic hydroxycinnamoyl-CoA:ω-hydroxy fatty acid transferase DPW2 plays a fundamental role in male reproduction via the biosynthesis of key components of the anther cuticle and pollen wall.
Abstract
Aliphatic and aromatic lipids are both essential structural components of the plant cuticle, an important interface between the plant and environment. Although cross links between aromatic and aliphatic or other moieties are known to be associated with the formation of leaf cutin and root and seed suberin, the contribution of aromatic lipids to the biosynthesis of anther cuticles and pollen walls remains elusive. In this study, we characterized the rice (Oryza sativa) male sterile mutant, defective pollen wall 2 (dpw2), which showed an abnormal anther cuticle, a defective pollen wall, and complete male sterility. Compared with the wild type, dpw2 anthers have increased amounts of cutin and waxes and decreased levels of lipidic and phenolic compounds. DPW2 encodes a cytoplasmically localized BAHD acyltransferase. In vitro assays demonstrated that recombinant DPW2 specifically transfers hydroxycinnamic acid moieties, using ω-hydroxy fatty acids as acyl acceptors and hydroxycinnamoyl-CoAs as acyl donors. Thus, The cytoplasmic hydroxycinnamoyl-CoA:ω-hydroxy fatty acid transferase DPW2 plays a fundamental role in male reproduction via the biosynthesis of key components of the anther cuticle and pollen wall.
In flowering plants, male reproductive development is a complex biological event that is essential to the survival and reproduction of the species. One distinctive feature of plant male development is the formation of the stamen, which consists of the anther and the supporting filament. The anther is composed of somatic anther wall layers and microspore mother cells (MMCs), which produces pollen grains via meiosis followed by two rounds of mitosis. After anther morphogenesis is complete, four centric layers are formed in the anther wall: the epidermis, endothecium, middle layer, and tapetum (McCormick, 1993; Goldberg et al., 1995; Scott et al., 2004; Ma, 2005; Zhang and Wilson, 2009; Chen et al., 2011; Zhang and Yang, 2014).
To protect pollen grains from environmental stresses, plants develop two barriers: One is the anther epidermis that is covered by a cutin matrix embedded and overlaid with waxes (Jeffree, 1996; Piffanelli et al., 1998; Nawrath, 2002; Jung et al., 2006; Li et al., 2006; Pollard et al., 2008; Shi et al., 2015). The second barrier is the pollen wall, which is indispensable for pollen-stigma communication during pollination (Piffanelli et al., 1997, 1998; Shi et al., 2015). Generally, the pollen wall includes three layers from the outer to inner side, namely the pollen coat, exine, and intine. The pollen coat is deployed on the surface of pollen and embedded in the cavity of the exine (Scott et al., 2004; Murphy, 2006; Blackmore et al., 2007; Grienenberger et al., 2009) and contributes to the defense of the pollen grain, pollen-pistil interaction, and pollen tube formation (Heslop Harrison, 1987; Edlund et al., 2004).
The chemically resistant sporopollenin represents the main constitutes of the exine, but its chemical composition is still unclear (Meuter-Gerhards et al., 1999; Ariizumi and Toriyama, 2011). Sporopollenin is thought to be composed entirely of polyalkyl and aromatic macromolecules as well as aliphatic and aromatic monomers, particularly ferulic and p-coumaric acids that are hinged together (Guilford et al., 1988; Wehling et al., 1989; Mösle et al., 1997; Blokker et al., 2005; De Leeuw et al., 2006; Watson et al., 2007). Intine, the innermost layer of the pollen wall, mainly consists of pectin, cellulose, hemicellulose (Kress and Stone, 1983; Edlund et al., 2004; Fang et al., 2008), and also of hydroxycinnamic acids (Meychik et al., 2006). In the past two decades, a number of genes associated with aliphatic or acyl-lipid-mediated male sterility have been identified (Li and Zhang, 2010; Ariizumi and Toriyama, 2011; Zhang et al., 2011; Jiang et al., 2013; Gong et al., 2015; Shi et al., 2015). However, our knowledge regarding the minor aromatic lipids in the anther cuticle and pollen wall is rather limited (Yoon et al., 2015); for example, how the aliphatic domain interlinks with the aromatic domain during the formation of the anther cuticle and pollen sporopollenin remains unknown.
Aromatic acids such as hydroxycinnamic acids often cross link with hydroxyl fatty acids and polysaccharides via BAHD acyltransferases, which belong to a large family of acyltransferases that utilize CoA thioesters and catalyze the formation of a diverse group of plant metabolites to form cutin, suberin, wax, lignin, phenolics, anthocyanins, and volatile esters (D’Auria, 2006; Yu et al., 2009; Tuominen et al., 2011; Bontpart et al., 2015; Molina and Kosma, 2015). These molecules can facilitate the adaptation of plants to fluctuating environments (Scott, 1994; Scott et al., 2004; Bontpart et al., 2015). BAHD acyltransferases were named after the first letter of each of the first four biochemically characterized enzymes of this family: BEAT (benzylalcohol O-acetyltransferase), AHCT (anthocyanin O-hydroxycinnamoyltransferase), HCBT (anthranilate N-hydroxycinnamoyl/benzoyltransferase), and DAT (deacetylvindoline 4-O-acetyltransferase; St-Pierre and De Luca, 2000). The BAHD proteins in plants are divided into five different clades. Clade I enzymes are involved in the malonylation of phenolic glucosides, clade II enzymes are associated with the elongation of very long-chain fatty acids, clade III enzymes are related to the synthesis of volatile esters, clade IV contains only one enzyme that is responsible for the acylation of a nitrogen to form amid, and clade V enzymes participate in the synthesis of volatile esters and the transfer of hydroxycinnamoyl- or benzoyl-CoAs as well (D’Auria, 2006). To date, five clade V BAHD/HXXXD (the consensus motifs in BAHD acyltransferase) acyltransferases have been implicated in alkyl hydroxycinnamate ester synthesis, mainly involved in the formation of feruloyloxy aliphatics in suberin, cutin, and wax polymers (Gou et al., 2009; Molina et al., 2009; Serra et al., 2010; Kosma et al., 2012; Rautengarten et al., 2012; Cheng et al., 2013). Arabidopsis (Arabidopsis thaliana) ASFT (aliphatic suberin feruloyl transferase)/RWP (reduced wall phenolics) functions as an ω-hydroxy fatty acid:feruloyltransferase and is involved in suberin synthesis in roots and seeds (Gou et al., 2009; Molina et al., 2009). Black cottonwood (Populus trichocarpa) FHT1 (hydroxyacid/fatty alcohol hydroxycinnamoyl transferase 1) takes p-coumaroyl and feruloyl-CoA as thioester donors to acylate ω-hydroxyacids and fatty alcohols in vitro; its ectopic expression in Arabidopsis increases phenolic esters in leaf cutin and in root and seed suberin (Cheng et al., 2013). Potato (Solanum tuberosum) FHT (fatty ω-hydroxyacid/fatty alcohol hydroxycinnamoyl transferase) conjugates ferulic acid with ω-hydroxyacid to make fatty alcohols for the biosynthesis of suberin and waxes that are required for the periderm maturation in potato (Serra et al., 2010). Arabidopsis FACT (fatty alcohol:caffeoyl-CoA caffeoyl acyltransferase) participated in the synthesis of alkyl hydroxycinnamates in root waxes using fatty alcohols as acyl acceptors (Kosma et al., 2012). Arabidopsis DCF (deficient in cutin ferulate) uses ω-hydroxy fatty acids as acyl acceptors and hydroxycinnamoyl-CoAs (preferentially feruloyl-CoA and sinapoyl-CoA) as acyl donors to form cutin polymers of aerial organs (Rautengarten et al., 2012). It seems that multiple functions of these BAHD/HXXXD acyltransferases contribute to these complex acyl donor/acceptor combinations. However, the role of BAHD/HXXXD acyltransferases in plant reproductive development remains largely unknown.
Here, we report the functional characterization of Defective Pollen Wall 2 (DPW2) from rice (Oryza sativa). dpw2 mutants have a higher accumulation of both aromatic and ω-hydroxylated fatty acids in anther cuticle and a decrease of phenolics in pollen grains, resulting in an abnormal anther cuticle and a defective pollen wall and leading to complete pollen abortion and male sterility. Recombinant DPW2 is capable of transferring hydroxycinnamic acid moieties in vitro, using ω-hydroxy fatty acids as acyl acceptors and hydroxycinnamoyl-CoAs as acyl donors. Thus, DPW2 is a BAHD/HXXXD acyltransferase (class V) with an essential role in synthesizing polymerized protective layers of anther cuticles and pollen walls.
RESULTS
Isolation and Phenotypic Analysis of dpw2
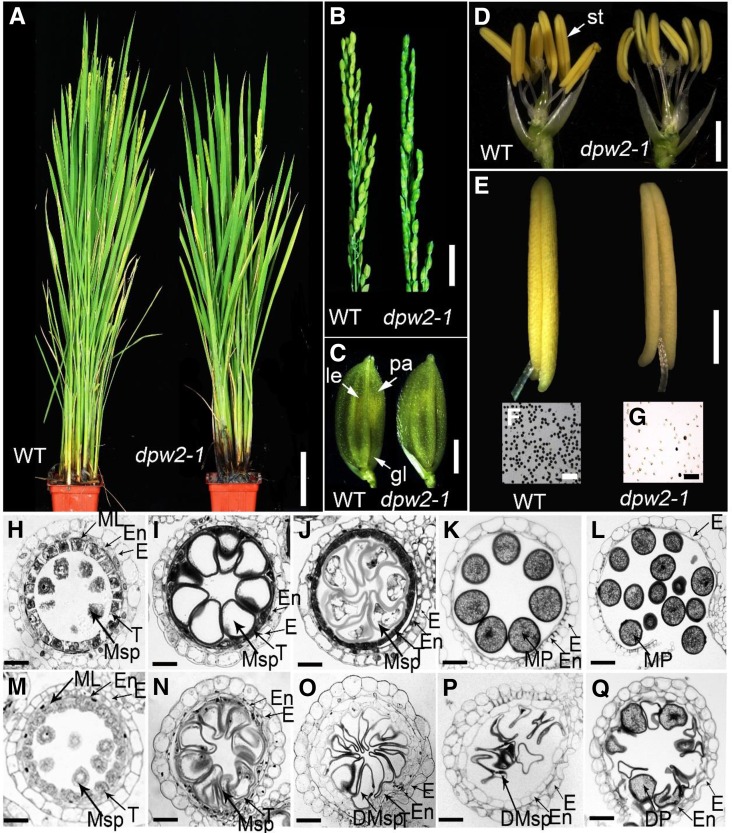
To reveal the molecular mechanism underlying rice male reproduction, two completely male sterile mutants, dpw2-1 and dpw2-2, were identified from our rice library generated with 60Coγ irradiation on the cultivar 9522 (O. sativa ssp. japonica; Chu et al., 2005; Liu et al., 2005). The mutants were named dpw2 because they displayed similar phenotypes (defective pollen wall and male sterility) to those of a known rice male sterile mutant, dpw (Shi et al., 2011). The two dpw2 mutants were allelic as evidenced by allelic test and showed similar phenotypes under the same growth conditions (Supplemental Table S1); thus, some of the subsequent analyses in this study were only performed on dpw2-1. All F1 plants of the backcross between the wild type and dpw2-1 were fertile, and the F2 plants had an approximately 3:1 ratio of phenotypic segregation (fertility: sterility = 72:22, χ2 = 0.1277, P > 0.05, χ2 test used), indicating that this mutation is a single nuclear recessive mutation. dpw2-1 showed normal vegetative development (Fig. 1A) and normal inflorescence as well as spikelet morphology (Fig. 1, B and C).By contrast, the anthers were smaller, and pale and pollen grains were unviable, as revealed by iodine potassium iodide (I2-KI) staining (Fig. 1, D–G).
Figure 1.
Phenotypic comparison between wild type and dpw2-1 mutant. A, Plants after bolting. B, Rice panicles at the heading stage. C, Spikelets at the heading stage. D, Spikelets after removal of the lemma and palea. E, Anthers at stage 13. F, Stage 13 wild-type pollen grains stained with 2% I2-KI solution. Mature pollen grains are stained. G, Stage 13 pollen grains of dpw2-1 stained with 2% I2-KI solution showing defects in most pollen grains. H to Q, Transverse section analysis of the anther development in the wild type (H–L) and dpw2-1 (M–Q) from stage 9 to stage 13. H and M, Stage 9; I and N, stage 10; J and O, stage 11; K and P, late stage 12; L and Q, stage 13. DMsp, Defective microspores; DP, defective pollen; E, epidermis; En, endothecium; gl, glume; le, lemma; ML, middle layer; MP, mature pollen; Msp, microspores; pa, palea; st, stamen; T, tapetal layer. Bars = 10 cm in A, 2 cm in B, 2 mm in C and D, 500 µm in E, 200 µm in F and G, and 15 µm in H to Q.
To characterize the anther defects in dpw2, we used semithin cross sections to examine defined anther developmental events in the wild type and the mutant (Zhang and Wilson, 2009; Chen et al., 2011). No significant differences between dpw2-1 and the wild type were detected at stage 9, where both anthers formed typical four anther wall layers and microsporocytes that are located at the center of each anther locule (Fig. 1, H and M). Meiosis in dpw2-1 was normal as indicated by 4′,6-diamidino-2-phenylindole (DAPI) staining (Supplemental Fig. S1). At stage 10, wild-type tapetal cells started to shrink and showed deep staining with toluidine blue, the middle layer became narrow and almost invisible, and the microspores became spherical with large vacuoles (Fig. 1I). By contrast, the dpw2-1 tapetal cells did not begin to condense and were only weakly stained by toluidine blue (Fig. 1N). At stage 11, the wild type nearly completed exine development (Fig. 1J). dpw2-1 microspores appeared slower in development with an abnormal appearance despite the presence of the two nuclei (Fig. 1O; Supplemental Fig. S1G). At stage 12, the wild-type tapetum had completely degenerated, and the mature pollen grains appeared round and full of storage starch and lipids (Fig. 1K). However, development of the dpw2-1 tapetum was still retarded, with tapetal layers still persistent and second mitosis failing to occur (Fig. 1P; Supplemental Fig. S1H). At stage 13, in contrast to the wild type with 94% of mature pollen grains stained by toluidine blue, only ∼8.4% of the mutant pollen grains showed relatively normal morphology, but less accumulation of storage starch (Fig. 1Q; Supplemental Fig. S2). At the same stage, wild-type anthers dehisced and mature pollen grains were released for pollination (Fig. 1L), while dpw2-1 displayed failed anther dehiscence, possibly due to the aborted pollen grains (Fig. 1Q). Consistent with the defect of dpw2-1 in pollen development, wild-type plants pollinated with mutant pollens failed to yield seeds due to the inviability of mutant pollens. These results suggest that DPW2 is indispensable for normal tapetal degeneration, second-round mitosis, and the accumulation of storage nutrients for pollen maturation.
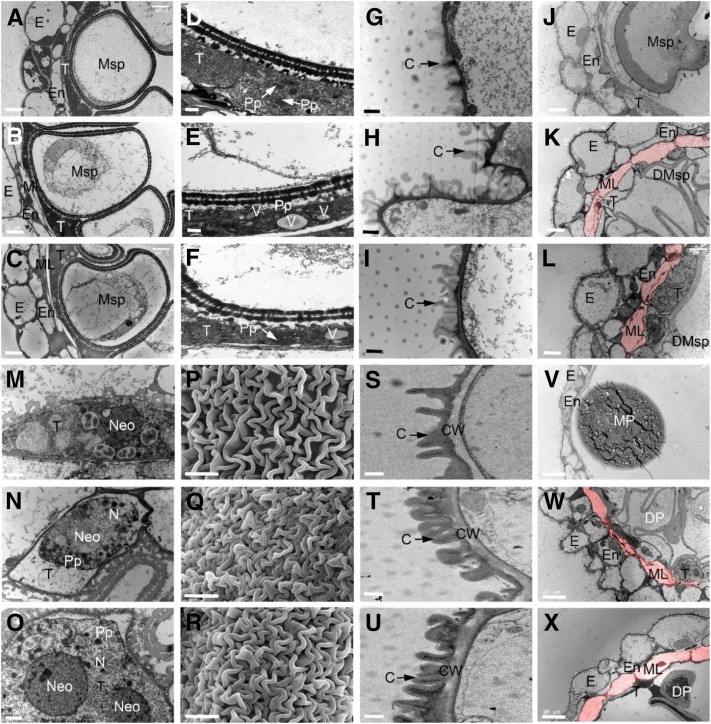
To more precisely characterize the differences between wild-type and dpw2 anthers, we used scanning electron microscopy (SEM) and transmission electron microscopy (TEM). During stage 9, microspores are normally released from the tetrads, and the tapetal cells are packed with enlarged ER stacks as well as fine fibrillar materials thought to be exine precursors (Owen and Makaroff, 1995; Scott et al., 2004; Li et al., 2012). TEM analysis showed that at stage 10 wild-type tapetal cells and middle layers continued to degenerate, whereby microspores were enlarged with expanded vacuoles (Fig. 2A). By contrast, in dpw2 the middle layers of the anther were intact (Fig. 2, B and C), and the tapetal cells contained less proplastids and numerous vesicles varying in size (Fig. 2, E and F), indicative of delayed degeneration. At stage 11, the knitted anther cuticle structure was obvious in the wild type but appeared denser in dpw2 (Fig. 2, G–I). At the same stage, the middle layer in the wild type disappeared, while in dpw2 mutant it remained present (Fig. 2, J–L). Moreover, the tapetum in the wild type continued to degenerate, with the nuclear envelope degrading and the concentrated nucleolus beginning to crack (Fig. 2M), whereas in dpw2 mutants, the nucleus was still intact, tapetal cells were enlarged, and proplastids were still present (Fig. 2, N and O). These results indicated that, starting from stage 11, tapetum degeneration was delayed in dpw2 mutants, resulting in delayed secretion of lipids and proteins for late pollen development. At stage 12, the wild-type anther cuticle was fully developed (Fig. 2S), while in the dpw2 mutants, the anther cuticle was denser (Fig. 2, T and U). In the wild type the tapetum and middle layer disappeared (Fig. 2V). By contrast, at this stage in dpw2, the middle layers were still intact and residues of the tapetum were still present (Fig. 2, W and X). SEM analysis also showed that at stage 12, the outer surface of dpw2 anthers had a denser cuticle layer compared to the wild type (Fig. 2, P–R). These results indicated that the mutation of DPW2 significantly affected tapetum degeneration, which, together with delayed degeneration of the middle layer, may contribute to impaired pollen wall formation.
Figure 2.
Electron microscopic examination of anther walls in wild type and dpw2 mutants. A to C, TEM images of anthers from the wild type (A), dpw2-1 (B), and dpw2-2 (C) at stage 10 showing the ring-vacuolate microspores. D to F, TEM images showing the morphology of tapetal cells of the wild type (D), dpw2-1 (E), and dpw2-2 (F) at stage 10. G to I, TEM images showing the outer region of anther epidermis in the wild type (G), dpw2-1 (H), and dpw2-2 (I) at stage 11. J to L, TEM images showing the anthers of the wild type (J), dpw2-1 (K), and dpw2-2 (L) at stage 11. M to O, TEM images showing the morphology of tapetal cells of the wild type (M), dpw2-1 (N), and dpw2-2 (O) at stage 11. P to R, SEM images showing the outer surface of anther at stage 12 in the wild type (P), dpw2-1 (Q), and dpw2-2 (R). S to U, TEM images showing the outer region of anther epidermis in the wild type (S), dpw2-1 (T), and dpw2-2 (U) at stage 12. V to X, TEM images showing the anthers of the wild type (V), dpw2-1 (W), and dpw2-2 (X) at stage 12. The cell layer highlighted in pink in K, L, W, and X represents the nondegraded middle layer in the mutant anther, which is absent in wild-type anthers at same stages. C, Cuticle; CW, cell wall; DMsp, degenerated microspore; DP, defective pollen; E, epidermis; En, endothecium; ML, middle layer; MP, mature pollen; Msp, microspores; N, nucleus; Neo, nucleolus; Pp, proplastid; T, tapetal layer; V, vacuole. Bars = 5 µm in A to C, J to L, and P to R; 1 µm in D to I, M to O, and S to U, and 10 µm in V to X.
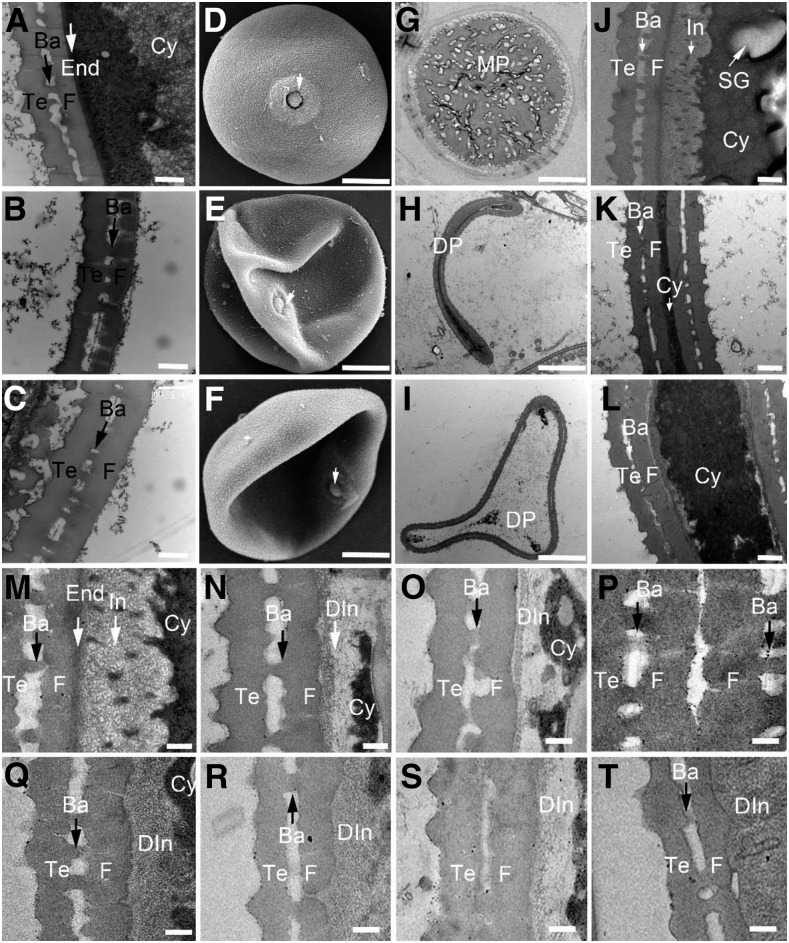
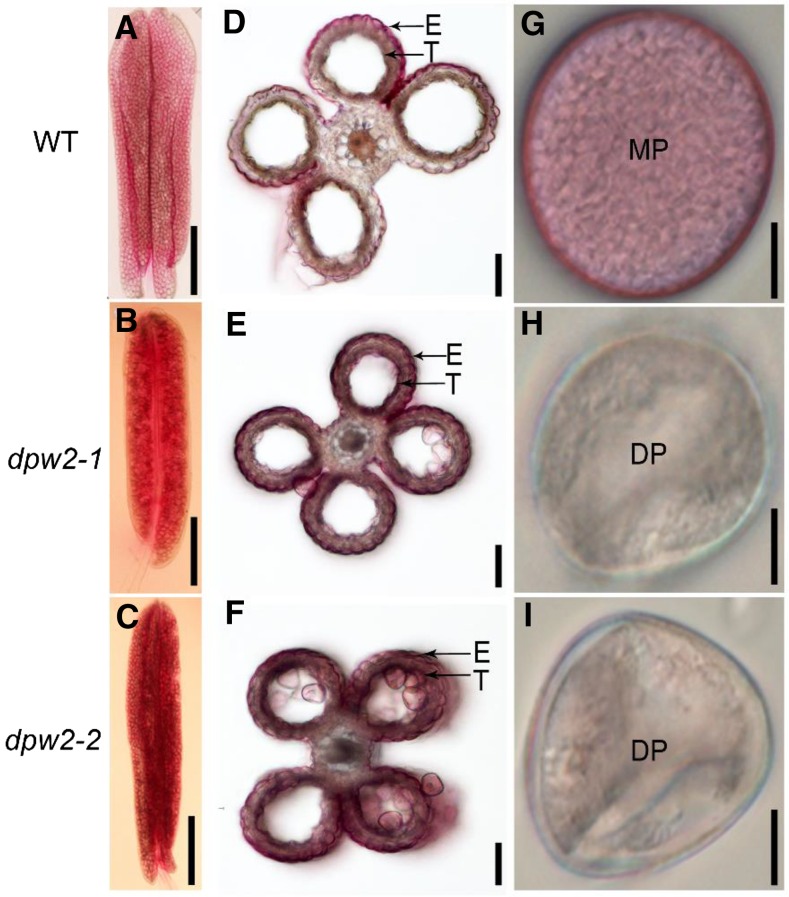
At stage 11, the wild-type pollen exine included extexine and endexine, the former include tectum, foot layer, and bacula linked between them, the latter is a thin and dark stained lamella located beneath the foot layer (Fig. 3A). In dpw2 mutants, the pollen exine skeleton looked similar to that of the wild type, but endexine and intine were almost invisible (Fig. 3, B and C). At stage 12, unlike the round and regular shaped wild-type pollen grains (Fig. 3D), the dpw2-1 mutant pollen showed irregular, collapsed, and shrunken pollen grains (Fig. 3, E and F). In agreement with the SEM observations, TEM images showed that wild-type pollen grains accumulated starch granules (Fig. 3G), whereas in the irregularly shaped pollen grains of dpw2, less starch granules were observed (Fig. 3, H and I). At this stage, wild-type exine structure was intact (Fig. 3, J and M), whereas dpw2 showed irregularly shaped exine that had a swollen foot layer (Fig. 3, K, L, N–Q, S, and T). Furthermore, the thin and deep stained lamella (endexine structure; Fig. 3M) under the foot layer, which is seen in wild-type pollen, was almost absent in dpw2 mutants (Fig. 3, N–Q, S, and T), and the cavities separated by tectum, foot layer, and bacula were smaller than those in the wild type (Fig. 3, K, L, O, Q, S, and T). Notably, dpw2 mutants showed different degrees of intine structure impairment (Fig. 3, K, L, N–Q, S, and T). These observations indicated that DPW2 may affect the formation of the anther cuticle and pollen wall at later stages of anther development.
Figure 3.
Electron microscopic examination of pollen wall in wild type and dpw2 mutants. A to C, TEM images showing pollen wall development of the wild type (A), dpw2-1 (B), and dpw2-2 (C) at stage 11. D to F, A higher magnification of SEM image showing a single pollen grain at stage 12 in the wild type (D), dpw2-1 (E), and dpw2-2 (F). The arrow indicates the germination pore. G to I, TEM images of stage 12 pollen grains in the wild type (G), dpw2-1 (H), and dpw2-2 (I). J to L, TEM images of the pollen wall from the wild type (J), dpw2-1 (K), and dpw2-2 (L) at stage 12. M to T, Higher magnification TEM images of stage 12 pollen wall showing the significant reduction of intine in dpw2-1 (N–P) and dpw2-2 (Q–T) in comparison with the wild type (M). Ba, Bacula; Cy, cytosol; DIn, defective intine; DP, defective pollen; End, endexine; F, foot layer; In, intine; MP, mature pollen; SG, starch granule; Te, tectum. Bars = 0.5 µm in A to C and J to L, 10 µm in D to I, and 0.2 µm in M to T.
Map-Based Cloning of DPW2 and Sequence Analysis
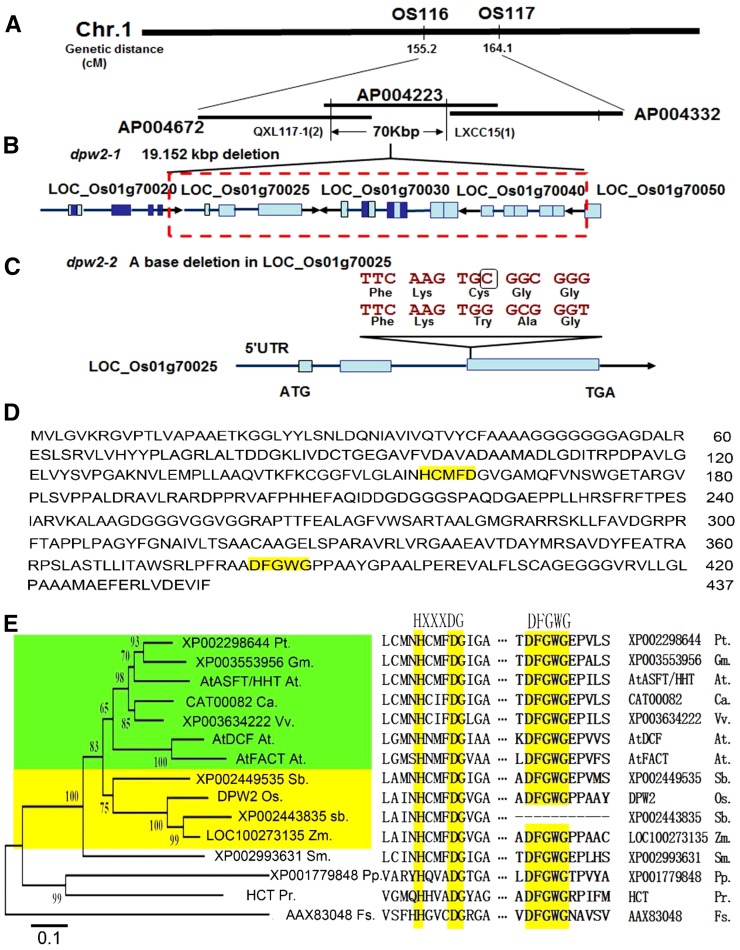
In order to identify the gene responsible for the mutation, a map-based cloning method was used by employing the F2 progeny derived from a cross between the wild type and dpw2-1. Based on initial mapping results, we mapped this gene to a genetic distance of 155.2 cM to 164.1 cM on chromosome 1 between markers OS116 and OS117 (Fig. 4A). After fine mapping, the dpw2 mutant gene was located to a 70-kb region between markers QXL117-1 and LXCC15 (Fig. 4A), which was on the BAC clone AP004223. Using PCR identification and DNA sequencing, we detected a DNA fragment deletion of 19.152 kb, which contained 6 bp of the 5′ terminus of Os01g70050, two putative transposons (Os01g70040 and Os01g70030), one predicted acyl transferase-encoding gene (Os01g70025), and the 3′-untranslated region of Os01g70020 (Fig. 4B). Analysis of the dpw2-2 allele identified a 1-bp deletion in the third exon of Os01g70025, causing a frame shift of the coding region (Fig. 4C).
Figure 4.
Map-based cloning and sequence analysis of DPW2. A, Fine mapping of the DPW2 gene. Names and positions of the markers are noted. B, A schematic representation of the deleted region in dpw2-1. The red dashed box shows the 19.152-kb deleted region. C, A schematic representation of the DPW2 gene, showing the mutation in dpw2-2. The black box indicates the base deletion that caused a frame shift. D, DPW2 protein sequence. The HXXXDG and DFGWG motifs are highlighted in yellow. E, Phylogenetic tree of DPW2 and its homologs from other species. A neighbor-joining phylogenetic tree was constructed using MEGA 3.1 to determine the evolutionary relationships among DPW2 and its homologs. The HXXXDG and DFGWG motifs are shown on the right. At, Arabidopsis thaliana; Ca, Coffea arabica; Fs, fungal species BT2; Gm, Glycine max; Os, Oryza sativa; Pp, Physcomitrella patens; Pr, Pinus radiata; Pt, Populus trichocarpa; Sb, Sorghum bicolor; Sm, Selaginella moellendorffii; Vv, Vitis vinifera; Zm, Zea mays. Bootstrap values are percentage of 1000 replicates.
To determine whether a mutation in, or the lack of, Os01g70025 is the cause of the defects observed in dpw2, we transformed dpw2-1 and dpw2-2 homozygous plants with a genomic DNA fragment consisting of a 1792-bp promoter region and a 2545-bp genomic DNA region of Os01g70025. Transgenic plants carrying the wild-type DNA restored normal anther development, pollen viability, and fertility in both mutants (Supplemental Fig. S3), confirming that Os01g70025 is DPW2.
To gain further information on the evolution of DPW2, we used the full-length DPW2 protein sequence to search for orthologous sequences in the public database National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/), using BLASTP. A total of 14 closely related sequences were retrieved from fungal, moss, pteridophyte, gymnosperm, and angiosperm species. Sequence comparison (Supplemental Fig. S4) showed that all 15 proteins contained the conserved domain HXXXD, and 14 proteins except sorghum (Sorghum bicolor) XP002443835 had the DFGWG domain (Fig. 4E). A neighbor-joining phylogenetic tree analysis grouped the 15 sequences into four clades. DPW2, together with its orthologs from Arabidopsis, black cottonwood, soybean (Glycine max), coffee (Coffea arabica), wine grape (Vitis vinifera), sorghum, and maize (Zea mays) were grouped in the first clade, which was further divided into two subclades. One subclade contained members from monocots such as rice, sorghum, and maize. The second subclade included those from dicots such as Arabidopsis, black cottonwood, soybean, coffee, and wine grape (Fig. 4E). This analysis indicated that DPW2 may represent a conserved and divergent acyltransferase member in monocots.
Spatiotemporal Expression Pattern of DPW2 and Protein Localization
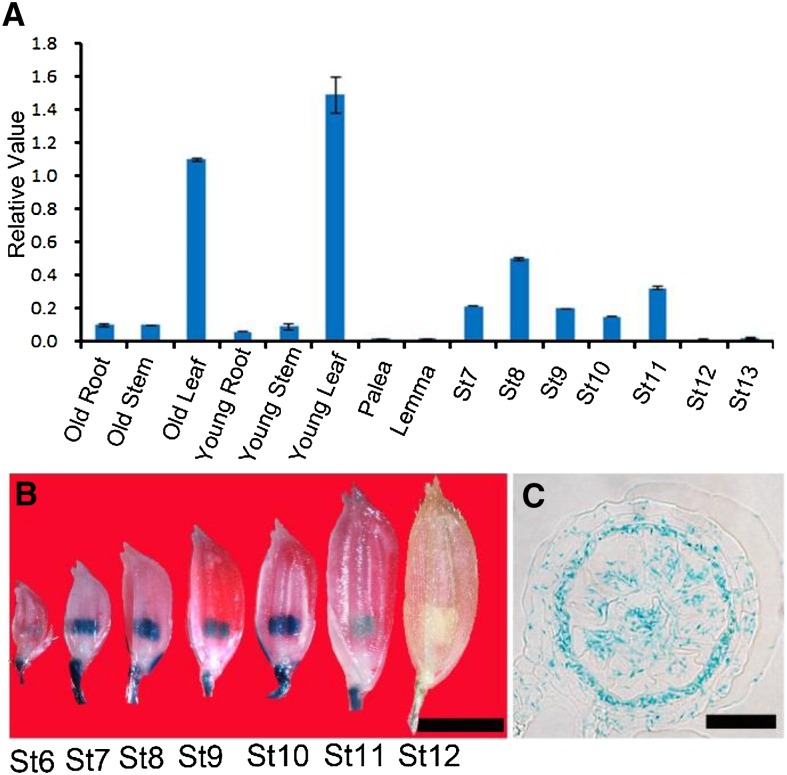
qPCR analysis of the DPW2 transcript revealed expression in root, stem, and leaf tissues, as well as in anthers from stage 7 to stage 13 (Fig. 5A). Transgenic plants expressing the GUS marker protein driven by the DPW2 promoter (DPW2pro:GUS) showed GUS staining in the anther from stage 6 to stage 12 (Fig. 5B), particularly in the tapetal layer and microspores (Fig. 5C). Although DPW2 is also expressed in vegetative tissues, the main morphological defects of dpw2 were restricted to anthers, indicating possible functional redundancy among DPW2 and other genes during vegetative growth.
Figure 5.
Expression analysis of DPW2. A, qPCR analysis of the DPW2 transcript. St7 to St13, Anther stages 7 to 13. Error bars indicate sd. Each reaction represents three biological repeats. B, GUS expression in spikelets at various anther stages in a representative DPW2pro:GUS transgenic line. St6 to St12, Anther stages 6 to 12. C, Cross section of stage 10 anther showing the presence of GUS signals in the tapetum and microspores. Bars = 3 mm in B and 15 µm in C.
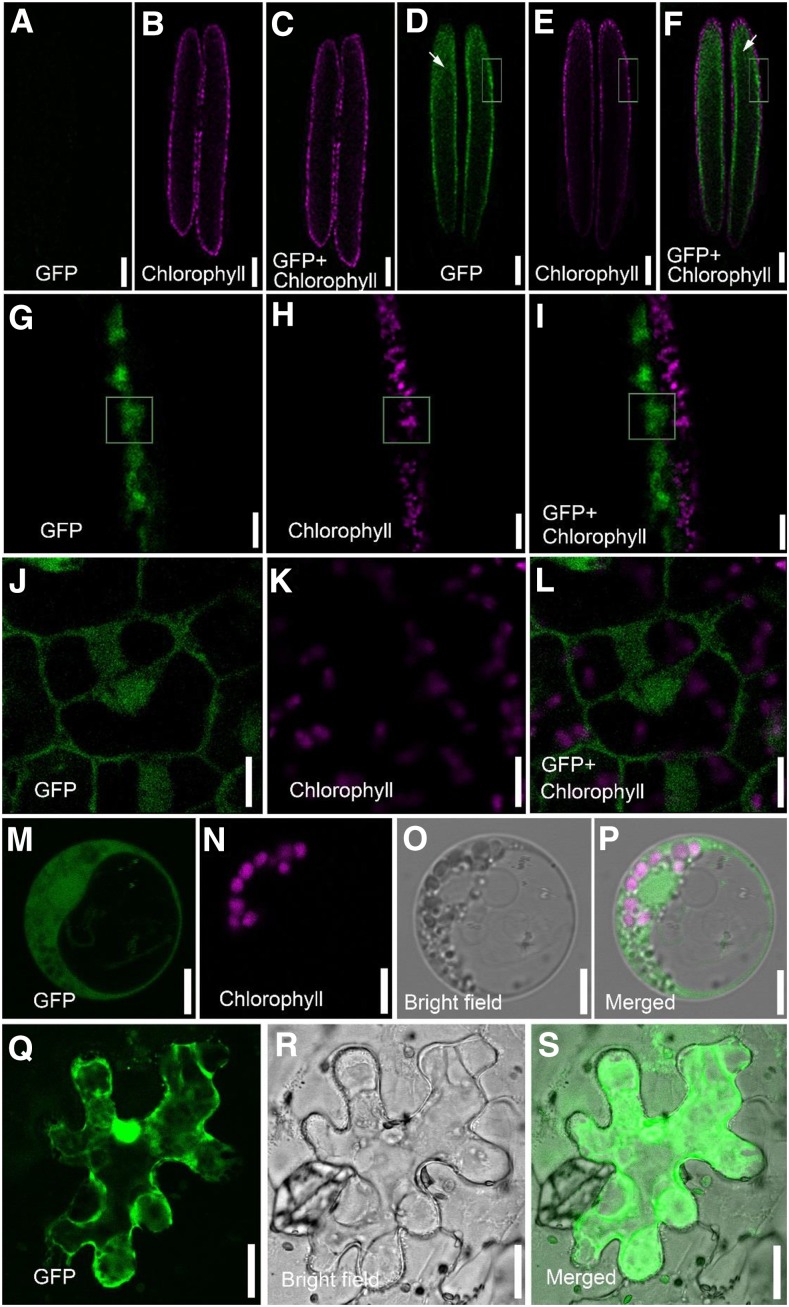
To further analyze the expression pattern of DPW2, a translational fusion of the full-length DPW2 coding region and GFP under the control of the native DPW2 promoter was created. Anthers of transgenic plants expressing DPW2pro:DPW2CDS-GFP showed a GFP signal at stage 8 (Fig. 6, A–I), particularly in the tapetum and microspores (Fig. 6, D and F). A close-up observation revealed the GFP signal mainly in the cytoplasm of tapetal cells (Fig. 6, J–L). In support of this data, subcellular localization analysis of DPW2 in rice protoplasts isolated from etiolated rice hypocotyls showed GFP signals mainly in the cytoplasm (Fig. 6, M–P). Moreover, we transiently expressed 35Spro:DPW2CDS-GFP in tobacco (Nicotiana benthamiana) epidermal cells and consistently detected GFP signals primarily originating from the cytoplasm (Fig. 6, Q–S).
Figure 6.
Localization of DPW2. A to C, Fluorescence micrographs of the control anther at stage 8. Images taken from the GFP (green; A), chlorophyll autofluorescence (magenta; B), and merged channels (C). D to F, Fluorescence micrographs of stage 8 anther in transgenic lines expressing DPW2pro:DPW2CDS-GFP. Images were taken from the GFP (green; D), chlorophyll autofluorescence (magenta; E), and merged channels (F). Arrows in D and F indicate the microspores located in the locule of the anther. G to I, Magnification of the boxed region in D, E, and F, respectively. J to L, Magnification of the cells from the tapetum in G, H, and I, respectively. M to P, A rice protoplast expressing pA7-DPW2CDS-GFP, showing images from GFP fluorescence (green; M), chlorophyll autofluorescence (magenta; N), bright field (O), and the merged image (P), respectively. Q to S, A tobacco epidermal cell expressing 35Spro:DPW2CDS-GFP, showing images from GFP fluorescence (green; Q), bright field (R), and the merged image (S). Bars = 100 µm in A to F, 10 µm in G to I, 5 µm in J to P, and 20 µm in Q to S.
Changes in the Composition of Cell Wall Components in dpw2 Anther and Pollen
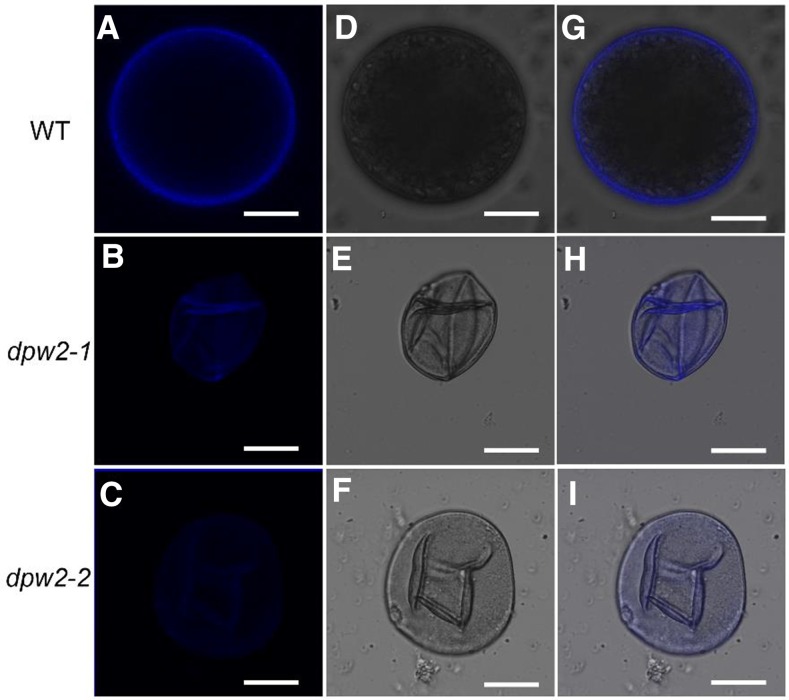
Given that DPW2 encodes a predicted acyl transferase and that dpw2 exhibited defective anther cuticles and pollen walls, we assumed that the chemical composition of these structures may be changed in mutant anthers. To test this hypothesis, we first stained the mature anther and pollen grains with Sudan Red 7B, a lipophilic dye (Brundrett et al., 1991). Compared to the wild type (Fig. 7, A and D), anthers of both dpw2-1 and dpw2-2 (Fig. 7, B and C) showed more intense staining in the epidermal and tapetal layers (Fig. 7, E and F). By contrast, the pollen wall of dpw2 mutants showed much weaker staining of Sudan Red 7B than the wild type (Fig. 7, G–I). The Sudan Red 7B staining results indicated that a disruption of DPW2 function leads to an increase of lipidic compounds in the anther epidermis but had an opposite effect on the pollen wall. The autofluorescence originating from phenolic compounds in pollen upon UV illumination (Driessen et al., 1989) was much weaker in mature pollen grains of dpw2 mutants compared to the wild type (Fig. 8, A–I), indicating a reduction in phenolic compounds in mutant pollen. Altogether, these results indicated that DPW2 plays multiple functions in the synthesis of essential components of the anther cuticle and pollen wall, such as phenolic compounds.
Figure 7.
Lipidic staining of the anther and pollen in wild type and dpw2 mutants. A to C, Comparison of Sudan Red 7B staining of anthers at stage 12. D to F, Cross sectioning of stage 12 anthers stained with Sudan Red 7B. G to I, Dissection of pollen grains from anthers after staining with Sudan Red 7B. DP, Defective pollen; E, epidermis; MP, mature pollen; T, tapetum. Bars = 500 µm in A to C, 50 µm in D to F, and 10 µm in G to I.
Figure 8.
Phenolic examination of pollen grains in wild type and dpw2 mutants at stage 12. A to C, Phenolic autofluorescence of pollens under UV illumination. D to F, Images of pollens in bright field. G to I, Merged images from phenolic autofluorescence under UV and bright field of pollen grains. Bars = 10 µm.
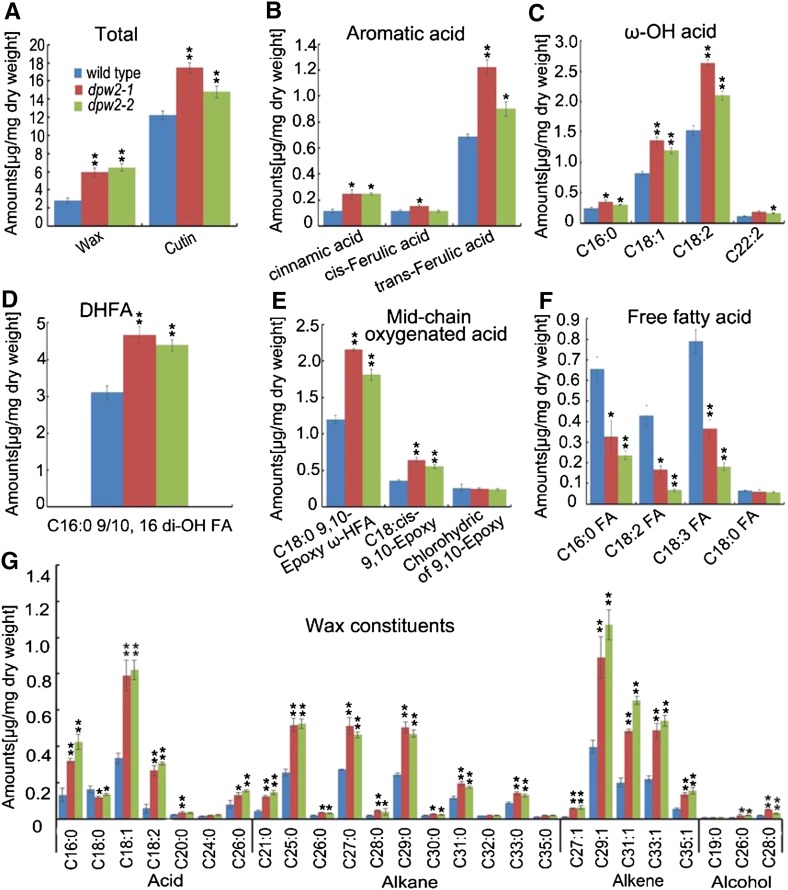
Due to the defects in anther, cuticle, and pollen wall formation in dpw2 mutants, we analyzed waxes, released by rapid chloroform extraction of dried anthers, and cutin monomers from transesterified cutin polymers of delipidated wild-type and dpw2 anthers by gas chromatography-mass spectrometry (GC-MS) and gas chromatography-flame ionization detection (Franke et al., 2005; Li et al., 2010a). The dpw2 mutants were found to contain increased levels of cutin monomers and wax components (Fig. 9A). The increased cutin monomers were attributed to increases of aromatic acids (cinnamic acid and ferulic acid), ω-OH acids (C16:0 ω-HFA acid, C18:1 ω-HFA acid, C18:2 ω-HFA acid, and C22:2 ω-HFA acid), C16:0 9/10, 16 di-OH FA, and midchain oxygenated acids (C18:0 9,10-epoxy ω-HFA, and C18:cis-9,10 epoxy; Fig. 9, B–E). By contrast, the amounts of detected free fatty acids (C16:0FA, C18:2FA, and C18:3FA) in dpw2 were significantly decreased (Fig. 9F). The increase of wax in dpw2 was mainly due to the increase of alkanes (C21, C25, C26, C27, C28, C29, C30, C31, and C33) and alkenes (C27:1, C29:1, C31:1, C33:1, and C35:1; Fig. 9G), free fatty acids (C16:0, C18:1, and C18:2; Fig. 9G), and long-chain fatty alcohols (C26:0 alcohol and C28:0 alcohol; Fig. 9G). These data indicated that the loss of function of DPW2 results in increased cross linking between the carboxyl moiety of the aromatic acids and the hydroxyl moiety of aliphatic acids in rice anthers.
Figure 9.
Anther cutin and wax profiles in wild type and dpw2 mutants. A, Total wax and cutin amount. B, Comparison of the exact compositions of aromatic acid changes. C, Comparison of the exact compositions of ω-OH acid changes. D, Comparison of the compositions of C16:0 9/10, 16 di-OH fatty acid changes. E, Comparison of the exact compositions of midchain oxygenated acid changes. F, Comparison of the exact compositions of free fatty acid changes. G, Comparison of the wax constituents in anthers. Data are presented as the mean of three biological replicates ± sd. Error bars in A to G indicate sd (n = 3). Compound names in C to F are abbreviated as follows: C16:0 ω-HFA, 16-hydroxy-hexadecanoic acid; C18:1 ω-HFA, 18-hydroxy-oleic acid; C18:2 ω-HFA, 18-hydroxy-linoleic acid; C22:0 ω-HFA, 22-hydroxy-docosanoic acid; C16:0 9/10,di-OH FA, 9(10),16-dihydroxypalmitic acid; C18:0 9,10-Epoxy ω-HFA, 9(10)-epoxy-18-hydroxy-stearic acid; C18: cis-9,10 Epoxy, cis-9(10)-epoxy-stearic acid; Chlorohydric of 9,10 Epoxy, chlorohydric of 9(10)-epoxy-18-hydroxy-stearic acid; C16:0 FA, palmitic acid; C18:2 FA, linoleic acid; C18:3 FA, linolenic acid; C18:0 FA, octadecanoic acid. Compound names in G are abbreviated as follows: C16:0, palmitic acid; C18:0, octadecanoic acid; C18:1, oleic acid; C18:2, linoleic acid; C20:0, eicosanoic acid; C24:0, tetracosanoic acid; C26:0, hexacosanoic acid. *P < 0.05; **P < 0.01.
DPW2 Has Hydroxycinnamoyl-CoA:ω-Hydroxy Fatty Acid Transferase Activity
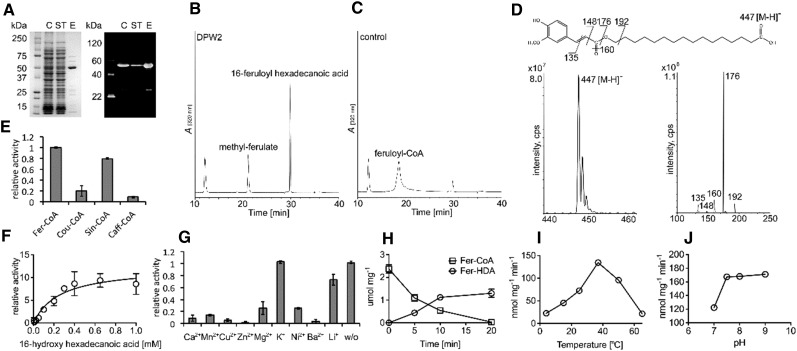
To determine whether DPW2 is a functional hydroxycinnamoyltransferase, we heterologously expressed DPW2 in Escherichia coli and affinity purified the corresponding protein. SDS-PAGE analysis revealed a band of about 50 kD in size (Fig. 10A). The purified recombinant DPW2 protein was incubated with feruloyl-CoA, which is regarded as the acyl-donor for BAHD family acyltransferase (Gou et al., 2009; Molina et al., 2009; Rautengarten et al., 2012), and 16-hydroxy-palmitic acid, a common type of ω-hydroxy fatty acid in cutin (Gou et al., 2009; Molina et al., 2009; Rautengarten et al., 2012). The resultant products were then analyzed by high-performance liquid chromatography (HPLC) and liquid chromatography-tandem mass spectrometry (LC-MS/MS). A major product with a UV absorption spectrum similar to 16-feruloylpalmitic acid and a major molecular ion of 447 [M-H]− was detected (Fig. 10, B and D), which was consistent with the single negatively charged molecular mass of 16-feruloylpalmitic acid of 447 D as reported previously (Rautengarten et al., 2012). Moreover, all detected diagnostic fragment ions of m/z = 135, m/z = 148, m/z = 160, m/z = 176, and m/z = 192 of this derived molecule were in accordance with each interrupted chemical structure, further confirming the existence of a feruloyl residue in the identified compound (Fig. 10D). A side product, methylferulate, was also detected, resulting from the reactivity of DPW2 with methanol, the solvent we used for 16-hydroxy-palmitic acid in our transferase reaction setup (Fig. 10B). In control reactions containing the heat-inactivated recombinant enzyme, only feruloyl-CoA was detected (Fig. 10C).
Figure 10.
In vitro enzymatic activity and characteristics of recombinant DPW2 protein. A, SDS-PAGE of purified recombinant DPW2 protein stained with Coomassie Brilliant Blue and the corresponding immunoblot analysis. C, Crude extract; ST, supernatant; E, eluate. B and C, HPLC chromatograms showing products formed after recombinant DPW2 (B) was incubated with feruloyl-CoA as acyl donor and 16-hydroxy-palmitic acid as acyl acceptor. C, The control reaction with heat-inactivated enzyme added. A, Relative A320. D, Mass spectra and chemical structures of the reaction products, ferulic acid methyl-ester, and 16-feruloyl-palmitic acid. E, Relative DPW2 activities with different acyl donors and 16-hydroxy-palmitic acid as acyl acceptor. F, Kinetics of recombinant DPW2 feruloyl-transferase activity. G, Relative DPW2 activities upon addition of different metal ion cofactors (2 mm). w/o, Without (no metal ions added). H, Time course profiles of recombinant DPW2’s feruloyl-transferase activity. I, Temperature profiles of recombinant DPW2’s feruloyl-transferase activity. J, pH profiles of recombinant DPW2’s feruloyl-transferase activity. The number of independent replicates analyzed in E to J is three.
The highest feruloyl transferase activity was found when DPW2 was incubated with ω-hydroxy fatty acids as acyl acceptors, in an order of C16 > C15 > C17 (Supplemental Table S2). No or little DPW2 activity was observed when longer chain (C20 and C22) ω-hydroxy fatty acids (Supplemental Table S2), short-chain primary alcohols, or fatty acids lacking a ω-hydroxy group, such as alkanes, amines, monolignols, and shikimic acid (Supplemental Table S2), were used as acceptors.
DPW2 was active with all other naturally occurring hydroxycinnamate-CoAs, in the order of feruloyl-CoA > sinapoyl-CoA > coumaroyl-CoA > caffeoyl-CoA (Fig. 10E), with the highest affinity for 16-hydroxy-palmitic acid when using feruloyl-CoA as the acyl donor with a substrate concentration of 0.26 (± 0.08) mm, giving rise to approximately half of the maximum activity (Fig. 10F). Whereas the addition of monovalent cations (K+, Li+) had no effect on enzymatic activity, the addition of divalent cations (Ca2+, Mn2+, Cu2+, Zn2+, Mg2+, Ni2+, Ba2+) severely inhibited the transferase reaction at 2 mm concentration (Fig. 10G). When a time course analysis was performed in a reaction containing 50 μm feruloyl-CoA and 1 mm saturated hydroxyhexadecanoic acid, the product 16-feruloylpalmitic acid was found to reach saturation state in 20 min (Fig. 10H). Temperature and pH profiles revealed that the DPW2 enzyme was mostly active at 37°C (Fig. 10I) and at a pH of 7.5 (Fig. 10J). Taken together, our in vitro enzymatic activity assays showed that DPW2 can cross link activated aromatic acids in the form of CoA (feruloyl-CoA, sinapoyl-CoA, caffeoyl-CoA, p-coumaroyl-CoA), with ω-hydroxy fatty acids as acyl acceptors.
DISCUSSION
DPW2 Is Required for Oligomer and Polymer Formation in the Rice Pollen Wall and Anther Wall
Aliphatic acids are essential components of plant biopolymers, such as the polymerized anther wall and pollen wall, yet current knowledge about the function of aromatic acids in the formation of these structures is very limited. In this study, we show that DPW2, a member of the BAHD/HXXXD family of acyltransferases that utilizes aromatic acids as carboxyl donors for cross linking, is required for the oligomer/polymer formation in the rice anther cuticle and pollen wall. The formation of these biopolymers likely requires the coordinated function of different acyltransferases to link the carboxyl moiety from aromatic acids with the hydroxyl moiety of hydroxylated aliphatic acids, fatty alcohols, glycerol, and polysaccharides (Meychik et al., 2006; Gou et al., 2009; Molina et al., 2009; Rautengarten et al., 2010; Serra et al., 2010; Kosma et al., 2012; Rautengarten et al., 2012; Cheng et al., 2013). Because loss of function of DPW2 alters the chemical profiles of both lipidic and phenolic precursors of anther cuticle and pollen wall exine and intine, DPW2 is essential for pollen development likely by acting on both lipidic and phenolic metabolism, two conserved pathways required for anther wall and pollen wall formation (Ariizumi and Toriyama, 2011; Shi et al., 2015). To the best of our knowledge, DPW2 is the first reported alkyl hydroxycinnamate ester acyltransferase of the BAHD/HXXXD family whose mutation interferes with male fertility in plants.
Aromatic lipids are minor but essential structural components of cutinized or suberized cell walls, such as those of the aerial organ surface, root, and seed coat (Bernards and Lewis, 1998; Franke et al., 2005; Panikashvili et al., 2009; Rautengarten et al., 2010). Phenolics such as ferulate and p-coumarate ester are also constituents of the anther (Yang et al., 2014; Zhao et al., 2015) and pollen walls (Wehling et al., 1989; Gubatz et al., 1993; Grienenberger et al., 2009; Negri et al., 2011). In suberized cell walls, the aromatic phenolics, mainly p-coumaric and ferulic acid, link the aliphatic domain to cell wall polysaccharides (Bernards, 2002). The molecular mechanisms underlying the involvement of aromatic phenolics in the formation of the anther wall and pollen exine are elusive. There is only one other report of a BAHD acyltransferase implicated in this process, namely the Arabidopsis spermidine hydroxycinnamoyl transferase (SHT), which cross links feruloyl-CoA and spermidine to produce hydroxycinnamoyl spermidines for pollen coat formation (Grienenberger et al., 2009).
It has been reported that the cell walls of monocotyledons (including grass) and the “core” Caryophyllales contain substantial amounts of the ester-linked hydroxycinnamic acids (Harris and Trethewey, 2010; Molinari et al., 2013), and both ferulic acids and coumaric acids are ester linked to adjacent arabinosyl units of glucuronoarabinoxylans in primary and secondary cell walls of grasses (Ishii, 1997; Lam et al., 2001; Ralph et al., 2004). In grasses, the ester-bound ferulate (including dehydro diferulates) and coumarate are required for the cross linking of adjacent polysaccharides (Harris and Trethewey, 2010; Grabber et al., 2004). By contrast, other angiosperms contain much lower amounts of ester-linked ferulic acids and coumaric acids as compared with grasses (Hatfield et al., 1999; Saulnier and Thibault, 1999). In addition, a rigorous analysis of cell wall fractions reports that ferulic acid is not esterified to polysaccharides in the Arabidopsis cell wall (Gou et al., 2009; Rautengarten et al., 2012). It was reported that pollen intine of lily contains similar structural polymers specific for the primary wall to those of plant somatic cells (Meychik et al., 2006). In lily pollen intine, the levels of hydroxycinnamic acids are significantly higher than those in the somatic cell walls, which are suggested to cross link polysaccharides in the intine as they do in the primary walls of somatic cells (Meychik et al., 2006). Due to technical limitation, it is very difficult to isolate the intine layer from the rice pollen for chemical analysis. But the dramatic decrease of aromatic compounds in the intine was evidenced by the observed weaker autofluorescence of the mutant pollen wall (Fig. 8).
Functional characterization of DPW2 in our study showed that aromatic acids are essential for the cross link reaction to form the pollen wall, particularly the pollen intine and anther wall. Thus, DPW2 emerges as a new player for plant pollen development, cross linking the aromatic moiety with hydroxyl moieties from various donors to form polymerized anther cuticles and pollen walls. While loss of function of rice DPW (Shi et al., 2011), CYP704B2 (Li et al., 2010a), or CYP703A3 (Yang et al., 2014) reduced the lipidic precursors for anther cuticle and pollen wall formation, mutation in DPW2 increased the cutinized lipid in anther, delayed the degeneration of the middle layer in anther, and impaired aromatic compounds accumulation in the pollen wall.
The Role of DPW2 in Rice Pollen Development
In plants, pathways for the biosynthesis of aromatic and aliphatic acids share the same substrate, acetyl-CoA (Li-Beisson et al., 2010; Shi et al., 2015; Fig. 11). The produced aromatic acids and ω-hydroxylated aliphatic acids are often cross linked to form phenolics for the biosynthesis of precursors of anther cutin and pollen exine (Wehling et al., 1989; Gubatz et al., 1993; Negri et al., 2011; Yang et al., 2014; Zhao et al., 2015; Zhang and Liang, 2016) and for pollen coat (Grienenberger et al., 2009). In this study, we provide morphological, chemical, histochemical, and in vitro enzyme activity data to suggest that DPW2 is mainly responsible for the polymerization of lipidic and phenolic pollen wall precursors. Due to technical difficulties in pollen wall lipidic and phenolic profiling, we were unable to provide direct evidence showing DPW2’s impact on pollen wall chemistry. However, chemical analysis performed on dpw2 anther samples (Fig. 9) provided indirect but substantial evidence that both aromatic acids (cinnamic and ferulic acid) and ω-hydroxylated fatty acids are increased in their cutin monomer profiles. It is most likely that loss of function of DPW2 breaks the balance of precursor allocation for the polymerization of cutin, sporopollenin, and cross linking of intine polysaccharides, and a feedback mechanism causes a more densely cutinized anther cuticle (Fig. 9) and a thicker pollen exine, but an impaired pollen intine (Fig. 3). Notably, the fact that dpw2 mutants exhibit obvious phenotypes associated with abnormal polysaccharide structures, such as defective intine (Fig. 3, B, C, and N–T) and delayed degeneration of the middle layer (Fig. 2, K, L, W, and X), indicated that DPW2 may be also associated with the cross linking of aromatic acids with polysaccharides to maintain the rigidity of pollen wall. Ferulate esters of pectin and other polysaccharides are reported to be important for cell wall recalcitrance in grasses (Scheller and Ulvskov, 2010). The spatial and temporal expression pattern of DPW2 as shown by GUS staining and qPCR (Fig. 5) and the localization of DPW2 to the tapetum and microspores (Fig. 6) corroborate the function of DPW2 in anther and pollen wall formation. These data highlighted the important contribution of phenolic acids such as ferulic and p-coumaric acid to anther cuticle (Reina and Heredia, 2001; Pollard et al., 2008; Li-Beisson et al., 2009) and pollen wall formation (Fig. 11). Disruption of intine formation frequently leads to the arrest of pollen development at later stage, which is characterized as lack of starch granule accumulation and collapse of mutant pollens (Drakakaki et al., 2006; Schnurr et al., 2006; Li et al., 2010b; Moon et al., 2013). Thus, we speculate that the reduced accumulation of starch granules in mutant pollen grains may be the indirect consequence of the intine deficiency.
Figure 11.
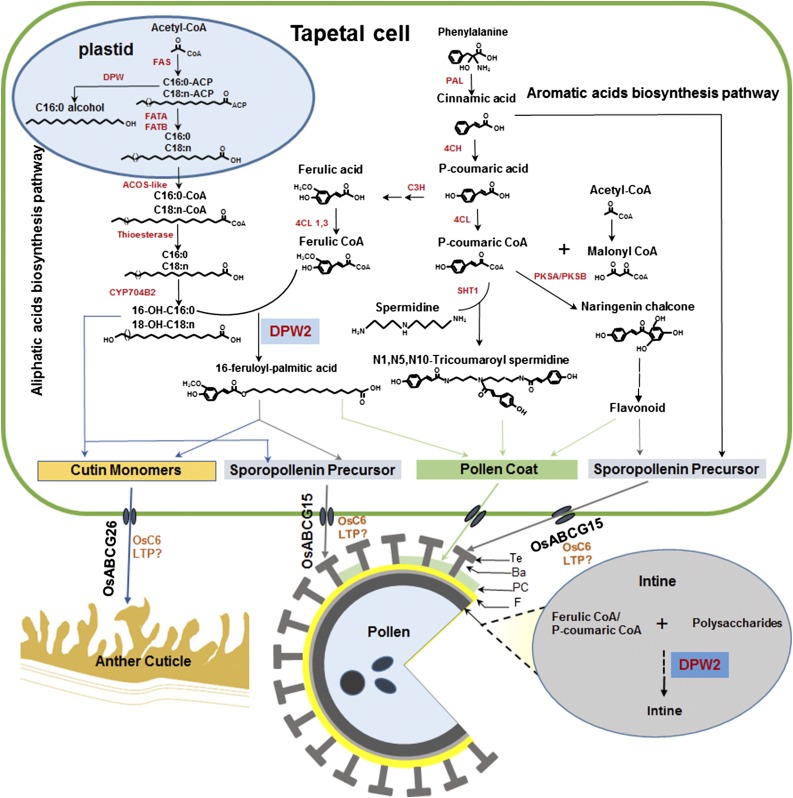
The proposed roles of DPW2 in anther cuticle and pollen wall development. In tapetal cells, fatty acids are synthesized de novo in plastids from acetyl-CoA to a maximum of C16 and C18 in length, followed by esterification to long-chain acyl carrier proteins (ACPs). Then the product is either reduced by DPW to fatty alcohols or further hydrolyzed by Acyl-ACP thioesterases (FATA and FATB) to release the free fatty acids and exported from the plastid to the cytoplasm, where they are modified via thioesterase and CYP450 hydroxylase. The resulting hydroxylated fatty acids are either used directly as the precursors for anther cutin and pollen sporopollenin or cross linked with aromatic acids generated from phenylpropanoid pathway by DPW2 to produce various precursors to be transported by ABCG (such as OsABCG15 and OsABCG26) or lipid transport proteins (LTP, such as OsC6). DPW2 may also cross link aromatic acids with polysaccharides for pollen intine biosynthesis. 4CL, 4-Coumarate:CoA ligase; ACOS-like, acyl-CoA synthase-like; C3H, coumaric acid 3-hydroxylase; C4H, cinnamate 4-hydroxylase; CYP704B2, cytochrome P450 of 704B2; DPW, defective pollen wall; FATA/FATB, fatty acyl thioesterase A (B); PAL, Phe ammonia-lyase; PKSA/PKSB, polyketide synthase a/polyketide synthase b; SHT1, spermidine hydroxycinnamoyl transferase 1; Ba, bacula; F, foot layer; PC, pollen coat; Te, tectum.
DPW2 Belongs to a Conserved BAHD Super Family HXXXD-Type Acyltransferase with Diversified Functions
The BAHD family members have been divided into five major phylogenetic clades, and each clade is further classified into several subclades based on their functions (Stewart et al., 2005; D’Auria, 2006; Tuominen et al., 2011). DPW2 belongs to hydroxycinnamoyl-CoA acyltransferases in clade V that transfer hydroxycinnamoyl-CoAs or benzoyl-CoAs (D’Auria, 2006; Yu et al., 2009; Tuominen et al., 2011; Bontpart et al., 2015; Molina and Kosma, 2015). In Arabidopsis, the biochemical function of some other hydroxycinnamoyl-CoA acyltransferases has been investigated. AtASFT/HHT is a hydroxycinnamoyltransferase involved in the synthesis of suberin aromatics by conjugating ferulate with 16-hydroxypalmitic acid (Gou et al., 2009; Molina et al., 2009). AtDCF feruloylates ω-hydroxy fatty acids for cutin polymers (Rautengarten et al., 2012). AtFACT, an Arabidopsis fatty alcohol:caffeoyl-CoA acyltransferase, cross links caffeoyl-CoA with dodecanoic acid (C12) for the synthesis of alkyl hydroxycinnamates in root waxes (Kosma et al., 2012). Our enzymatic activity assays showed that the recombinant DPW2 protein has hydroxycinnamoyl-transferase activity, with the preference for feruloyl-CoA and sinapoyl-CoA as acyl donors, and using 16-hydroxy-palmitic acid as acyl acceptors (Fig. 10). This activity is similar to that of AtASFT/HHT and AtDCF, which affect the levels of aromatic acids in various polymerized layers when mutated (Gou et al., 2009; Molina et al., 2009; Rautengarten et al., 2012). For example, suberin ferulate and stem cutin ferulate were strongly decreased in asft/rwp (Gou et al., 2009; Molina et al., 2009), leaf and stem cutin ferulate were both reduced in atdcf (Rautengarten et al., 2012), and alkyl caffeates and alcohols of suberin wax were decreased significantly in atfact (Kosma et al., 2012). Thus, DPW2 was likely evolved to participate in similar biosynthetic pathways for suberin, cutin, and wax in plant reproductive organs. Until now, to our knowledge, no study has reported the existence of suberin in anthers. Given that sporopollenin, suberin, and cutin share similar aromatic and aliphatic constituents and polymerization mechanisms (Domínguez et al., 1999; Li et al., 2010a; Shi et al., 2011; Yang et al., 2014), DPW2 may have evolved to function specifically within the anther, particularly in the pollen wall (exine and intine) and the anther wall (Fig. 11). The reason that mutants of the Arabidopsis ortholog of DPW2 do not show any visible male sterile phenotypes (Gou et al., 2009; Molina et al., 2009; Kosma et al., 2012; Rautengarten et al., 2012) may be explained by the difference in the composition of hydroxycinnamic acids in their pollen walls.
CONCLUSION
DPW2 encodes a BAHD superfamily HXXXD-type hydroxycinnamoyl-CoA:ω-hydroxy fatty acid acyltransferase with roles in cross linking the carboxyl moiety of aromatic acids with the hydroxyl moiety of ω-hydroxylated aliphatic acids, phenolics, or polysaccharides, polymerizing those molecules together for proper formation of anther cuticle, anther middle layer, and pollen exine and intine. Loss of function of DPW2 results in abnormal anther surface, delayed degenerated middle layer, impaired pollen wall, and ultimately complete male sterility. This study provides new insights into the cross linking of aromatic acids in plant development, particularly the formation of pollen grains.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Molecular Cloning of DPW2
Rice (Oryza sativa) plants were grown in the paddy field of Shanghai Jiao Tong University, at Minghang (31.03°N, 121.45°E), Shanghai. F2 mapping populations were generated from crosses between the dpw2-1 or dpw2-2 (9522 background, japonica) with Guan Lu Ai 4 (indica). Male sterile plants were selected for gene mapping (Chu et al., 2005; Liu et al., 2005). For fine mapping of the DPW2 locus, bulked segregant analysis was used and indel molecular markers were designed based on the sequence difference between japonica and indica cultivars (Chu et al., 2005; Liu et al., 2005), and we first mapped DPW2 between the indel molecular markers OS116 and OS117 on chromosome 1. After that, 600 F2 segregants from the mapping cross were generated, and DPW2 was eventually mapped to a 70-kb region between QXL117-1 and LXCC15 (Fig. 4A). All primers used in the mapping are listed in Supplemental Table S3.
Phenotypic Characterization of dpw2
Photos of whole plants and panicles were taken by a Nikon E995 digital camera. Fresh rice spikelets, flowers, and dehiscent anthers were photographed with a Leica M205A microscope. For pollen viability analysis, both wild-type and dpw2-1 anthers were immersed into I2-KI solution (2% potassium iodide and 0.2% iodine in water) at room temperature and then photographed with a Nikon Eclipse 80i microscope. Semithin cross sections of developing anthers were performed with the Technovit Embedding Kits (Heraeus Kulzer) as described previously (Hong et al., 1995; Li et al., 2006).
SEM and TEM analyses were done as described previously (Li et al., 2006; Zhou et al., 2011). For SEM analysis, the materials were fixed in formaldehyde-acetic acid solution that contained a 5:5:63:27 (volume) mixture of 37% formalin, acetic acid, ethanol, and water, dehydrated with an alcohol gradient (70%, 80%, 95%, and 100%), critical point dried using a Leica EM CPD300 critical point drier, covered with 5-nm layer of gold using a Leica EMSCD050 vacuum coater, and observed with a scanning electron microscope (Hitachi S3400N). For TEM analysis, spikelets of different stages were fixed in 2.5% glutaraldehyde, postfixed by 1% OsO4, dehydrated, embedded in Epon812 resin, and ultrathin sectioned, before being examined with a Tecnai G2 spirit Biotwin transmission electron microscope.
Histochemical Analysis
Pollen grains were separated from anthers, stained with 0.2% DAPI and observed under a microscope, as reported previously (Tan et al., 2012). For lipidic staining, anthers and pollen grains at stage 12 were stained with Sudan Red 7B solution (0.1% [w/v] Sudan Red 7B, 50% [v/v] PEG-400, 45% [v/v] glycerol, and 5% [v/v] water) for 1 h, washed three times with 1% SDS and double distilled water (Höfer et al., 2008), mounted on slides with a drop of water, and observed under a microscope, as described previously (Brundrett et al., 1991; Beisson et al., 2007). For fluorescence microscopy, pollen were deposited between a microscope slide and a coverslip in a droplet of water and observed with a laser scanning confocal microscope (Leica TCS SP5) with the excitation wavelength in the range of 330 to 380 nm and emission wavelength of 405 to 450 nm.
Mutant Complementation Analysis
Genomic DNA of the wild type (9522) was used as a template to amplify the 4337-bp genomic DNA fragment of DPW2, which contained a 1792-bp 5′ upstream region, 1887-bp genic region (including introns), and 658-bp 3′ downstream region, using a primer pair of P70025-gDNA-F and P70025-gDNA-R (Supplemental Table S4). The amplified fragment was subcloned into the vector pCAMBIA1301:GUS by In-Fusion (Takara Bio). The resulting plasmid containing DPW2pro:DPW2gDNA was introduced into dpw2-1 and dpw2-2 mutants by the Agrobacterium tumefaciens-mediated transformation method.
Sequence and Phylogenetic Analysis
The full-length amino acid sequence of DPW2 and 14 most similar sequences were retrieved with BLASTP (http://www.ncbi.nlm.nih.gov/), and conserved motifs were searched in SMART (http://smart.embl-heidelberg.de/). Sequence alignment was done using Clustal X and GENEDOC tool (Nicholas et al., 1997; Kohli and Bachhawat, 2003), and the resultant alignment (Supplemental Fig. S4) was used to construct a neighbor-joining tree with MEGA 3.1 (Kumar et al., 2004), using Poisson correction, pairwise deletion, and 1000 bootstrap replicates (Fig. 4E).
GUS Staining
A 1792-bp DNA fragment upstream of the transcriptional start codon of DPW2 was amplified as the promoter of DPW2, using primers 70025Promoter-GUS-F and 70025Promoter-GUS-R (Supplemental Table S4) from genomic DNA and introduced into pCAMBIA1301:GUS digested with EcoRI and NcoI by In-Fusion. The DPW2pro:GUS construct was introduced into wild-type rice calli by A. tumefaciens transformation. GUS activity was determined by staining spikelets at different developmental stages of transgenic lines as described previously (Jefferson et al., 1987).
qPCR Analysis
Total RNA was isolated from various rice organs including roots, shoots, leaves, lemmas/paleas, pistils, and anthers at different stages, using TRIzol reagent (Thermos Fisher Scientific). Stages of anthers were defined according to the spikelet length (Feng et al., 2000). Quality and concentration of total RNA was analyzed using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Reverse transcription of total RNA into cDNA was done with PrimeScript RT reagent kit with gDNA Eraser (Perfect Real Time). PCR reactions were carried out with iQ SYBR Green Supermix (Bio-Rad) with the primer pair dpw2-RT-F, 5′-TGCCCCTCCTCGCAGCTCAGGT-3′ and dpw2-RT-R, 5′-GGACGGAGAGCGGCACGCCCCT-3′, and the qPCR was performed using a C1000 CFX96 Real-Time PCR system (Bio-Rad) with the following program: 95°C for 2 min, 40 cycles of two-step amplification (95°C for 5 s, and 55°C for 35 s). PCR was performed in three biological replicates, each with three technique repeats. ACTIN was used as an internal control gene, and expression levels were calculated using a relative quantitation method (comparative CT) to quantify the relative expression level of the target genes.
Subcellular Localization of DPW2
For transient expression in rice protoplasts, the full-length DPW2 CDS was amplified using primers PA7-CDS70025-GFP-F and PA7-CDS70025-GFP-R (Supplemental Table S4) and introduced into pA7-GFP digested by XhoI and SpeI to generate pA7-DPW2CDS-GFP. The resulting plasmid was then transformed into rice protoplasts isolated from etiolated hypocotyl of rice (Bart et al., 2006) by polyethylene glycol-mediated transformation as described previously (Bart et al., 2006; Miao and Jiang, 2007).
For transient expression in leaf epidermis of tobacco (Nicotiana benthamiana) plants, full-length DPW2 CDS was PCR amplified using 70025CDS-GFP-F and 70025CDS-GFP-R (Supplemental Table S4), and introduced into 1301-GFP vector digested with BglII to generate 35Spro:DPW2CDS-GFP. The GFP construct was transformed into Agrobacterium strain GV3101 and infiltrated into leaves of 4-week-old tobacco plants (Sparkes et al., 2006).
For stable expression in rice, the promoter region of DPW2 was amplified using 70025Promoter-F1 and 70025Promoter-R1 (Supplemental Table S4), the 35Spro:DPW2CDS-GFP plasmid was digested with EcoRI and BglII, and the promoter of DPW2 was subcloned into 35Spro:DPW2CDS-GFP to form DPW2pro:DPW2CDS-GFP by In-Fusion.
Rice protoplasts transformed with pA7-DPW2CDS-GFP were mounted in W5 solution (Bart et al., 2006), while tobacco leaves with 35Spro:DPW2CDS-GFP and stage 8 anthers with DPW2pro-DPW2CDS-GFP were mounted in water and observed under a confocal microscope (Leica TCS SP5). GFP fluorescent signals were imaged at the excitation wavelength of 488 nm and emission wavelength of 520 to 580 nm. Red autofluorescence of chlorophyll was imaged at 514-nm excitation wavelength and emission wavelength in the range of 640 to 750 nm.
Determination of Wax and Cutin Composition
Wax and cutin of anthers were analyzed as described previously (Jung et al., 2006; Li et al., 2010a). To extract wax, 5 to 10 mg of freeze-dried anthers were submerged in 1 mL of hot (55°C) chloroform for 1 min. Extraction was repeated once, and the resulting chloroform extracts were combined, 10 μg of C24 Alkan (tetracosane) was added as an internal standard and organic solvent was evaporated under nitrogen. Compounds containing free hydroxyl and carboxyl groups were converted to their trimethylsilyl ethers with 20 μL of BSTFA [bis-(trimethylsilyl)trifluoroacetamide; Sigma-Aldrich] and 20 μL of pyridine for 40 min at 70°C before analyses by gas chromatography-flame ionization detection (Agilent Technologies) and GC-MS (Agilent gas chromatograph coupled to an Agilent 5973N quadrupole mass selective detector). The residual anther sample was delipidated and depolymerized before cutin analyses as described previously (Franke et al., 2005; Shi et al., 2011).
Prokaryotic Expression and Protein Purification
The CDS of DPW2 was amplified using 70025ORF-F and70025ORF-R (Supplemental Table S4) and subcloned into the pET-28a(+) protein expression vector that had been digested by BamHI and EcoRI. Protein was expressed in Escherichia coli strain BL21(DE3) and affinity purified as previous described (Rautengarten et al., 2012). Protein purity and integrity was verified by SDS-PAGE and immune blotting using an anti-His antibody (Qiagen), and the recombinant protein was stored at −20°C in 20 mm Tris buffer, pH 7.5, containing 20% (v/v) glycerol.
Hydroxycinnamoyl-CoA-Transferase Activity Analysis and Mass Spectrometry
For in vitro DPW2 activity assays, 1 μg of recombinant protein was incubated in 50 mm Tris buffer containing 50 μm hydroxycinnamate-CoA and 1 mm acyl acceptor. The mixture (total volume of 50 μL) was incubated at 30°C for 10 min, and the reactions were terminated by incubating for 10 min at 95°C. After the addition of 50 μL of methanol, 20 μL of reactions were subjected to HPLC analysis as previously described (Rautengarten et al., 2012). Mass spectrometry was undertaken by direct infusion of the enzyme reaction with 1:1 (v/v) acetonitrile at 20 µL/min using a 4000 QTRAP liquid chromatography-tandem mass spectrometry system (Sciex) equipped with a TurboIonSpray ion source and operated in negative ion mode. For the determination of precursor ion mass [M-H]− the instrument scan type was enhanced MS (EMS). Specific parameters were declustering potential (DP) of −40, entrance potential (EP) of −10, collision cell exit potential (CXP) was −15. The ion spray voltage was set at −4200 V, source temperature (TEM) at 425°C, collision gas (CAD) was set to High, source gas 1 (GS1) and 2 (GS2) were both set to 20, and a dynamic fill time was used. For analysis of product ions, the instrument scan type was enhanced product ion with the above parameters and a collision energy of −54. All data were acquired using Analyst 1.5.1 Build 5218 (Sciex).
Accession Numbers
Sequence data from this article for the cDNA and genomic DNA of DPW2 can be found in the GenBank/EMBL/Gramene data libraries under accession number LOC_Os01g70025.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. DAPI staining of dpw2-1 microspores.
Supplemental Figure S2. The ratio of pollen grains in a normal round shape in the semithin section of the wild type and dpw2-1 mutant at heading stage.
Supplemental Figure S3. Complementation of dpw2-1 and dpw2-2 mutants by DPW2 genomic DNA.
Supplemental Figure S4. Sequence alignment of DPW2 and 14 DPW2-related proteins.
Supplemental Table S1. Allelic test of dpw2-1 and dpw2-2.
Supplemental Table S2. In vitro substrate specificities of recombinant DPW2 protein with different acyl acceptors.
Supplemental Table S3. List of the primers used for DPW2 mapping.
Supplemental Table S4. Primers used for DPW2 vector construction.
Supplementary Material
Acknowledgments
We thank Dr. Guorun Qu for his assistance in anther cuticle analysis and Jie Xu, Wanwan Zhu, and Duoxiang Wang for their help in TEM observation. We thank Zhijin Luo, Mingjiao Chen, and Zibo Chen for mutant screening, allelic test, and generation of F2 populations for the mapping.
Glossary
- DAPI
4′,6-diamidino-2-phenylindole
- GC-MS
gas chromatography-mass spectrometry
Footnotes
This work was supported by funds from the National Key Basic Research Developments Program, Ministry of Science and Technology, China (2013CB126902); National Transgenic Major Program (2016ZX08009003-003-007); the National Natural Science Foundation of China (31430009, 31322040, 31271698); China Innovative Research Team, Ministry of Education, and the Programme of Introducing Talents of Discipline to Universities (111 Project, B14016); the Science and Technology Commission of Shanghai Municipality (grant no. 13JC1408200); Leading Scientist in Agriculture of Shanghai Municipality; and the German Research Foundation (DFG; to L.S.). The work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
Articles can be viewed without a subscription.
References
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park C-J, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA. (2002) Demystifying suberin. Can J Bot 80: 227–240 [Google Scholar]
- Bernards MA, Lewis NG (1998) The macromolecular aromatic domain in suberized tissue: a changing paradigm. Phytochemistry 47: 915–933 [DOI] [PubMed] [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR (2007) Pollen wall development in flowering plants. New Phytol 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Blokker P, Yeloff D, Boelen P, Broekman RA, Rozema J (2005) Development of a proxy for past surface UV-B irradiation: a thermally assisted hydrolysis and methylation py-GC/MS method for the analysis of pollen and spores. Anal Chem 77: 6026–6031 [DOI] [PubMed] [Google Scholar]
- Bontpart T, Cheynier V, Ageorges A, Terrier N (2015) BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol 208: 695–707 [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Kendrick B, Peterson CA (1991) Efficient lipid staining in plant material with Sudan Red 7B or Fluoral Yellow 088 in polyethylene glycol-glycerol. Biotech Histochem 66: 111–116 [DOI] [PubMed] [Google Scholar]
- Chen W, Yu X-H, Zhang K, Shi J, De Oliveira S, Schreiber L, Shanklin J, Zhang D (2011) Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol 157: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A-X, Gou J-Y, Yu X-H, Yang H, Fang X, Chen X-Y, Liu C-J (2013) Characterization and ectopic expression of a populus hydroxyacid hydroxycinnamoyltransferase. Mol Plant 6: 1889–1903 [DOI] [PubMed] [Google Scholar]
- Chu H-W, Liu H-S, Li H, Wang H-M, Wei J-L, Li N, Ding S-Y, Huang H, Ma H, Huang C-F, et al. (2005) Genetic analysis and mapping of the rice leafy-hull mutant Oslh. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 31: 594–598 [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- De Leeuw JW, Versteegh GJM, van Bergen PF (2006) Biomacromolecules of algae and plants and their fossil analogues. Plant Ecol 182: 209–233 [Google Scholar]
- Domínguez E, Mercado JA, Quesada MA, Heredia A (1999) Pollen sporopollenin: degradation and structural elucidation. Sex Plant Reprod 12: 171–178 [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N (2006) Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol 142: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen M, Willemse M, Van Luijn J (1989) Grass pollen grain determination by light-and UV-microscopy. Grana 28: 115–122 [Google Scholar]
- Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16(Suppl): S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K, Wang Y, Yu T, Zhang L, Baluška F, Šamaj J, Lin J (2008) Isolation of de-exined pollen and cytological studies of the pollen intines of Pinus bungeana Zucc. Ex Endl. and Picea wilsonii Mast. Flora 203: 332–340 [Google Scholar]
- Feng J, Lu Y, Liu X, Xu X (2000) Pollen development and its stages in rice (Oryza sativa L.). Chinese J Rice Sci 15: 21–28 [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L (2005) Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Sanders PM, Beals TP (1995) A novel cell-ablation strategy for studying plant development. Philos Trans R Soc Lond B Biol Sci 350: 5–17 [DOI] [PubMed] [Google Scholar]
- Gong F, Wu X, Wang W (2015) Diversity and function of maize pollen coat proteins: from biochemistry to proteomics. Front Plant Sci 6: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Yu XH, Liu CJ (2009) A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc Natl Acad Sci USA 106: 18855–18860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Lapierre C, Barrière Y (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. C R Biol 327: 455–465 [DOI] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Gubatz S, Rittscher M, Meuter A, Nagler A, Wiermann R (1993) Tracer experiments on sporopollenin biosynthesis. Grana (Suppl 1) 32: 12–17 [Google Scholar]
- Guilford WJ, Schneider DM, Labovitz J, Opella SJ (1988) High resolution solid state C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol 86: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PJ, Trethewey JA (2010) The distribution of ester-linked ferulic acid in the cell walls of angiosperms. Phytochem Rev 9: 19–33 [Google Scholar]
- Hatfield RD, Ralph J, Grabber JH (1999) Cell wall cross-linking by ferulates and diferulates in grasses. J Sci Food Agric 79: 403–407 [Google Scholar]
- Heslop-Harrison J. (1987) Pollen germination and pollen-tube growth. Int Rev Cytol 107: 1–78 [Google Scholar]
- Hong SK, Aoki T, Kitano H, Satoh H, Nagato Y (1995) Phenotypic diversity of 188 rice embryo mutants. Dev Genet 16: 298–310 [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R (2008) The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot 59: 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T. (1997) Structure and function of feruloylated polysaccharides. Plant Sci 127: 111–127 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffree CE. (1996) Structure and ontogeny of plant cuticles. In Kerstiens G, ed, Plant Cuticles: An Integrated Functional Approach. BIOS Scientific Publishers, Oxford, UK, pp 33-82 [Google Scholar]
- Jiang J, Zhang Z, Cao J (2013) Pollen wall development: the associated enzymes and metabolic pathways. Plant Biol (Stuttg) 15: 249–263 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, Lee YS, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim YW, Hwang I, An G (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli DK, Bachhawat AK (2003) CLOURE: Clustal Output Reformatter, a program for reformatting ClustalX/ClustalW outputs for SNP analysis and molecular systematics. Nucleic Acids Res 31: 3501–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Molina I, Ohlrogge JB, Pollard M (2012) Identification of an Arabidopsis fatty alcohol:caffeoyl-Coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiol 160: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Stone DE (1983) Pollen intine structure, cytochemistry and function in monocots. In Mulcahy DL, Ottaviano E, eds, Pollen: Biology and Implications for Plant Breeding. Elsevier, New York, pp 159–163 [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lam TB, Kadoya K, Iiyama K (2001) Bonding of hydroxycinnamic acids to lignin: ferulic and p-coumaric acids are predominantly linked at the benzyl position of lignin, not the β-position, in grass cell walls. Phytochemistry 57: 987–992 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. The Arabidopsis Book 8: e0133, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010a) Cytochrome P450 family member CYP704B2 catalyzes the omega-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang D (2010) Biosynthesis of anther cuticle and pollen exine in rice. Plant Signal Behav 5: 1121–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu M, Geng LL, Zhao J (2010b) The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J 64: 482–497 [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Zhu J, Yang J, Zhang GR, Xing WF, Zhang S, Yang ZN (2012) Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Mol Plant 5: 131–142 [DOI] [PubMed] [Google Scholar]
- Liu H, Chu H, Li H, Wang H, Wei J, Li N, Ding S, Huang H, Ma H, Huang C, et al. (2005) Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (OsMS-L) mutant. Chin Sci Bull 50: 122 [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- McCormick S. (1993) Male gametophyte development. Plant Cell 5: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuter-Gerhards A, Riegert S, Wiermann R (1999) Studies on sporopollenin biosynthesis in Cucurbita maxima (DUCH.)—II. The involvement of aliphatic metabolism. J Plant Physiol 154: 431–436 [Google Scholar]
- Meychik NR, Matveyeva NP, Nikolaeva YI, Chaikova AV, Yermakov IP (2006) Features of ionogenic group composition in polymeric matrix of lily pollen wall. Biochemistry (Mosc) 71: 893–899 [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang L (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Molina I, Kosma D (2015) Role of HXXXD-motif/BAHD acyltransferases in the biosynthesis of extracellular lipids. Plant Cell Rep 34: 587–601 [DOI] [PubMed] [Google Scholar]
- Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M (2009) Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol 151: 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari HB, Pellny TK, Freeman J, Shewry PR, Mitchell RA (2013) Grass cell wall feruloylation: distribution of bound ferulate and candidate gene expression in Brachypodium distachyon. Front Plant Sci 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Kim S-R, Zhao G, Yi J, Yoo Y, Jin P, Lee S-W, Jung KH, Zhang D, An G (2013) Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol 161: 663–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösle B, Finch P, Collinson ME, Scott AC (1997) Comparison of modern and fossil plant cuticles by selective chemical extraction monitored by flash pyrolysis-gas chromatography-mass spectrometry and electron microscopy. J Anal Appl Pyrol 40: 585–597 [Google Scholar]
- Murphy DJ. (2006) The extracellular pollen coat in members of the Brassicaceae: composition, biosynthesis, and functions in pollination. Protoplasma 228: 31–39 [DOI] [PubMed] [Google Scholar]
- Nawrath C. (2002) The biopolymers cutin and suberin. The Arabidopsis Book 1: e0021, doi/10.1199/tab.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri G, Teixeira EW, Alves ML, Moreti AC, Otsuk IP, Borguini RG, Salatino A (2011) Hydroxycinnamic acid amide derivatives, phenolic compounds and antioxidant activities of extracts of pollen samples from Southeast Brazil. J Agric Food Chem 59: 5516–5522 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. EMBnet.News 4: 1–4 [Google Scholar]
- Owen HA, Makaroff C (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A (2009) The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol 151: 1773–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Ross JH, Murphy DJ (1997) Intra- and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J 11: 549–562 [DOI] [PubMed] [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ (1998) Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod 11: 65–80 [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Ralph J, Bunzel M, Marita JM, Hatfield RD, Lu F, Kim H, Schatz PF, Grabber JH, Steinhart H (2004) Peroxidase-dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem Rev 3: 79–96 [Google Scholar]
- Rautengarten C, Baidoo E, Keasling JD, Scheller HV (2010) A simple method for enzymatic synthesis of unlabeled and radiolabeled hydroxycinnamate-CoA. Bioenerg Res 3: 115–122 [Google Scholar]
- Rautengarten C, Ebert B, Ouellet M, Nafisi M, Baidoo EEK, Benke P, Stranne M, Mukhopadhyay A, Keasling JD, Sakuragi Y, et al. (2012) Arabidopsis Deficient in Cutin Ferulate encodes a transferase required for feruloylation of ω-hydroxy fatty acids in cutin polyester. Plant Physiol 158: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina JJ, Heredia A (2001) Plant cutin biosynthesis: the involvement of a new acyltransferase. Trends Plant Sci 6: 296. [DOI] [PubMed] [Google Scholar]
- Saulnier L, Thibault JF (1999) Ferulic acid and diferulic acids as components of sugar-beet pectins and maize bran heteroxylans. J Sci Food Agric 79: 396–402 [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Schnurr JA, Storey KK, Jung H-JG, Somers DA, Gronwald JW (2006) UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta 224: 520–532 [DOI] [PubMed] [Google Scholar]
- Scott R. (1994) Pollen exine: the sporopollenin enigma and the physics of pattern. In Scott RJ, Stead MA, eds, Molecular and Cellular Aspects of Plant Reproduction. University Press, Cambridge, UK, pp 49–81 [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell (Suppl) 16: S46–S60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Hohn C, Franke R, Prat S, Molinas M, Figueras M (2010) A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. Plant J 62: 277–290 [DOI] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim Y-J, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Shi J, Tan H, Yu XH, Liu Y, Liang W, Ranathunge K, Franke RB, Schreiber L, Wang Y, Kai G, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V (2000) Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. In Romeo JT, Ibrahim R, Varin L De Luca V, eds, Recent Advances in Phytochemistry, Vol 34 Elsevier Science, Oxford, pp 285–315 [Google Scholar]
- Stewart C Jr, Kang BC, Liu K, Mazourek M, Moore SL, Yoo EY, Kim BD, Paran I, Jahn MM (2005) The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J 42: 675–688 [DOI] [PubMed] [Google Scholar]
- Tan H, Liang W, Hu J, Zhang D (2012) MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Dev Cell 22: 1127–1137 [DOI] [PubMed] [Google Scholar]
- Tuominen LK, Johnson VE, Tsai CJ (2011) Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JS, Sephton MA, Sephton SV, Self S, Fraser WT, Lomax BH, Gilmour I, Wellman CH, Beerling DJ (2007) Rapid determination of spore chemistry using thermochemolysis gas chromatography-mass spectrometry and micro-Fourier transform infrared spectroscopy. Photochem Photobiol Sci 6: 689–694 [DOI] [PubMed] [Google Scholar]
- Wehling K, Niester C, Boon JJ, Willemse MT, Wiermann R (1989) p-Coumaric acid— a monomer in the sporopollenin skeleton. Planta 179: 376–380 [DOI] [PubMed] [Google Scholar]
- Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, et al. (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56: 979–994 [DOI] [PubMed] [Google Scholar]
- Yoon J, Choi H, An G (2015) Roles of lignin biosynthesis and regulatory genes in plant development. J Integr Plant Biol 57: 902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XH, Gou JY, Liu CJ (2009) BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression. Plant Mol Biol 70: 421–442 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liang W (2016) Improving food security: using male fertility for hybrid seed breeding. In Pushing the Boundaries of Scientific Research: 120 Years of Addressing Global Issues. Science/AAAS, Washington, DC, pp 45–48
- Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang D, Wilson ZA (2009) Stamen specification and anther development in rice. Chin Sci Bull 54: 2342–2353 [Google Scholar]
- Zhang D, Yang L (2014) Specification of tapetum and microsporocyte cells within the anther. Curr Opin Plant Biol 17: 49–55 [DOI] [PubMed] [Google Scholar]
- Zhao G, Shi J, Liang W, Xue F, Luo Q, Zhu L, Qu G, Chen M, Schreiber L, Zhang D-B (2015) Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol 169: 2064–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Wang Y, Li W, Zhao Z, Ren Y, Wang Y, Gu S, Lin Q, Wang D, Jiang L, et al. (2011) Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. Plant Cell 23: 111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.