Two putative GPI-anchored aspartic proteases, A36 and A39, are involved in pollen and ovule development as well as in directional growth of the pollen tube toward ovules in Arabidopsis.
Abstract
Aspartic proteases are a class of proteolytic enzymes with conserved aspartate residues, which are implicated in protein processing, maturation, and degradation. Compared with yeast and animals, plants possess a larger aspartic protease family. However, little is known about most of these enzymes. Here, we characterized two Arabidopsis (Arabidopsis thaliana) putative glycosylphosphatidylinositol (GPI)-anchored aspartic protease genes, A36 and A39, which are highly expressed in pollen and pollen tubes. a36 and a36 a39 mutants display significantly reduced pollen activity. Transmission electron microscopy and terminal-deoxynucleotidyl transferase-mediated nick end labeling assays further revealed that the unviable pollen in a36 a39 may undergo unanticipated apoptosis-like programmed cell death. The degeneration of female gametes also occurred in a36 a39. Aniline Blue staining, scanning electron microscopy, and semi in vitro guidance assays indicated that the micropylar guidance of pollen tubes is significantly compromised in a36 a39. A36 and A39 that were fused with green fluorescent protein are localized to the plasma membrane and display punctate cytosolic localization and colocalize with the GPI-anchored protein COBRA-LIKE10. Furthermore, in a36 a39, the abundance of highly methylesterified homogalacturonans and xyloglucans was increased significantly in the apical pollen tube wall. These results indicate that A36 and A39, two putative GPI-anchored aspartic proteases, play important roles in plant reproduction in Arabidopsis.
In higher flowering plants, the life cycle from the generation of haploid gametophytes to the generation of diploid sporophytes is essential to reproduction (Dumas and Mogensen, 1993). Through meiosis and mitosis, the male and female gametophytes, also called the pollen grain and the embryo sac, respectively, are generated from anther tissue and ovules. The pollen grain germinates on the stigma, forming a long pollen tube, which grows rapidly through the maternal tissue and accurately reaches the female gametes. Then, the pollen tube ruptures and releases the two sperm cells to complete double fertilization with the egg and central cell (Palanivelu and Tsukamoto, 2012). Therefore, successful reproduction relies on normal male and female gametogenesis as well as on precise communication between the pollen tube and the ovule.
Aspartic protease (EC 3.4.23), one of the four major types of proteolytic enzymes, exist widely in bacteria, fungi, animals, and plants (Barrett et al., 1998). These enzymes contain two Asp residues within the conserved Asp-Thr/Ser-Gly motifs at the active site and are activated at acidic pH and specifically inhibited by pepstatin A (Takahashi et al., 2008). Aspartic proteases have been widely studied in animals, including β-secretase, pepsin, cathepsin D, etc. (Ingram, 1951; Vassar et al., 1999; Bach et al., 2015). However, research on plant aspartic proteases remains limited.
The first well-studied plant aspartic protease was phytepsin from barley (Hordeum vulgare), which possesses a plant-specific insert, is localized to the vacuole, and may function in the programmed cell death (PCD) of tracheary elements and sieve cells (Runeberg-Roos and Saarma, 1998; Kervinen et al., 1999). Rice (Oryza sativa) oryzasin also contains a plant-specific insert domain, is highly expressed 2 or 3 weeks after flowering, and may be involved in the processing of storage proteins in seeds (Asakura et al., 1995, 2000). Faro and Gal (2005) found 51 genes that may encode putative aspartic proteases in the Arabidopsis (Arabidopsis thaliana) genome and divided them into three types, typical aspartic proteases, nucellin-like aspartic proteases, and atypical aspartic proteases, according to their putative domains and active sites. Takahashi et al. (2008) enlarged the aspartic protease family to approximately 69 members.
In the last decade, emerging evidence has revealed important roles for aspartic proteases in leaf senescence, pathogen resistance, stress responses, and reproduction. Tobacco (Nicotiana tabacum) CND41 is a DNA-binding aspartic protease expressed in chloroplasts, which plays an essential role in leaf aging by degrading the Rubisco protein (Kato et al., 2005). In Arabidopsis and rice, the apoplastic aspartic protease CONSTITUTIVE DISEASE RESISTANCE1 (CDR1) is involved in salicylic acid-mediated disease resistance (Xia et al., 2004; Prasad et al., 2009). Furthermore, the overexpression of ASPARTIC PROTEASE INGUARD CELL1 increased abscisic acid sensitivity in guard cells, conferring drought tolerance in Arabidopsis (Yao et al., 2012). Recently, aspartic protease APCB1 was revealed to be involved in the processing of Bcl-2-ASSOCIATED ATHANOGENE6 to trigger autophagy and defense resistance (Li et al., 2016).
Aspartic proteases also play vital roles in plant sexual reproduction, such as tapetum degeneration and gamete development. The proper timing of PCD in tapetum cells is crucial for pollen formation. AtMYB80 and ETERNAL TAPETUM1 in rice are transcription factors that control PCD in the tapetum and target the downstream aspartic protease genes AtUNDEAD and OsAP25/OsAP37, respectively. Silencing the expression of AtUNDEAD caused the tapetal PCD to occur earlier, leading to pollen abortion (Phan et al., 2011), and overexpressing OsAP25 or OsAP37 induced cell death in both plants and yeast (Niu et al., 2013). In addition, many aspartic proteases also are involved in gametophyte development. In rice, S5 participates in indica-japonica hybrid fertility and could stimulate endoplasmic reticulum (ER) stress, giving rise to PCD in the embryo sac (Chen et al., 2008; Yang et al., 2012). OsAP65 was shown to be essential for pollen germination and tube growth (Huang et al., 2013). In Arabidopsis, PROMOTION OF CELL SURVIVAL1 (PCS1) encodes an ER-localized aspartic protease, and loss function of PCS1 causes the degeneration of gametophytes and embryos (Ge et al., 2005). Therefore, plant aspartic proteases may be implicated in the regulation of PCD in plant reproduction, although the underlying mechanism is not clear. Moreover, the functions of the majority of Arabidopsis aspartic proteases are still unknown.
In this study, we characterized A36 and A39, two putative glycosylphosphatidylinositol (GPI)-anchored aspartic proteases that are preferentially expressed in the pollen and pollen tube in Arabidopsis. Loss function of A36 and A39 results in the apoptosis-like PCD of microspores, the degeneration of female gametophytes, and compromised pollen tube guidance. Moreover, A36-GFP and A39-GFP are colocalized with COBRA-LIKE10 (COBL10), which is a GPI-anchored protein involved in pollen tube guidance. Our results indicated that A36 and A39 play important roles in plant fertility.
RESULTS
A36 and A39 Are Highly Expressed in Pollen and Pollen Tube and Display Aspartic Proteolytic Enzyme Activity
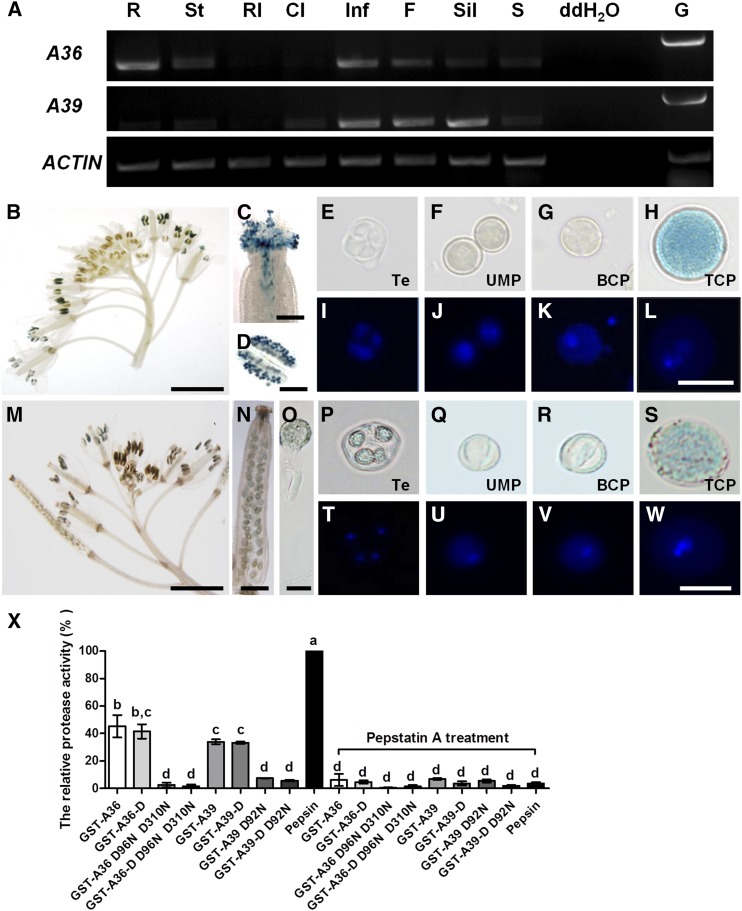
Our preliminary work on proteomic analyses of proteins in germinating Arabidopsis pollen had identified 71 differentially expressed proteins (Ge et al., 2011). In this study, two aspartic proteases, A36 (At5G36260) and A39 (At1G65240), were selected for further investigation. Evolutionary analysis indicates that A36 shares high similarity with several proteins from different model organisms but also displays relatively clear differentiation (Supplemental Fig. S1A). In addition, A39 shares 73% amino acid identity with A36, both contain N-terminal signal peptide and hydrophobic C terminus (http://smart.embl-heidelberg.de/; http://www.cbs.dtu.dk/services/SignalP-2.0/), and both are predicted to be GPI-anchored proteins by big-PI Predictor (http://mendel.imp.ac.at/gpi/gpi_server.html; Eisenhaber et al., 1998; Supplemental Fig. S1B). Based on protein sequence alignments, A36 shows 23% and 22% identity with yeast (Saccharomyces cerevisiae) ScYps1p and human BACE1, respectively. To explore the detailed functions of A36 and A39 in plant growth, particularly in male gametophyte development, reverse transcription (RT)-PCR was performed to examine the expression pattern of A36 and A39 in different organs, including roots, stems, rosette leaves, cauline leaves, inflorescence, open flowers, siliques, and seedlings (Fig. 1A). As shown in Figure 1A, both A36 and A39 are highly expressed in inflorescence and open flowers. A36 mRNA also is present at relatively high levels in the roots and has moderate expression in the stems, siliques, and seedlings, while A39 transcripts were highly detected in the siliques and showed little expression in the cauline leaves and seedlings. To further explore the expression profiles of A36 and A39, pA36::GUS and pA39::GUS transgenic plants were generated. In T3 transgenic lines, A36 is highly expressed exclusively in the pollen (Fig. 1, B and D) and growing pollen tubes (Fig. 1C; Supplemental Fig. S2E). During microspore development, the GUS signal of A36 could be found only in tricellular pollen and not in uninuclear or bicellular pollen (Fig. 1, E–L; Supplemental Fig. S2, H–K). The GUS signal of A36 was not detected in the seedlings, leaves, stems, siliques, or ovules (Supplemental Fig. S2, A–D and P). Compared with A36, A39 shows a similar but distinct expression profile. Similar to A36, A39 also is highly expressed in the inflorescence and pollen tubes (Fig. 1, M and O), and its GUS signal is not detected in the seedlings or leaves (Supplemental Fig. S2, F and G). However, different from A36, A39 was expressed from tetrad to tricellular pollen (Fig. 1, P–W; Supplemental Fig. S2, L–O), young siliques (Fig. 1N), and developing ovules (Supplemental Fig. S2Q). Therefore, both A36 and A39 are expressed preferentially in tricellular pollen and pollen tubes, although A39 also is expressed in the developing microspores, ovules, and young siliques.
Figure 1.
A36 and A39 are highly expressed in mature pollen grains and growing pollen tubes and display aspartic proteolytic activity in vitro. A, Expression patterns of A36 and A39 in wild-type plants by RT-PCR. Total RNA was isolated from wild-type plants. ACTIN7 was taken as an internal control. Amplification was performed for 25 cycles for ACTIN7 and for 30 cycles for A36 (1,206 bp) and A39 (1,258 bp). R, Roots; St, stems; Rl, rosette leaves; Cl, cauline leaves; Inf, inflorescences; F, flowers; Sil, siliques; S, seedlings. Genomic DNA (G) as a positive control and distilled, deionized water (ddH2O) as a negative control also are presented. B to D, Expression pattern of A36 revealed by a promoter assay, showing GUS signals in inflorescence (B), stigma (C), and anther (D). Bars = 200 μm for B, 5 mm for C, and 20 μm for D. E to L, Developing male gametophytes by 4′,6-diamidino-2-phenylindole (DAPI) and GUS staining. Te, Tetrad microspore (E and I); UMP, uninuclear microspore (F and J); BCP, bicellular pollen (G and K); TCP, tricellular pollen (H and L). Bar = 20 μm. M to O, Expression pattern of A39 revealed by a promoter assay, showing GUS signals in inflorescence (M), silique (N), and pollen tube (O). Bars = 5 mm for M, 500 μm for N, and 20 μm for O. P to W, Developing male gametophytes by DAPI and GUS staining. Te, Tetrad microspore (P and T); UMP, uninuclear microspore (Q and U); BCP, bicellular pollen (R and V); TCP, tricellular pollen (S and W). Bar = 20 μm. X, Semiquantitative assay of proteolytic activity. The GST-A36, GST-A36-D, GST-A39, GST-A39-D, and site-directed mutants GST-A36 D96N D310N, GST-A36-D D96N D310N, GST-A39 D92N, and GST-A39-D D92N were expressed in E. coli and purified in vitro. FITC-casein was used as the substrate. Equal amounts (375 ng) of GST fusion proteins and pork pepsin (positive control) were used, and the reaction was carried out at pH 5.4 and 22°C. Data were collected from three independent experiments. Error bars show se, calculated using one-way ANOVA; P < 0.05 by Tukey’s method.
To clarify whether A36 and A39 share aspartic protease activity, we first performed a sequence alignment of their active site regions with the corresponding regions from PCS1 (A35) and CDR1 (A3; Supplemental Fig. S1C). Like other eukaryotic aspartic proteases, both A36 and A39 contain two catalytic Asp residues within two conserved active sites with the sequence Asp-Thr/Ser-Gly-Ser/Thr. Next, we made four glutathione S-transferase (GST) fusion protein constructs: GST-A36-D (106–1,347 bp of the coding sequence), GST-A36 (full coding sequence), GST-A39-D (88–1,275 bp of the coding sequence), and GST-A39 (full coding sequence; Supplemental Fig. S3). In addition, four mutations of A36 and A39 were generated by site-directed mutagenesis: D96N and D310N in A36 and A36-D (two Asp residues at positions 96 and 310 were replaced by Asn) and D92N (a single Asp residue at position 92 was replaced by Asn) in A39 and A39-D (Supplemental Fig. S3). All of these GST fusion proteins were expressed in Escherichia coli and purified in vitro. Because additional bands existed in purified GST fusion proteins, we used different contents of bovine serum albumin (BSA) and the corresponding mean Gray value to make a standard curve to measure the concentration of purified full-length GST fusion proteins (Supplemental Fig. S3). A semiquantitative assay was performed using fluorescein isothiocyanate (FITC)-labeled casein as the substrate (Fig. 1X). The same amounts of proteins were used to measure the proteolytic activity. At pH 5.4, the proteolytic activity of GST-A36 was 45.6% of that of pork pepsin, while GST-A36-D showed 41.5% relative activity (Fig. 1X). GST-A39 and GST-A39-D displayed proteolytic activities that were 34% and 33.2% of those of pork pepsin, respectively (Fig. 1X). However, GST-A36 D96N D310N, GST-A36-D D96N D310N, GST-A39 D92N, and GST-A39-D D92N had hardly any proteolytic activity (Fig. 1X). The proteolytic activity of all GST fusion proteins was significantly inhibited by the aspartic protease inhibitor pepstatin A (Fig. 1X). Thus, both A36 and A39 show aspartic proteolytic activity in vitro.
Disruption of A36 Results in Decreased Male Transmission Efficiency
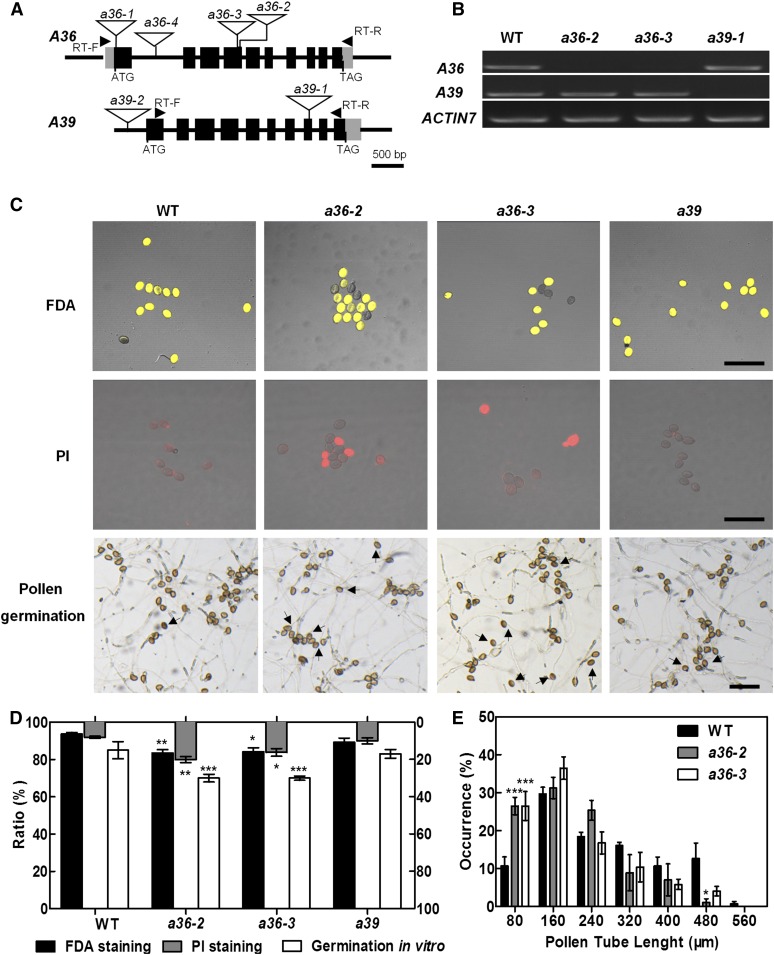
To explore the biological functions of A36 and A39 in plant growth, particularly in male gametophyte fertility, we obtained four different T-DNA insertion alleles for A36 (a36-1, SALK_149238C; a36-2, SAIL_697_C09; a36-3, SALK_065978; and a36-4, SALK_080509C) and two for A39 (a39-1, SALK_018723C; and a39-2, SALK_010730) from the Arabidopsis Biological Resource Center (ABRC; Fig. 2A). Transcript analyses of A36 and A39 revealed that a36-2, a36-3, and a39-1 (hereafter abbreviated as a39) are homozygous knockout mutants (Fig. 2B), which were used for further investigation.
Figure 2.
a36 displays reduced pollen grain activity and pollen germination ratio in vitro. A, Genomic organization of the intronless A36 and A39 genes and positions of a36-2, a36-3, and a39-1 T-DNA insertions. The positions of the primers used for RT-PCR are indicated by arrowheads. Exons are represented by black boxes and introns by lines. The locations of the T-DNA insertions are indicated with triangles. B, RT-PCR analysis from open-flower cDNAs shows no A36 transcripts in a36-2 and a36-3 and no A39 transcripts in a39-1. ACTIN7 was used as an internal control. Amplification was performed for 25 cycles for ACTIN7 and for 30 cycles for A36 (1,206 bp) and A39 (1,258 bp). C, Pollen viability of the wild type (WT), a36-2, a36-3, and a39 by FDA (top row) and PI (middle row) staining. FDA-positive (viable) and PI-positive (unviable) pollen are indicated in yellow and red, respectively. The bottom row shows the pollen germination assay in vitro for 8 h. Pollen of the wild type and a39 germinated very well and produced actively growing pollen tubes. By contrast, pollen germination in a36-2 and a36-3 was impaired. Arrows designate pollen that did not germinate. Bars = 100 μm. D, Statistical analysis of pollen viability by FDA and PI staining and germination rates of pollen grains from wild-type, a36-2, a36-3, and a39 plants in vitro. Data were collected from three independent experiments (n > 900). E, Statistical analysis of pollen tube length distribution after incubation for 4 h in vitro. Data were collected from three independent experiments (n = 60). For D and E, error bars show se and asterisks indicate values that differ significantly from those of the wild type (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, calculated using two-way ANOVA).
During vegetative growth, the a36-2, a36-3, and a39 single mutants exhibit normal vegetative development like wild-type plants (Supplemental Fig. S4A). However, the siliques of a36-2 and a36-3 show slightly higher abortion ratios than the wild type, whereas a39 exhibits normal seed setting (Supplemental Fig. S4, B and C). We then performed genetic analyses to investigate the function of gametophytes in the a36-2, a36-3, and a39 mutants. The progeny of the self-pollinated heterozygous a36-2/+ and a36-3/+ plants segregated at a ratio of 1(136):1.63(222):0.72(98) and 1(140):1.65(231):0.71(100) for wild type:heterozygote:homozygote, respectively (Table I), which deviated significantly from the classic Mendelian segregation ratio of 1:2:1. Then, transmission efficiency analyses were conducted in which a36/+ plants were used as the male or female parent in crosses with wild-type plants. When using a36-2/+ or a36-3/+ as the male parent, the male transmission efficiency was decreased markedly to 68% and 68.3%, respectively (Table I). When taking a36-2/+ or a36-3/+ as the female parent, the female transmission efficiency also was reduced to 81.2% and 78.7%, respectively (Table I). Therefore, both male and female gametophyte transmission efficiency is reduced significantly in the a36 mutant, while the self-segregation ratio of a39/+ was in line with the classic Mendelian segregation ratio and a39 had normal male and female transmission efficiency (Table I).
Table I. Segregation of self progeny and the genetic transmission of a36/+ and a39/+.
Reciprocal crosses were performed between a36-2, a36-3, and a39 heterozygous plants and wild-type plants. The transmission efficiency (TE) of gametes was calculated as follows: TE = number of progeny with the a36-2, a36-3, and a39 allele/number of wild-type progeny × 100%. The observed TE was compared with the expected TE for normal gametes (100%). TEF, Female transmission efficiency; TEM, male transmission efficiency. χ2 indicates the χ2 test (degrees of freedom = 1, P < 0.05, χ2 > 3.84). NA, Not applicable.
| Parental Genotype |
Progeny Genotype |
TEF | TEM | χ2 | Expected |
|||
|---|---|---|---|---|---|---|---|---|
| Female × Male | +/+ | +/− | −/− | TE | χ2 | |||
| a36-2/+ Y | 136 | 222 | 98 | NA | NA | 6.65 | NA | 5.99 |
| ♀Wild type × ♂a36-2/+ | 372 | 253 | NA | NA | 68.0% | 22.28 | 100% | 3.84 |
| ♀a36-2/+ × ♂wild type | 367 | 298 | NA | 81.2% | NA | 6.95 | ||
| a36-3/+ Y | 140 | 231 | 100 | NA | NA | 6.97 | NA | 5.99 |
| ♀Wild type × ♂a36-3/+ | 161 | 110 | NA | NA | 68.3% | 9.22 | 100% | 3.84 |
| ♀a36-3/+ × ♂wild type | 179 | 141 | NA | 78.7% | NA | 4.28 | ||
| a39/+ Y | 119 | 202 | 127 | NA | NA | 4.60 | NA | 5.99 |
| ♀Wild type × ♂a39/+ | 223 | 198 | NA | NA | 88.8% | 1.37 | 100% | 3.84 |
| ♀a39/+ × ♂wild type | 133 | 165 | NA | 124.0% | NA | 3.22 | ||
Pollen Activity and Pollen Germination Rate Are Reduced in a36
Next, the viability of mature pollen grains in the a36-2, a36-3, and a39 mutants was examined by scanning electron microscopy (SEM), DAPI staining, and Alexander staining (Supplemental Fig. S5, A–L). However, compared with wild-type plants, no obvious differences were detected in the mature pollen grains of the a36-2, a36-3, and a39 mutants (Supplemental Fig. S5, M–O). Successful Alexander staining in pollen only indicates that the cells have cell wall and intact cytoplasm. The fluorescein diacetate (FDA) staining assay could test the integrity of membranes and some esterase activity in cells. Therefore, we performed FDA staining of mature pollen, and the viable pollen rate in the a36-2 and a36-3 mutants was significantly lower than that of the wild type (Fig. 2C). Compared with 6.4% of pollen grains in the wild type, approximately 16% of pollen grains from the a36-2 (16.5%) or a36-3 (15.8%) mutant could not be stained by FDA (Fig. 2D). Then, propidium iodide (PI) was used to stain (red spot) the dead pollen (Fig. 2C). Coincidentally, 20.2% and 15.9% of pollen grains from a36-2 and a36-3 mutants showed positive PI staining, respectively (Fig. 2D). These results suggested that a portion of pollen grains may be dead or unviable in the a36-2 or a36-3 mutant. In addition, no significant difference was detected between the pollen grains collected from a39 mutants and wild-type plants by FDA or PI staining.
Then, we performed pollen germination assays in vitro (Fig. 2C). The pollen germination ratios of the a36-2 and a36-3 mutants, but not the a39 mutant, were decreased significantly at 8 h after germination in vitro (Fig. 2D). The ungerminated pollen in the a36-2 and a36-3 mutants may correspond to the inactive pollen detected via FDA or PI staining. Furthermore, the length of the pollen tube was analyzed at 4 h after germination in vitro. The wild-type pollen tubes reached a length of 160 to 320 μm at 4 h after germination (n > 20), whereas the a36-2 and a36-3 mutant pollen tubes only reached a length of 80 to 160 μm (Fig. 2E). Next, to explore whether the mutant phenotype of a36 is caused by a sporophytic or gametophytic defect, we analyzed the pollen grain activity and pollen germination ratio of a36-2/+ heterozygous plants by FDA staining and pollen germination assays in vitro (Supplemental Fig. S6A). We found that the pollen grain activity and pollen germination ratio in a36-2/+ plants are comparable to those of the wild type and significantly higher than those in a36-2 plants (Supplemental Fig. S6, B and C). These results indicate that the mutant phenotype of a36 is caused by sporophytic defects.
We also examined the growth rate of a36-2, a36-3, and a39 mutant pollen tubes in the wild-type style and transmitting tract in vivo (Supplemental Fig. S7A). But no significant difference was found between the mutant and wild-type plants (Supplemental Fig. S7B). The transmission efficiency of female gametophytes also was reduced slightly in the a36-2 and a36-3 mutants. We also examined the morphology of the female gametophyte stage 7 (FG7) embryo sac via differential interference contrast (DIC) and confocal laser scanning microscopy (CLSM) analyses (Supplemental Fig. S8, A and B), and no obvious defects were found (Supplemental Fig. S8C).
Complementation of the a36 Mutant
To confirm whether the mutant phenotype in a36-2 resulted from the disruption of A36, we generated a construct containing the full-length genomic DNA sequence of A36 (4 kb from the 5′ untranslated region to the 3′ untranslated region and 2 kb of the 5′ native promoter) in the pCAMBIA1300 vector. This construct was then introduced into homozygous a36-2 plants. Thirty-two of 38 independent T2 transgenic lines were recovered to wild-type plants. Then, three homozygous T3 lines were used to perform detailed phenotypic analyses, including pollen viability and pollen germination in vitro (Supplemental Fig. S9A). All of these transgenic lines resembled wild-type plants (Supplemental Fig. S9B), which demonstrated that the A36 gene fully rescued the mutant phenotypes.
a36 a39 Double Mutants Display Notable Seed Abortion
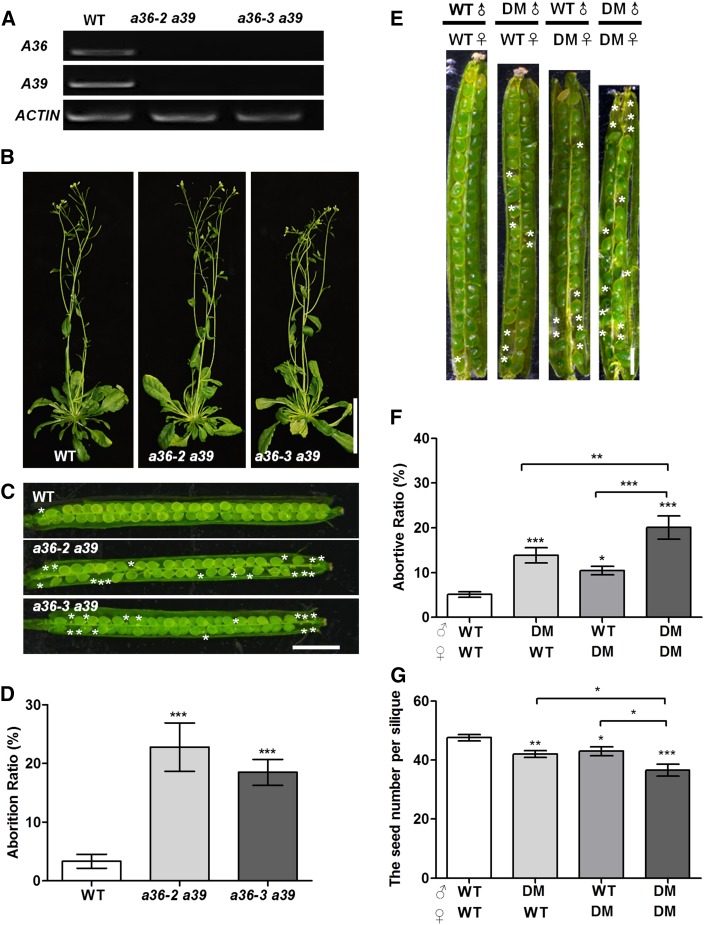
Bioinformatics analyses indicated that A36 and A39 are predicted to share 73% amino acid identity. Through genetic crosses, we obtained a36-2 a39 and a36-3 a39 double mutants (Fig. 3A). During vegetative growth, no significant defects were detected for a36-2 a39 or a36-3 a39 double mutants (Fig. 3B). However, a36-2 a39 and a36-3 a39 plants produced siliques with 22.2% and 18.4% abortion, respectively (Fig. 3, C and D).
Figure 3.
Phenotype analysis of a36 a39 double mutants. A, RT-PCR analysis from open-flower cDNAs shows that no A36 and A39 transcripts are detected in a36-2 a39 and a36-3 a39. ACTIN7 was used as an internal control. Amplification was performed for 25 cycles for ACTIN7 and for 30 cycles for A36 (1,206 bp) and A39 (1,258 bp). B, Six-week-old wild-type (WT) and a36 a39 plants. Bar = 5 cm. C, The a36-2 a39 and a36-3 a39 mutants have reduced seed set. Asterisks designate unfertilized ovules in the silique. Bar = 1 mm. D, Statistical analysis of the abortion ratios of seed set from the wild-type, a36-2 a39, and a36-3 a39. The values are based on three biological replicates (n = 300). E, In vivo reciprocal cross-pollination of double mutant (DM; a36-2 a39) to wild-type plants. Siliques were then dissected for the examination of fertilized ovules. Asterisks designate unfertilized ovules in the silique. Bar = 1 mm. F, Statistical analysis of the seed number per mature silique. The values are based on three biological replicates (n = 120 per cross). G, Statistical analysis of the abortion ratios of the seed set. The values are based on three biological replicates (n = 120 per cross). For D, F, and G, error bars show se and asterisks indicate values that differ significantly from those of the wild-type plant (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, calculated using Student’s t test).
Then, in order to dissect the cause of the silique abortion in the double mutants, we performed reciprocal crosses using wild-type plants and the a36-2 a39 double mutant as parents (Fig. 3E) and examined the abortion ratio of siliques (Fig. 3F) and the seed number (Fig. 3G). When the wild type and a36-2 a39 mutants were self-pollinated by hand pollination, the abortion ratios were 5.1% and 20.1% (Fig. 3F), respectively, which is close to that of natural self-pollination. When a36-2 a39 was used as the male and the wild type as the female, the abortion ratio of siliques was 13.8%, significantly different from that of the wild type and a36 a39 by self-crossing. While when a36-2 a39 was used as female and the wild type as male, the abortion ratio of siliques was 10.4% (Fig. 3F), which was also remarkably different from that of the wild type and a36 a39 by self-crossing. A statistical analyses of the seed numbers of siliques gave similar results (Fig. 3G). Thus, the functions of male and female in a36 a39 are both defective.
Next, to investigate whether embryogenesis has any defects in the a36 a39 double mutant, we observed embryo development in the wild type and a36-2 a39 in artificial self-crossing (Supplemental Fig. S10, A–L). We found that 90.8% and 79.7% of the early embryos in the wild type and a36-2 a39, respectively, reached the globular stage at 3 and 4 d after hand pollination and the heart-shaped and torpedo-shaped embryo stages at 5 and 6 d after pollination (Supplemental Fig. S10, A–H and M). Consistent with the abortion ratio of siliques in self-pollinated a36-2 a39 by hand pollination, approximately 20.3% of a36-2 a39 ovules were unfertilized and no defects in embryogenesis were found (Supplemental Fig. S10, I–M). Thus, these results indicate that the embryogenesis in a36 a39 is not impaired and that the seed abortion in the a36 a39 double mutant results from unfertilized ovules. Therefore, the above data also suggest that the defects in both the male and female of the double mutant may be responsible for the notable seed abortion in the siliques.
We then performed a transmission efficiency assay to test the function of the male and female gametophytes in a36 a39. Reciprocal crosses between the wild-type plant and a36/+ a39/− were made first (Table II). When the a36-2/+ a39/− and a36-3/+ a39/− pistils were hand pollinated with wild-type pollen (the progeny produced the genotypes a36/+ a39/+ and a39/+, respectively), we found that 67.4% and 73.3% of the a36 a39 alleles were transmitted via the female gametophytes, respectively (Table II). In the reverse combination, when a36-2/+ a39/− and a36-3/+ a39/− were used as the male and the wild type as female, respectively, only 10.3% and 15.9% of the transmission occurred via the male gametophytes, respectively (Table II). Then, reciprocal crosses between the wild-type plant and a36/− a39/+ were performed. We found that 118% and 93.8% of a36-2 a39 and a36-3 a39 were transmitted via the female gametophytes, respectively (Table II), while only 22.4% and 27.3% of the transmission occurred via the male gametophytes, respectively (Table II). These results imply that the male transmission in the a36 a39 mutant is severely compromised, while the female transmission is affected only slightly.
Table II. Transmission efficiency analysis using reciprocal crosses of a36/+ a39/+.
Reciprocal crosses were performed between a36-2 a39 and a36-3 a39 heterozygous plants and wild-type plants. The transmission efficiency (TE) of gametes was calculated as follows: TE = number of progeny with a36-2 a39 and a36-3 a39 allele/number of wild-type progeny × 100%. The observed TE was compared with the expected TE for normal gametes (100%). TEF, Female transmission efficiency; TEM, male transmission efficiency. χ2 indicates the χ2 test (degrees of freedom = 1, P < 0.05, χ2 > 3.84; degrees of freedom = 2, P < 0.05, χ2 > 5.99). NA, Not applicable.
| Parental Genotype |
Progeny Genotype |
TEF | TEM | χ2 | Expected |
||
|---|---|---|---|---|---|---|---|
| Female × Male | ++/+− | +−/+− | TE | χ2 | |||
| ♀Wild type × ♂a36-2/+ a39/− | 495 | 51 | NA | 10.3% | 359.4 | 100% | 3.84 |
| ♀a36-2/+ a39/− × ♂wild type | 273 | 184 | 67.4% | NA | 16.9 | ||
| ♀Wild type × ♂a36-3/+ a39/− | 207 | 33 | NA | 15.9% | 124.7 | 100% | 3.84 |
| ♀a36-3/+ a39/− × ♂wild type | 135 | 99 | 73.3% | NA | 5.2 | ||
| ♀Wild type × ♂a36-2/− a39/+ | 420 | 94 | NA | 22.4% | 205.4 | 100% | 3.84 |
| ♀a36-2/− a39/+ × ♂wild type | 170 | 202 | 118% | NA | 2.6 | ||
| ♀Wild type × ♂a36-3/− a39/+ | 454 | 124 | NA | 27.3% | 187.2 | 100% | 3.84 |
| ♀a36-2/− a39/+ × ♂wild type | 163 | 153 | 93.8% | NA | 0.5 | ||
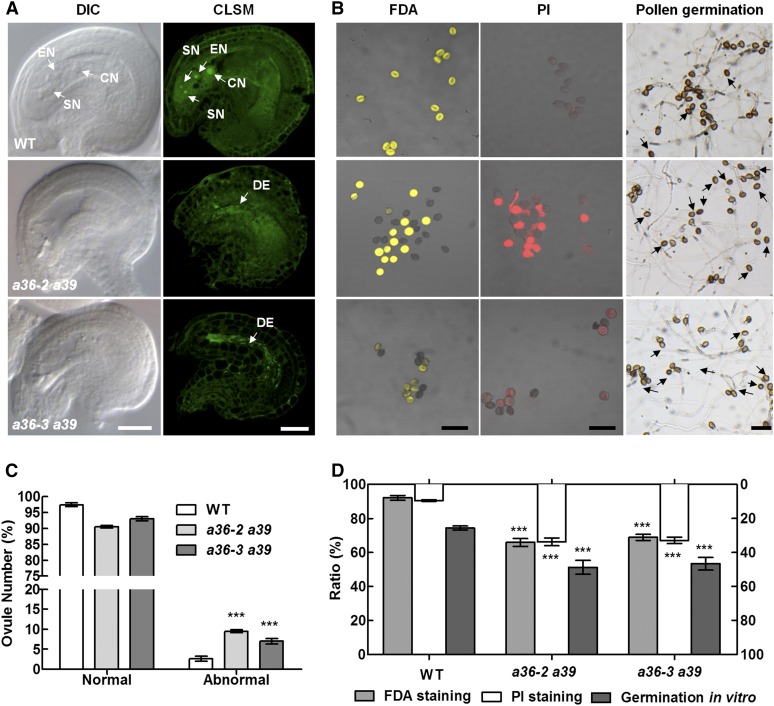
Then, we examined the morphology of the FG7 embryo sac by DIC imaging (Fig. 4A). In the wild-type plants, the structure of the embryo sac is intact and contains one egg cell, two synergids, and a central cell (Fig. 4A). However, in a36 a39, approximately 10% of the ovules exhibit a collapsed embryo sac (Fig. 4C), which is consistent with the abortion ratio of siliques when a36 a39 was used as female and the wild type was used as male (Fig. 3G). We also performed CLSM and found that the mature embryo sac of the wild type contains four cells (Fig. 4A). In a36 a39, no clear nuclear structure was observed in the embryo sac, which was replaced by strong irregular linear fluorescence signals (Fig. 4A). These results indicate that the development of female gametophytes was impaired in the a36 a39 mutant. To clarify whether the female gametophyte defects are controlled by the gametophyte or sporophyte, we observed the morphology of the FG7 embryo sac in trans-heterozygous a36-2/+ a39/− and a36-2/− a39/+ plants (Supplemental Fig. S11, A–F). However, the embryo sac in a36-2/+ a39/− and a36-2/− a39/+ has no significant difference from that of the wild-type plant (Supplemental Fig. S11G). These data imply that the female gametophyte defects in a36 a39 are caused by sporophytic defects. We next performed genetic crosses using a wild-type male and wild-type, a36-2, trans-heterozygous a36-2/- a39/+, and a36-2 a39 as the female (Supplemental Fig. S12A). When trans-heterozygous a36-2/- a39/+ was used as the female, the abortion ratio of the siliques was comparable to that in the wild type and a36-2 single mutant as female but was significantly different from that of the a36 a39 double mutant as female (Supplemental Fig. S12B). Similar results were obtained from a statistical analysis of the seed number per silique (Supplemental Fig. S12C). Therefore, these data support that female gametophyte defects in a36 a39 are caused by sporophytic defects.
Figure 4.
Pollen grains and embryo sac are defective in a36 a39. A, Ovules at FG7 were observed by DIC and CLSM in plants. Ovules with a seven-celled embryo sac at stage FG7 in wild-type (WT), a36-2 a39, and a36-3 a39 pistils contain a two-synergid nucleus (SN), the egg cell nucleus (EN), the large central cell nucleus (CN), and a degenerated embryo sac (DE). Bars = 20 μm. B, Pollen viability and pollen germination in the wild type, a36-2 a39, and a36-3 a39. FDA-positive (viable) and PI-positive (unviable) pollen are indicated in yellow and red, respectively. The right column shows pollen germinated for 8 h after incubation in vitro. The pollen of the wild type and a36 germinated very well and produced actively growing pollen tubes. By contrast, pollen germination in a36-2 a39 and a36-3 a39 was impaired. Arrows designate pollen that did not germinate. Bars = 100 μm. C, Statistical analysis of normal and abnormal ovules at FG7. The values are based on three biological replicates (n = 200). D, Statistical analysis of pollen viability by FDA and PI staining and germination ratio of pollen grains from wild-type, a36-2 a39, and a36-3 a39 plants. Data were collected from three independent experiments (n < 300). For C and D, error bars show se and asterisks indicate values that differ significantly from those of the wild type (***, P <; 0.001, calculated using two-way ANOVA).
Microspores Undergo Unexpected Apoptosis-Like PCD in the a36 a39 Mutant
To further explore the male defects in a36 a39, SEM, DAPI, and Alexander’s staining assays were performed to examine the mature pollen grains from a36 a39 and wild-type plants (Supplemental Fig. S13, A–I). The pollen grains of a36 a39 were comparable to those of the wild type in these assays (Supplemental Fig. S13, J–L). However, in FDA staining assay, 34.1% (a36-2 a39) and 31.1% (a36-3 a39) of pollen could not be stained (Fig. 4, B and D). Correspondingly, 33.8% and 32.9% of pollen was PI positive in a36-2 a39 and a36-3 a39, respectively (Fig. 4, B and D). In addition, in pollen germination assays in vitro, 74.3% (n > 300) of wild-type pollen grains were able to germinate at 8 h after germination, whereas only 56.6% (a36-2 a39; n > 300) and 57.3% (a36-3 a39) of the pollen grains germinated successfully (Fig. 4, B and D). Therefore, compared with the a36 single mutant, microspore development is seriously defective in the a36 a39 double mutant.
To further investigate the decline in pollen vitality, we explored the progress of pollen development in a36 a39 and wild-type plants by examining semithin sections of the anthers. According to published morphological characteristics, anther development can be divided into 14 well-ordered stages in Arabidopsis (Sanders et al., 1999). However, from anther stage 4 to 13, there were no obvious differences between wild-type and a36 a39 anthers, including tapetum development and degradation, as well as microsporogenesis (Supplemental Fig. S14).
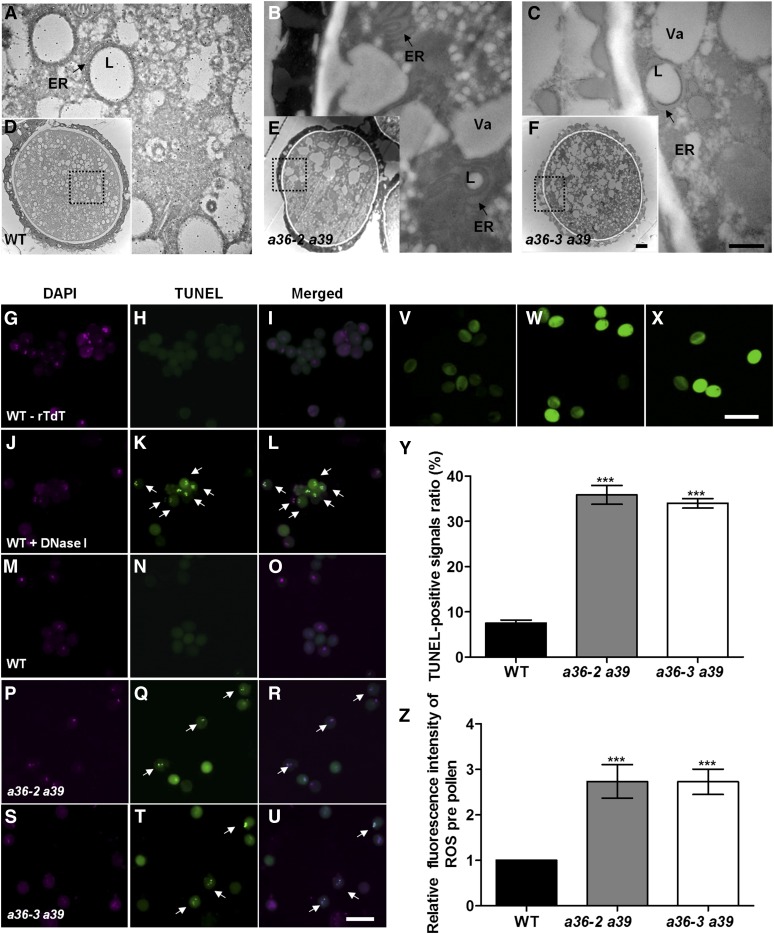
Then, transmission electron microscopy (TEM) was performed to compare the ultrastructures of mature pollen from anther stage 12 in wild-type and a36 a39 plants (Fig. 5, A–F). a36 a39 pollen exhibited a similar pollen exine pattern to wild-type pollen (Fig. 5, D–F). However, 35.7% of pollen from a36-2 a39 and 35.3% of pollen from a36-3 a39 displayed abnormal intracellular ultrastructures (Fig. 5, A–C and Y). In the wild type, the pollen cytoplasm contained many small vacuoles and lipid bodies (Fig. 5, A and D); however, in a36 a39 double mutants, mature pollen grains contained fewer but larger vacuoles and lipid bodies (Fig. 5, B, C, E, and F). In wild-type mature pollen, lipid bodies were surrounded by normal rough ER (Fig. 5A), whereas the structure of the ER lumen was expanded significantly in a36 a39 double mutants (Fig. 5, B and C).
Figure 5.
Apoptosis-like PCD occurs in the mature pollen grains of a36 a39. A to F, TEM analysis of mature pollen grains in wild-type (WT) and a36 a39 plants. A, B, and C are enlarged views of the dotted boxes in D, E, and F. The ultrastructure of pollen grains is shown in wild-type (A and D), a36-2 a39 (B and E), and a36-3 a39 (C and F) plants at anther stage 12. Arrows in A and C indicate the ER, and arrows in B and C indicate the expanded ER. L, Lipid body; Va, vacuole. Bars = 500 nm in A to C and 2 μm in D to F. G to U, Fluorescence microscopy of DNA fragmentation detected using TUNEL assay of pollen grains in the wild type treated without terminal-deoxynucleotidyl transferase (rTdT) as a negative control (G–I), the wild type treated with DNase I as a positive control (J–L), the wild type (M–O), a36-2 a39 (P–R), and a36-3 a39 (S–U). Arrows indicate the TUNEL-positive signal in the pollen grains. Bar = 20 μm. V to X, Mature pollen grains of the wild type (V), a36-2 a39 (W), and a36-3 a39 (X) incubated with H2DCF-DA staining. Green fluorescence intensity indicates the level of ROS. Bar = 50 μm. Y, Statistical analysis of TUNEL-positive signal ratios in the wild type, a36-2 a39, and a36-3 a39. Data were collected from three independent experiments (n = 300). Z, Relative levels of ROS measured by quantification of the fluorescence from each pollen grain. The values are averages of three measurements (n = 200). For Y and Z, error bars show se and asterisks indicate values that differ significantly from those of the wild type (***, P < 0.001, calculated using one-way ANOVA).
The expansion of the ER is a characteristic morphological feature of cells undergoing apoptosis-like PCD (Lodish, 2003). Terminal-deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) staining is a classic method for detecting apoptosis via the enzymatic incorporation of fluorescein-12-dUTP at the 3′-OH ends of fragmented DNA in situ. Thus, TUNEL assays were performed on the mature pollen released from open flowers in double mutant and wild-type plants (Fig. 5, G–U and Z). DNA fragmentation was detected using TUNEL assays with wild-type pollen grains in the absence of terminal-deoxynucleotidyl transferase as a negative control (Fig. 5, G–I) and following DNase I treatment as a positive control (Fig. 5, J–L). In the testing group, TUNEL signals were not detected in either sperm or vegetative nuclei in wild-type pollen grains (Fig. 5, M–O). However, in a36-2 a39 and a36-2 a39, nearly 35.8% and 34.1% of TUNEL-positive signals, respectively, were observed in sperm and vegetative nuclei at this stage (Fig. 5, P–U and Z). Reactive oxygen species (ROS) are involved in PCD progress (Lam, 2004; Gechev and Hille, 2005; Gechev et al., 2006; De Pinto et al., 2012). We detected the relative fluorescence intensity levels of intracellular ROS in the pollen grains via H2DCF-DA staining (Fig. 5, V–X). ROS relative fluorescence intensity level in a36 a39 was significantly higher than that in the wild type (Fig. 5Z).
To investigate whether the reduced pollen activity of a36 a39 is caused by gametophytic or sporophytic defects, we first tested the mature pollen viability of trans-heterozygous a36-2/− a39/+ plants by FDA and PI staining (Supplemental Fig. S15, A–H). We found that a36-2/− a39/+ displays reduced pollen grain activity in vitro, showing a significant difference from that of a36 a39 but no significant difference from that of the a36-2 single mutant (Supplemental Fig. S15B). Additionally, we also performed a TUNEL assay using the mature pollen from a36-2/− a39/+ plants (Supplemental Fig. S16, A–R). The ratio of apoptosis-like PCD that occurs in the pollen from a36-2/− a39/+ was reduced significantly compared with that of a36 a39, but there was no significant difference from that of the a36-2 single mutant (Supplemental Fig. S16S). Therefore, these data indicate that the pronounced defects in the microspore development of a36 a39 are caused by sporophytic defects.
Pollen Tubes in a36 a39 Plants Display Compromised Micropylar Guidance
That 33.8% inactive pollen ratio in the a36 a39 mutant (Fig. 4, B and D) could not explain why, when excess double mutant pollen was used to artificially pollinate wild-type-pistils, the abortion ratio of siliques was still markedly higher than that of wild-type plants (Fig. 3, E–G). Therefore, we observed pollen tube growth in vivo using Aniline Blue staining (Supplemental Fig. S17A). Wild-type and a36 a39 double mutant pollen tubes reached the bottom of the transmitting tract at 12 h after pollination (Supplemental Fig. S17A). The length of the pollen tube at 4, 8, 12, and 24 h after pollination was measured, but no defects in growth speed were detected for a36 a39 pollen tubes growing in vivo (Supplemental Fig. S17B).
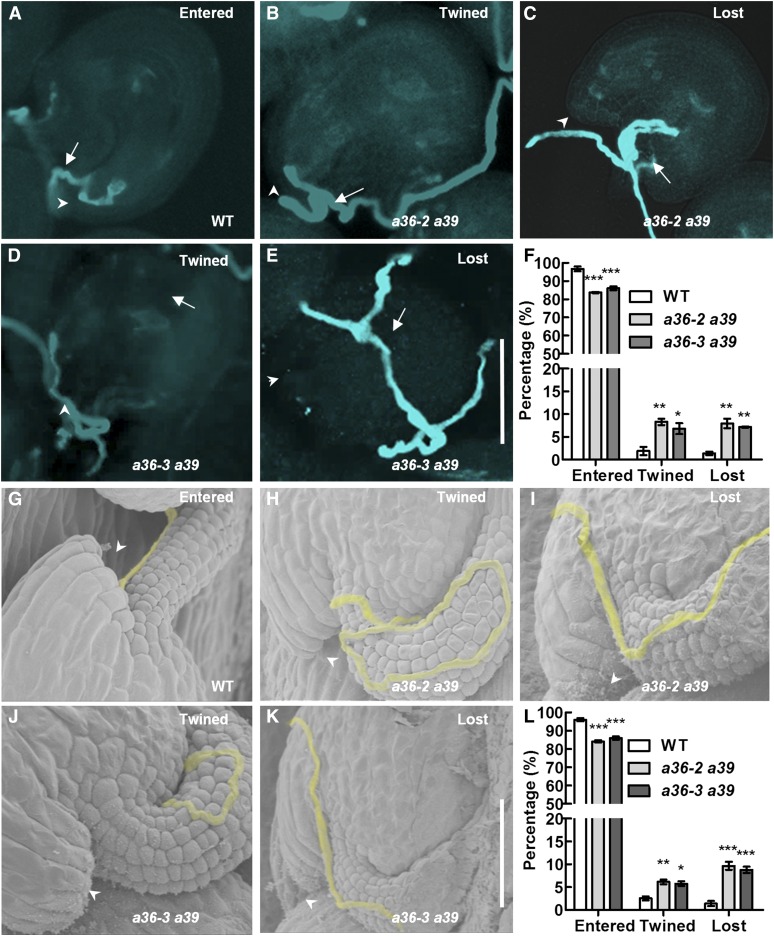
To determine whether pollen tube guidance was compromised, we tracked the path of pollen tube growth at 24 h after limited pollination using Aniline Blue staining. Compared with 96.7% (entered; n = 331; Fig. 6, A and F) in the wild type, about 84.1% (entered; n = 342; a36-2 a39; Fig. 6F) and 86.1% (entered; n = 311; a36-3 a39; Fig. 6F) of pollen tubes grew normally and entered the micropyles directly in a36-2 a39 and a36-3 a39 double mutants. About 15.9% (n = 342; a36-2 a39; Fig. 6, B, C, and F) and 13.9% (n = 311; a36-3 a39; Fig. 6, D–F) of pollen tubes showed abnormal guidance: twined or lost. In detail, 8.3% (twined; n = 342; a36-2 a39; Fig. 6, B and F) and 6.8% (twined; n = 311; a36-3 a39; Fig. 6, D and F) of pollen tubes were twisted around the micropyles; 7.6% (lost; n = 342; a36-2 a39; Fig. 6, C and F) and 7.1% (lost; n = 311; a36-3 a39; Fig. 6, E and F) of pollen tubes were growing on the ovule surface or twisting around the funicular surface, ultimately missing the micropyles.
Figure 6.
Defective navigation of a36 a39 mutant pollen tubes in a limited pollination assay. A to E, Pollen tubes were stained with Aniline Blue and visualized by fluorescence microscopy. A, Wild-type (WT) pistils were pollinated with a limited number of pollen grains from the wild type. Wild-type pollen tubes enter the micropyle directly. B to E, Wild-type pistils were pollinated with a limited number of pollen grains from a36-2 a39 and a36-3 a39 plants. The pollen tubes were twisted around the micropyle (B and D), were growing on the ovule surface, or were twisted around the funicular surface, ultimately missing the micropyle (C and E). Bar = 50 μm. Arrows indicate the pollen tubes, and arrowheads indicate the micropyle. F, Statistical analysis of pollen tubes with different situations in limited pollination after 24 h by Aniline Blue staining. G to K, SEM analysis 24 h after pollination with a limited number of pollen grains. Pollen tubes are highlighted in yellow. Arrowheads indicate the micropyle. G, The normal pollen tube enters the micropyle after growth along the funiculus. H and J, The pollen tube was twisted around the micropyle. I and K, The pollen tube was growing on the integument, ultimately missing the micropyle. L, Statistical analysis of the different behaviors in pollen tubes with limited pollination after 24 h by SEM. For F and L, values are based on three biological replicates, error bars show se, and asterisks indicate values that differ significantly from those of the wild type (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, calculated using two-way ANOVA).
Furthermore, SEM analysis was performed to detect pollen tube growth (Fig. 6, G–K). Wild-type pistils were pollinated with a limited number (n < 45) of pollen grains from wild-type or a36 a39 plants. In the wild type, 96.1% (entered; n = 301; Fig. 6, G and L) of pollen tubes grew normally and entered the micropyles directly, but only 84.2% of pollen tubes (entered; n = 310; Fig. 6L) and 85.9% (entered; n = 307; Fig. 6L) of pollen tubes from the a36-2 a39 and a36-3 a39 double mutants grew normally and entered the micropyles directly. By contrast, about 15.8% (n = 310; a36-2 a39; Fig. 6, H, I, and L) and 14.1% (n = 307; a36-3 a39; Fig. 6, J–L) of pollen tubes in double mutants showed defects in micropylar entry at 24 h after pollination. Among them, 6.2% (twined; n = 310; Fig. 6, H and L) in a36-2 a39 and 5.8% (twined; n = 307; Fig. 6, J and L) in a36-3 a39 pollen tubes twisted around the micropyles, and 9.6% (lost; n = 310; Fig. 6, I and L) in a36-2 a39 and 8.3% (lost; n = 307; Fig. 6, K and L) in a36-3 a39 pollen tubes missed the micropyles. Together, these results suggest that funicular guidance was normal, but micropylar guidance was significantly compromised, in a36 a39 double mutant pollen tubes.
To confirm that the pollen tubes of a36 a39 mutants have defective micropylar responses, we performed a semi in vivo pollen tube guidance assay (Supplemental Fig. S18). Pollen from wild-type and a36 a39 plants was hand pollinated in wild-type pistils. Then, we cut the styles from the pistils and placed them on agarose medium surrounded by excised ovules. At 4 h after pollination, pollen tubes emerged from the cut end of the wild-type styles and targeted the ovules. After being washed with water, over 45% ± 34.5% (sd; n = 120) of the ovules were targeted by a wild-type pollen tube (Supplemental Fig. S18, A, D, and G). Only 8.3% ± 14.4% and 9.7% ± 15.2% ovules were targeted by pollen tubes from a36-2 a39 (Supplemental Fig. S18, B, E, and H) and a36-3 a39 (Supplemental Fig. S18, C, F, and I), respectively. Therefore, the micropylar guidance is hampered in the pollen tube of a36 a39.
Subcellular Locations of A36 and A39
A36 and A39 are predicted GPI-anchored proteins that harbor both N-terminal signal peptides (SPs) and hydrophobic C-termini (Supplemental Fig. S1B). Thus, fusion tags at either terminal may affect protein processing and sorting. Therefore, we constructed pA36:SP:GFP:A36 and pA39:SP:GFP:A39, in which GFP was inserted behind the N-terminal signal peptide of the A36/A39, to study the subcellular localization of these proteins (Fig. 7A). These constructs were each transformed into a36-2 a39 and successfully complemented the a36-2 a39 mutant phenotype for both pollen germination and pollen tube guidance (Supplemental Fig. S19). These results indicated that both the A36-GFP and A39-GFP fusion proteins were functional in plants.
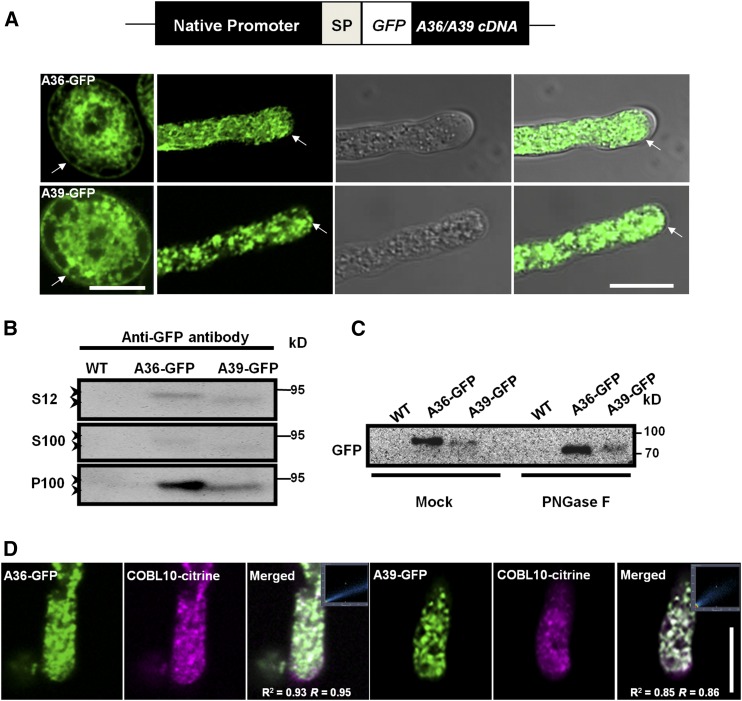
Figure 7.
Subcellular location analysis of A36-GFP and A39-GFP. A, The pA36::SP::GFP::A36cDNA and pA39::SP::GFP::A39cDNA constructs and the GFP (white box) gene insert in frame downstream of the A36 and A39 signal peptide sequences. Confocal images show A36-GFP and A39-GFP localization in an Arabidopsis pollen grain and pollen tube. Arrows point to the membranes of the pollen grains and pollen tubes. Bars = 10 μm. B, Protein western blot probed with anti-GFP antibody, showing that A36-GFP and A39-GFP proteins are detected in the total proteins (S12), microsomal fraction (P100), and soluble fraction (S100). A36-GFP and A39-GFP proteins are indicated on the left with arrowheads, and markers of molecular mass are indicated on the right. C, Microsomal membrane proteins were treated with PNGase F and buffer solution (mock) from the wild type (WT), and pA36::SP::GFP::A36cDNA/a36-2 a39 and pA39::SP::GFP::A39cDNA/a36-2 a39 transgenic plants were probed with the anti-GFP antibody. PNGase F removed N-glycans from A36-GFP and A39-GFP. D, Colocalization analyses of A36-GFP, A39-GFP, and COBL10-citrine were conducted in pollen tubes. These images are of individual slices. The images in the squares show colocalization analysis by the Pearson correlation coefficient and Manders’ overlap coefficient for A36-GFP/A39-GFP and COBL10-citrine using Zeiss software. For A36-GFP and COBL10-citrine, r2 = 0.93 and r = 0.95; for A39-GFP and COBL10-citrine, r2 = 0.85 and r = 0.86. Bar = 10 μm.
Consistent with the results revealed by GUS staining, the A36-GFP and A39-GFP signals were present mainly in the pollen grain and pollen tube. In pollen grains, A36-GFP and A39-GFP were detected mainly at the plasma membrane and reticular structures in the cytosol (Fig. 7A). In pollen tubes, A36-GFP and A39-GFP displayed punctate localization in the cytosol and also occurred weakly at the plasma membrane (Fig. 7A). We also extracted membrane-associated and soluble proteins from open flowers of pA36:SP:GFP:A36 and pA39:SP:GFP:A39 transgenic plants and explored subcellular localization by western blotting (Fig. 7B). Most A36-GPF and A39-GFP protein appeared in the microsome fraction (100,000g pellet [P100]), with less being detected in the soluble fraction (100,000g supernatant [S100]).
A36-GFP and A39-GFP migrated slower than expected on SDS-PAGE (A36-GFP is 95 kD instead of the predicted 79 kD, and A39-GFP is 95 kD instead of the predicted 78 kD). This effect may be due to glycosylation, because GPI-anchored proteins are reported to be glycosylated (Eisenhaber et al., 1998; Sedbrook et al., 2002; Cheung et al., 2014). Next, crude microsomal fraction extracts from open flowers were treated with peptide N-glycosidase F (PNGase F), and the mobility of A36-GFP and A39-GFP was measured again. As shown in Figure 7C, A36-GFP and A39-GFP in the PNGase F-treated sample migrated faster than in the mock-treated sample. These results suggest that A36-GFP and A39-GPF are glycosylated, membrane-associated proteins.
Previous studies showed that ABNORMAL POLLEN TUBE GUIDANCE1 (APTG1) encodes a putative mannosyltransferase that is implicated in the biosynthesis of GPI anchors (Dai et al., 2014). Therefore, we wondered whether the subcellular localization of A36-GFP was altered in the aptg1/+ mutant background. In pA36:SP:GFP:A36/a36-2 a39 aptg1/+ plants, A36-GFP lost its plasma membrane and cytoplasmic punctate localization and displayed a reticular localization (Supplemental Fig. S20, A–E).
Recent studies showed that COBL10 is a GPI-anchored protein that localizes to the apical plasma membrane and cytoplasm in growing pollen tubes (Li et al., 2013). Similar to the a36 a39 double mutant, the pollen tubes in the colb10 mutant also exhibit compromised guidance toward micropyles. To investigate whether COBL10 colocalizes with A36-GFP and A39-GFP, we crossed two transgenic plants expressing A36-GFP and A39-GFP with transgenic plants expressing COBL10-citrine. Then, colocalization analyses of A36-GFP, A39-GFP, and COBL10-citrine were conducted in the pollen tubes. As shown in Figure 7D, A36-GFP colocalized well with COBL10-citrine both at the plasma membrane and in the cytoplasm in the growing pollen tubes (Pearson’s correlation coefficient [r2] is 0.93 and Manders’ overlap coefficient [r] is 0.95). A39-GFP also showed colocalization with COBL10-citrine in the plasma membrane and cytoplasm (r2 is 0.85 and r is 0.86).
Highly Methylesterified Homogalacturonans and Xyloglucans Were Increased Significantly in the Tip Region of the Pollen Tube Wall in a36 a39
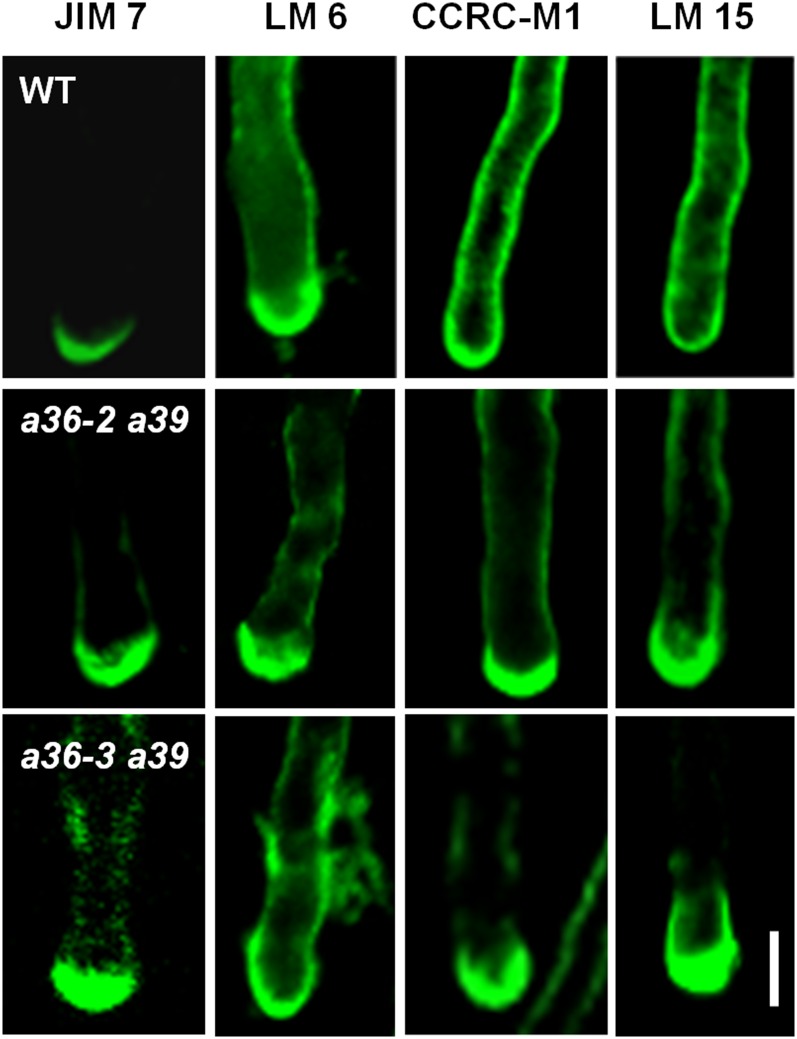
Many GPI-anchored proteins in plants, including COBL10, were reported to take part in cell wall organization or remodeling. Therefore, the pollen tube cell wall was examined by immunolabeling with specific cell wall-polysaccharide antibodies. First, the distribution of pectins was analyzed. JIM7 and JIM5 recognize highly and lowly methylesterified homogalacturonan (HG) in the cell wall, respectively. As shown in Figure 8, JIM7 staining was concentrated in the apical region of the wild-type pollen tube cell wall. A similar pattern was observed in a36 a39 double mutants, but the signal intensity was much stronger (Fig. 8). There was no significant difference between the wild type and a36 a39 when labeling with JIM5 (Supplemental Fig. S20, F and G). The wild type and a36 a39 double mutants also exhibited similar patterns when probing with LM6 (Fig. 8), which specifically recognizes rhamnogalacturonan I (RGI). Epitopes with LM6 were found mainly in the tip region of the pollen tube and less so in the shank. We then detected RGI with CCRC-M1 (Fig. 8), which labels RGI and fucosylated xyloglucans. CCRC-M1 staining was uniform along the cell wall of the growing pollen tubes in the wild type. However, in the a36 a39 double mutants, CCRC-M1 strongly labeled the apical wall, with weaker staining along the shank wall, implying that the distribution of fucosylated xyloglucans was altered significantly in the a26 a39 double mutants. Furthermore, we used LM15 to label the nongalactosylated xyloglucans (Fig. 8). In the wild-type pollen tubes, LM15 uniformly labeled the whole pollen tube wall, whereas in the a36 a39 double mutants, as with the LM15 labeling, brighter fluorescence was observed in the tip region of the pollen tube, with weaker staining along the shank wall. Moreover, crystalline cellulose was labeled using cellulose-binding module 3a (Supplemental Fig. S20, H and I); however, no obvious difference was observed. These data indicate that the abundances of highly methylesterified HG, fucosylated xyloglucans, and nongalactosylated xyloglucans were increased significantly in a36 a39 double mutants in the apical cell wall of the pollen tube.
Figure 8.
Immunolabeling of pollen tube wall polysaccharides in wild-type (WT) and a36 a39 plants. Fluorescence micrographs show median optical sections of wild-type, a36-2 a39, and a36-3 a39 tubes by individual slices. Immunofluorescence labeling was performed using JIM7 for highly methylesterified HG, LM6 for RGI, CCRC-M1 for RGI and fucosylated xyloglucans, and LM15 for nongalactosylated xyloglucans. Representative pollen tubes of three replicate experiments (n = 200) are shown for each labeling. Bar = 5 μm.
DISCUSSION
GPI modification is an important posttranslational modification in eukaryotes (Maeda and Kinoshita, 2011; Cheung et al., 2014). GPI-anchored proteins play roles in cell recognition and adhesion, signal transduction, and cell-surface enzymatic reactions (Rudd et al., 1997; Ahmed et al., 1999; Hwa, 2001). Among the 69 aspartic proteases in Arabidopsis, four are predicted to be GPI-anchored proteins (Lalanne et al., 2004), but none of them have been characterized. In this study, we identified and characterized two putative GPI-anchored aspartic protease genes, A36 and A39, which are highly expressed in pollen and the pollen tube (Fig. 1; Supplemental Fig. S2). A36 and A39 exhibit aspartic proteolytic activity in vitro (Fig. 1X). The pollen of a36 a39 undergoes unexpected apoptosis-like PCD, and the female gametophyte also experiences degradation (Figs. 4A and 5, A–U). Moreover, the pollen tube guidance is compromised toward the micropylar (Fig. 6), and the distribution of the pollen tube cell wall components is altered in the apical pollen tube of the a36 a39 plant (Fig. 8). Thus, A36 and A39 play important roles in reproduction in Arabidopsis.
A36 and A39 Play Vital Roles in Pollen and Ovule Development
Many aspartic proteases in plants have been reported to be involved in sexual reproduction (Ge et al., 2005; Chen et al., 2008; Yang et al., 2012; Huang et al., 2013; Niu et al., 2013). In this study, RT-PCR and GUS staining assays revealed that both A36 and A39 are specifically expressed in pollen grains and in growing pollen tubes (Fig. 1, A–W ; Supplemental Fig. S2). The male transmission efficiency is reduced significantly in the a36 single mutant (Table I) and in a36 a39 double mutants (Table II). Then, we found that the pollen grain activity and pollen germination ratio in a36 are reduced significantly (Fig. 2, A–D). While the a39 single mutant had normal growth and fertility (Fig. 2, A–D), in the a36 a39 double mutant the ratio of dead pollen grains was increased significantly (Fig. 4, B and D), although these pollen grains still displayed normal external morphology and nuclei (Supplemental Fig. S13). These observations imply that the death of pollen occurred at the tricellular pollen stage. TEM and TUNEL staining further revealed that a portion of mature pollen in a36 a39 experienced unexpected apoptosis-like PCD (Fig. 5, A–U and Y). Besides, ROS commonly serve as a signal to induce plant PCD, which plays important roles in environmental responses and reproductive development (Cominelli et al., 2013; Rosenzweig et al., 2014; Xie et al., 2014). Higher ROS accumulation was detected in the pollen grains of a36 a39 mutants compared with wild-type plants (Fig. 5, V–X and Z).
To our surprise, the decreased pollen viability in a36 is caused by sporophytic defects upon examining the phenotype of a36-2/+ (Supplemental Fig. S6). The reduced pollen vitality in a36 a39 also is controlled by the sporophyte, as revealed by FDA staining, PI staining, and the TUNEL assay in trans-heterozygous a36-2/− a39/+ (Supplemental Figs. S15 and S16). These results are beyond our speculation, because A36 and A39 are highly expressed in the pollen and pollen tube (Fig. 1, B–W) and the male transmission efficiency is reduced significantly in a36 and a36 a39 (Tables I and II). In flowering plants, though, the successful development of pollen grains is not only dependent on the microsporocyte or the microspore itself, it also requires the surrounding anther tissues. We observe that A39, not A36, is expressed in the anther tissue in the GUS staining assay (Supplemental Fig. S2L). The possibility that A36 also may be expressed in the anther tissue could not be excluded due to the sensitivity limits of the GUS staining and RT-PCR assay.
In addition, A39 but not A36 shows faint expression in the development of ovules in the GUS staining assay (Supplemental Fig. S2, P and Q). No obvious defect of the ovules was found in the a36 or a39 single mutant (Supplemental Fig. S11, B–E), but collapsed embryo sacs were clearly observed in a36 a39 (Supplemental Fig. S11F). By observing the ovules of the trans-heterozygous a36-2/− a39/+ or a36-2/+ a39/− and genetic crosses, we found that the defective female gametophyte development in a36 a39 also is caused by sporophytic defects (Supplemental Fig. S12, A–C). Therefore, A36 and A39 are important for ovule development in Arabidopsis.
Among the aspartic proteases reported to date in Arabidopsis, the phenotype of pcs1 shares some similarity with that of a36 a39. The loss of function of aspartic protease PCS1 also causes the degeneration of male gametophytes (Ge et al., 2005). The mutation in PCS1 also impaired embryonic development, which results in no homozygous mutants (Ge et al., 2005). In addition, ectopic expression of PCS1 causes a failure in anther dehiscence by preventing the normal PCD in specified cells (Ge et al., 2005). Unlike putative GPI-anchored A36 and A39, PCS1 is an ER-localized protein. However, PCS1, A36, and A39 all play a negative role in cell death. In addition, UNDEAD also is a negative regulator of cell death (Phan et al., 2011). The knocking down of UNDEAD results in premature tapetal PCD and leads to the abortion of microspores in Arabidopsis (Phan et al., 2011). How do these aspartic proteases function at the molecular level? PCS1 is supposed to process and activate unknown survival factors. We propose that the putative GPI-anchored A36 and A39 may be involved in the proteolysis and activation of extracellular survival signals in anther and ovule to sustain successful gametophyte development.
However, most reported plant aspartic proteases induce PCD, such as NtCDN41, nucellin, OsS5, AtCDR1, OsAP25/37, and APCB1 (Chen and Foolad, 1997; Xia et al., 2004; Kato et al., 2005; Chen et al., 2008; Niu et al., 2013; Li et al., 2016). These findings suggest that plant aspartic proteases have different, even contrary, functions in PCD, which may be due to the specific substrates of these aspartic proteases in different tissues or cell types. To date, only the substrate of APCB1 has been found, and APCB1-mediated BAG processing is required for autophagy (Li et al., 2016). Therefore, future efforts to identify the physiological substrates of A36 and A39 during microspore development will shed light on the molecular mechanisms that block cell death or promote cell survival in plants.
A36 and A39 Are Implicated in the Micropyle Guidance of the Pollen Tube
Cell surface factors in the pollen tube are thought to directly sense female signals to regulate pollen tube competency (Higashiyama and Takeuchi, 2015). Several crucial male factors located on the cell surface have been identified to be involved in pollen tube guidance. Two receptor-like kinases, LOST IN POLLEN TUBE GUIDANCE1 (AtLIP1) and AtLIP2 (Liu et al., 2013), which localize to the plasma membrane of the pollen tube, are essential for micropylar guidance and sensing the female attractant AtLURE1. More recently, Wang et al. (2016) found that Leu-rich repeat receptor-like kinase MDIS1-MIK heteromers directly perceive the AtLURE1 signal, and Arabidopsis sister species transformed with AtMDIS1 could more efficiently target the Arabidopsis ovules.
Distinct from transmembrane motifs, GPI anchors provide an additional way to target proteins to the outer leaflet of the plasma membrane. COBL10, a GPI-anchored protein, plays an important role in pollen tube growth and guidance (Li et al., 2013). In the plasma membrane of the pollen tube, A36-GFP and A39-GFP show good colocalization with COBL10-citrine (Fig. 7D). In a36 a39, a fraction of the pollen tubes fail to target the female gametophyte (ovule). According to Aniline Blue staining, SEM, and semi in vivo assays, pollen tubes in a36 a39 mutants show significant defects in micropylar guidance (Fig. 6), which are similar to the phenotype of cobl10 mutants. However, in a36 a39 double mutants, the growth rate of the pollen tube was comparable to that of the wild type (Supplemental Fig. S17), which differs from the compromised pollen tube growth seen in cobl10, suggesting different roles for these proteins. GPI-anchored proteins are thought to accumulate in lipid rafts, which serve as a platform for protein trafficking and cell signaling (Maeda and Kinoshita, 2011). Hence, A36/A39 may localize to lipid rafts to decipher cues from female gametophytes by hydrolyzing and activating unknown peptides. In addition, transmembrane proteins are considered to be candidate partners of GPI-anchored proteins in mediating extracellular-to-intracellular signals. Recently, the GPI-anchored proteins LORELEI (LRE) and LRE-like GPI-AP1 were shown to function as chaperones and coreceptors for the receptor kinase FERONIA in mediating signal transduction (Li et al., 2015). Thus, whether A36 and A39 have some relationship with reported receptor-like kinases such as AtLIP1/2 and MDIS1-MIK requires further investigation.
In S. cerevisiae, the Yapsin family contains five GPI-anchored aspartic proteases, which are required for cell wall integrity via glucan homeostasis (Krysan et al., 2005). In Candida glabrata, 11 GPI-anchored aspartic proteases are crucial for cell wall remodeling (Kaur et al., 2007; Bairwa et al., 2014). Moreover, many GPI-anchored proteins in plants also have been implicated in cell wall integrity (Sedbrook et al., 2002; Shi et al., 2003; Gillmor et al., 2005). The abundances of JIM7-labeled highly methylesterified HG, fucosylated xyloglucans, and nongalactosylated xyloglucans are increased markedly in a36 a39 (Fig. 8). In contrast, the pollen tube cell wall of cobl10 displays decreased highly methylesterified HG labeling by JIM7 and cellulose observed by TEM.
During pollen germination, highly methylesterified HGs are delivered to the elongating cell wall at the apex by exocytosis. Simultaneously, pectin methylesterase deesterifies the highly methylesterified HGs to form the low methylesterified HGs. Calcium causes the low methylesterified HGs into a stiffer material to prevent any further expansion behind the tip region. The degree of pectin methylesterification from high to low methylesterified pectin is essential to help the pollen tube complete the highly polarized growth (Bosch and Hepler, 2005; Palin and Geitmann, 2012). On the other side, xyloglucans are thought to play a major role in the cross link with cellulose microfibrils and also can be covalently linked to pectin (Doblin et al., 2001; Cosgrove, 2005; Cavalier et al., 2008; Dardelle et al., 2010; Chebli et al., 2012; Xiao et al., 2016). The fucosylated xyloglucans were not detected in some regions of pollen tubes, where the tubes seem to change direction or diameter in pollen tube growth (Chebli et al., 2012). Therefore, xyloglucans are considered to be essential for pollen tube growth and interaction with female tissues (Dardelle et al., 2015). In a36 a39, the increased levels of highly methylesterified HGs and xyloglucans may have no significant influence on pollen tube growth in the transmitting tract, because the growth rate of the pollen tube is comparable to that of wild-type plants. But this alteration in the pollen tube cell wall may dramatically impair the tight control of pollen tube guidance toward the micropyle in a36 a39.
In conclusion, this work uncovers two previously unknown putative GPI-anchored aspartic proteases, A36 and A39, which are expressed preferentially in the pollen and the pollen tube and play important roles in pollen and ovule development as well as pollen tube guidance in Arabidopsis. These findings highlight the importance of aspartic proteases in antagonizing PCD and in precisely controlling the pollen tube guidance toward the micropyle of the ovules.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines in background ecotype Columbia-0 were supplied by the ABRC (http://www.arabidopsis.org/) at Ohio State University. Seeds were sterilized (1 min in 75% [v/v] ethanol, two rinses in sterile water, 12 min in 2.5% bleach, and four rinses in sterile water), planted on growth medium (Murashige and Skoog basal salt mixture, 4.33 g L−1 supplied with 0.75% agar; purchased from PhytoTechnology Laboratories), and then cold treated for 3 to 5 d. The medium plates were then transferred to a growth chamber (Percival) at 22°C and 50% humidity under a long photoperiod (16 h of light/8 h of dark) for approximately 14 d prior to planting in soil.
Phylogenic and Sequence Analyses
Using MEGA6.06 software (http://www.megasoftware.net/; Tamura et al., 2013), a rectangular phylogenetic tree was constructed with full-length sequences by the neighbor-joining method with 1,000 replicates for bootstrap and default settings for other options. Potential domains and motifs were by SMART (http://smart.embl-heidelberg.de/; Schultz et al., 1998; Letunic et al., 2015) and CBS Prediction Server (http://www.cbs.dtu.dk/services/; Blom et al., 1999), respectively.
Alexander’s, FDA, DAPI, and H2DCF-DA Staining
Pollen was photographed with a Zeiss digital camera. Flower and plant images were taken with a Leica dissection microscope. Alexander solution (Alexander, 1969) and pollen viability were determined using FDA (Heslop-Harrison and Heslop-Harrison, 1970), and DAPI staining was performed as described (Ross et al., 1996). For H2DCF-DA staining, the pollen was vacuum infiltrated with 5 μm H2DCF-DA staining solution for 10 min and then incubated at 25°C for 2 h (Xie et al., 2014). Fluorescence imaging of the H2DCF-DA-stained pollen was performed with a Zeiss LSM-710 confocal microscope at the same parameters without reaching signal saturation in any sample for comparative quantification. H2DCF-DA was excited at 488 nm, and emissions were collected at 493 to 634 nm. These images were analyzed by Zeiss software for fluorescence intensity, and the number of pollen grains was calculated in each image. The fluorescence intensity of each pollen grain = the fluorescence intensity of each image/the number of total pollen grains in each image. The calculation of relative fluorescence intensity is based on the fluorescence intensity of the wild-type pollen as 1.
SEM, Cross Section, and TEM
For SEM, pollen grains from newly opened flowers were dipped on stubs over carbon double-sided tape (Nisshin) and coated with gold particles using a sputter coater. Specimens were observed with an S-4800 field emission scanning electron microscope (Hitachi).
For cross section, wild-type and a36 a39 double mutant inflorescences were fixed in 2.5% (v/v) glutaraldehyde in 0.1 m phosphate-buffered saline (PBS; pH 7.4), embedded in Spurr’s resin as described (Sanders et al., 1999), and sectioned (2–3 μm) with a microtome (Ultratome V; LKB). Anther sections were stained in 1% Toluidine Blue. Bright-field photographs of the cross sections were taken using a Zeiss microscope.
For pollen grain ultrastructural analysis, pollen grains from opened flowers were fixed in 4% (v/v) glutaraldehyde in 0.1 m PBS (pH 7.4) on ice, rinsed in 0.1 m PBS (pH 7.4), and postfixed in 1% OsO4 (dissolved in 0.1 m PBS, pH 7.4), then embedded in Spurr’s resin as described for the cross-section procedure. Ultrathin sections (80 nm) were cut using a diamond knife on a Leica Ultracut ultramicrotome. Sections were double stained with saturated uranyl acetate and lead citrate and examined with a transmission electron microscope (H-7650; Hitachi).
To observe pollen tube growth in pistils, pollinated pistils were fixed with 3% (v/v) glutaraldehyde in 0.1 m PBS (pH 7) as described (Dai et al., 2014). Then, the dry pistils were observed with the S-4800 field emission scanning electron microscope (Hitachi) and photographed.
GUS Staining
Tissues of pA36::GUS and pA39::GUS plants were stained in a solution of 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-GlcA (Goldbo Com), 10 mm EDTA·2Na, 0.5 mm K3FeC6N6, 0.5 mm K4Fe(CN)6·3H2O, 50 mm NaH2PO4, 50 mm Na2HPO4, and 0.1% (v/v) Triton X-100 under 37°C after treating in 90% precooling acetone. After GUS staining, chlorophyll was removed using absolute ethanol, and the tissues were treated with 2% (v/v) HCl in 20% (v/v) methanol and 7% (w/v) NaOH in 60% (v/v) ethanol (Yang et al., 1999).
Construction and Expression of the A36 and A39 Fusion Protein in Escherichia coli
The full-length coding sequences of A36 and A39 were amplified by PCR and subcloned into the pGEX-4T-1 expression vector to generate pGEX-A36 and pGEX-A39, in which A36 and A39 are fused at the C terminus of GST, respectively. The partial coding sequences of A36 and A39 were amplified by PCR and subcloned into the pGEX-4T-1 expression vector to generate pGEX-A36-D and pGEX-A39-D, in which A36-D and A39-D are fused at the C terminus of GST, respectively. For the construction of A36 and A36-D site-directed mutants, pGEX-A36 and pGEX-A36-D were used as templates for PCR. For D96N and D310N in A36 and A36-D, in which the Asp residues at positions 96 and 310 were replaced by Asn, PCR was carried out using the forward primer 5′-GTTCAAGTTAATACAGGAAGTGATATACTTTG-3′ and the reverse primer 5′-CAAAGTATATCACTTCCTGTATTAACTTGAAC-3′ (D96N) and the forward primer 5′-GAGGAACGATAATCAATAGTGGTACAACTT-3′ and the reverse primer 5′-AAGTTGTACCACTATTGATTATCGTTCCTC-3′ (D310N), respectively. For the construction of A39 and A39-D site-directed mutants, pGEX-A39 and pGEX-A39-D were used as templates for PCR. For D92N, in which the Asp at position 92 was replaced by Asn, A39 mutation was carried out using the forward primer 5′-TCATGTACAAGTTAATACAGGAAGTGATATAC-3′ and the reverse primer 5′-GTATATCACTTCCTGTATTAACTTGTACATGA-3′ (D92N). The position of mutagenesis was the GAT codon for Asp being replaced by the AAT codon for Asn. The PCR products were digested by DpnI (New England Biolabs; R0176S) at 37°C for 2 to 3 h, the products were introduced into DH5α for plasmid amplification, and then these plasmids were introduced into BL21. When E. coli strain BL21 cells carrying the expression construct grew to an optical density at 600 nm of 0.6, 0.1 mm (final concentration) isopropyl-β-d-thiogalactopyranoside was added to induce expression, with further incubation at 16°C for 9 h (Xia et al., 2004). Purification of the GST fusion proteins was performed on a glutathione-Sepharose 4B column according to the manufacturer’s instructions. All proteins were electrophoresed on SDS-PAGE gels, and different contents (200, 400, 800, 1,000, and 2,000 ng) of BSA and the corresponding mean Gray values were used to prepare a standard curve to measure the concentrations of purified full-length GST fusion proteins. The standard curve was calculated by Linear. The quantitative analysis of the purified protein was performed with the software Image Lab 5.2.1.
Protease Activity Assays
Protease activity assays were performed according to the product information (FITC-casein; Sigma; C3777) with slight modifications. Twenty microliters of 0.5% (w/v) FITC-casein substrate solution was added to 20 μL of 20 mm sodium phosphate buffer with 150 mm sodium chloride (pH 5.4 at 22°C). The mixture above was then mixed and equilibrated to 22°C before adding 150 μL of 2.5 μg mL−1 (total, 375 ng) purified GST-A36, GST-A36-D, GST-A36 D96N D310N, GST-A36-D D96N D310N, GST-A39, GST-A39-D, GST-A39 D92N, and GST-A39-D D92N and the positive control pork pepsin (Sigma; P7125). In the pepstatin A treatment groups, equal amounts of the above proteins were treated with 5 μm (final concentration) pepstatin A for 15 min at 37°C and then incubated at 22°C for 120 min. Then, 150 μL of 20% (v/v) TCA solution (Sigma; T6399) was added and incubated at 22°C for 120 min to terminate the reaction. Centrifugation was performed for 15 min, and 100 μL of the supernatants was added into 5 mL of 500 mm Tris-HCl solution (pH 8.5 at 22°C). The solutions were then transferred to suitable cuvettes. The fluorescence intensity was recorded at the excitation wavelength of 490 nm and the emission wavelength of 525 nm for all the containers using a suitable fluorometer at 25°C (model F-2500; Hitachi). The conversion of relative protease activity (%) was calculated based on the positive control (pork pepsin) as 100%.
Pollen Germination in Vitro and in Vivo
Pollen germination in vitro was performed as described elsewhere (Boavida and McCormick, 2007). The Aniline Blue staining of the pollen tubes in vivo was conducted as described elsewhere (Ishiguro et al., 2001). The pollinated pistils were collected at different time points and fixed in Carnoy’s fluid (ethyl alcohol:acetic acid [v/v], 3:1) for 2 h followed by three washes in 0.1 m PBS buffer for 20 min. Then, 8 m sodium hydroxide was added for 4 h, followed by three washes in 0.1 m PBS buffer for 20 min. Finally, the pollinated pistils were stained with Aniline Blue (Ishiguro et al., 2001) for 3 h. The stained pistils were observed and photographed using a Zeiss LSM-710 confocal microscope. The Aniline Blue staining was excited at 405 nm, and the emissions were collected at 410 to 585 nm.
Protein Extraction, Western-Blot, and Glycosylation Analyses
Flowers from wild-type and A36::SP::GFP::A36cDNA/a36-2 a39 and A39::SP::GFP::A39cDNA/a36-2 a39 transgenic plants were collected and ground in liquid nitrogen to a fine and smooth powder and then ground in an extraction buffer containing 20 mm Tris-HCl, pH 8.8, 150 mm NaCl, 1 mm EDTA, 20% glycerol, 20 mm NaF, 50 nm microcystin, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail (Roche). The mixture was centrifuged at 1,000g for 25 min at 4°C, and the supernatant (S1) was filtered through four layers of Miracloth (Calbiochem). The filtered supernatant was centrifuged at 12,000g for 30 min, and then the supernatant (S12) was transferred to a new ultracentrifuge tube and subsequently centrifuged at 100,000g for 2 h at 4°C. The supernatant (S100) was collected, and the pellet (P100) was rinsed several times with cold extraction buffer. The membrane protein was resuspended in 1 mL of resuspension buffer (10 mm Tris-HCl, pH 7.3, 150 mm NaCl, 1 mm EDTA, and 10% glycerol) by pipetting. Triton X-100 was added to a final concentration of 0.5% to solubilize the membrane protein. All the samples were mixed with SDS-PAGE loading buffer and boiled before gel fractionation (Li et al., 2011). Western blotting was performed using anti-GFP antibody (TongBio Pacific). For glycosylation analysis, crude flower microsomal fraction extracts from wild-type and A36::SP::GFP::A36cDNA/a36-2 a39 and A39::SP::GFP::A39cDNA/a36-2 a39 transgenic plants were digested by PNGase F (New England Biolabs; 0704S) according to the manufacturer’s instructions.
TUNEL Assay
Pollen from opened flowers was collected and fixed in 4% (w/v) paraformaldehyde in 0.1 m PBS (pH 7.4) for 30 min and then washed with 0.1 m PBS (pH 7.4) at room temperature, and TUNEL was performed using the Dead End Fluorometric TUNEL system (Promega; G3250) according to the manufacturer’s instructions. Samples were analyzed with an inverted fluorescence microscope (Zeiss). The fluorescent filter was set to view the green fluorescence of fluorescein at 500 to 540 nm and the magenta filter was set to view the fluorescence of DAPI at 460 nm. When there was a TUNEL-positive signal in the cell nucleus, the pollen was counted as a TUNEL-positive pollen grain. The TUNEL-positive signal ratio was calculated as follows: TUNEL-positive signal ratio = the number of pollen grains with TUNEL-positive signals/the total number of pollen grains.
Genetic Methods
Transgenic plants were generated via Agrobacterium tumefaciens (strain GV3101)-mediated flower dip transformation (Clough and Bent, 1998) and selected on Murashige and Skoog solid medium containing 25 mg L−1 hygromycin B (Calbiochem) for segregation screening.
For plant genotyping, leaf genomic DNA was extracted via a rapid preparation method as described with some modifications (Edwards et al., 1991). PCR genotyping was performed using three primers, one primer annealing to the T-DNA insertion site (LBb1.3/LB2) and a pair of primers designed to amplify the fragment of DNA spanning the insertion site, which were obtained from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/). Genetic crosses were conducted by applying pollen from newly dehiscing flowers onto the stigmas of flowers that had been surgically emasculated 1 d before just prior to dehiscence.
Total RNAs were extracted using RNAiso Plus (TaKaRa) as described by the supplier. The first-strand cDNAs were synthesized from 1 μg of total RNA by the PrimeScript RT Reagent Kit (TaKaRa) according to the manufacturer’s instructions.
CLSM and Confocal Microscopy
All confocal images were captured using a Zeiss 710 confocal laser scanning microscope with a 40× or 63× water-immersion objective.
For subcellular location of A36-GFP and A39-GFP in transgenic plants, pollen grains were immersed in distilled, deionized water, and pollen tubes were immersed in pollen germination buffer and captured on a confocal microscope. A36-GFP and A39-GFP were excited at 488 nm, and emissions were collected at 505 to 530 nm.
For colocalization, COBL10-citrine was crossed with rescue lines expressing A36-GFP and A39-GFP. Then, the progeny pollen tubes were analyzed for colocalization to image the coexpression of GFP and yellow fluorescent protein, and settings matched the previous report to avoid channel cross talk (Brandizzi et al., 2002). All collected images were of individual slices. Colocalization was analyzed by Pearson’s correlation coefficient and Manders’ overlap coefficient using Zeiss software.
Immunofluorescence Labeling of Pollen Tubes
Pollen tube growth in vitro was examined as described. Immunofluorescence labeling was performed with JIM7, JIM5, CCRC-M1, CBM3a, and LM15 as well as LM6 as described (Dardelle et al., 2010; Chebli et al., 2012). For fluorescence labeling, the pollen tubes were fixed in 4% (w/v) freshly prepared paraformaldehyde in 0.1 m PBS (pH 7.4) for 15 min, followed by three washes in 0.1 m PBS buffer for 5 min, and then blocked with 1% (w/v) BSA in 0.1 m PBS (pH 7.4) for 30 min, followed by three washes in 1% (w/v) BSA in 0.1 m PBS (pH 7.4) for 5 min. All antibodies were diluted in 0.1 m PBS (pH 7.4) buffer with 0.1% (w/v) BSA, and incubations were done overnight at 4°C or for 2 h at 37°C (covered with Parafilm for each slide and placed in a wet box with filter paper soaked in 0.1 m PBS [pH 7.4] with 0.1% [w/v] BSA). The slides should be balanced for several minutes at room temperature before the next step and then washed in 0.1 m PBS (pH 7.4) buffer with 0.1% (w/v) BSA three times for 10 min, incubated with a secondary antibody (Alexa Fluor 594 anti-mouse IgG; 1:100; Invitrogen) in 0.1 m PBS (pH 7.4) buffer with 0.1% (w/v) BSA for 2 h at 37°C (covered with Parafilm for each slide and placed in a wet box with filter paper soaked in 0.1 m PBS [pH 7.4] buffer with 0.1% [w/v] BSA), and again washed in 0.1 m PBS (pH 7.4) with 0.1% (w/v) BSA three times for 10 min. Finally, the slide was covered with a cover slip and polyvinyl alcohol mounting medium with DABCO antifading agent (Sigma; 10981-100 ML) and stored at 4°C until being observed. Using a Zeiss LSM-710 confocal microscope, the cells were excited at 488 nm, and emissions were collected at 493 to 634 nm. For JIM5, JIM7, CCRC-M1 (diluted 1:20; Complex Carbohydrate Research Center, University of Georgia), and CBM3a (diluted 1:20; Plant Probes), they were followed by a monoclonal mouse anti-poly-His antibody (diluted 1:20; Sigma). Subsequently, all were incubated with the secondary antibody Alexa Fluor 594 anti-mouse IgG (diluted 1:100; Molecular Probes). For LM6 and LM15 (diluted 1:20; Plant Probes; LM6-050 and LM15-050), they were incubated subsequently with secondary antibody (Alexa Fluor 594 anti-rat IgG; 1:100; Invitrogen) in 0.1 m PBS (pH 7.4) with 0.1% (w/v) BSA.
Molecular Manipulation
For expression pattern analysis using GUS staining, a genomic DNA fragment containing the nucleotide sequence from 2,190 to 2,149 bp upstream of the A36/A39 start codon with a 60-/180-bp open reading frame was amplified by PCR from the genomic DNA of Arabidopsis ecotype Columbia-0 plants and cloned into pCAMBIA1300-Pro-35S:GFPXB-4 binary vector using PstI/SmaI to generate ProA36:GUS and ProA39:GUS, which hold the GUS coding sequence behind the promoter. To generate the A36 complementation construct, the A36 promoter obtained as described above was inserted into the pCAMBIA1300-Pro35S:TerNOS binary vector by PstI/SacI to take the place of the 35S promoter, and then three fragments of A36 genomic DNA, which were amplified by PCR from the genomic DNA of Arabidopsis ecotype Columbia-0 plants using specific primers, were cloned into the plasmid at the same time by the combination of SacI/BamHI and BamHI/KpnI.
For subcellular localization, to generate pA36:SP:GFP:A36, the A36 promoter obtained as described above was cloned into the pCAMBIA1300-Pro35S:TerNOS binary vector by PstI/BamHI, eGFP with SacI not a stop codon was amplified by PCR from pCAMBIA1300-Pro-35S:GFPBS-2 and placed in frame within SacI sites in the A36 gene just downstream of the nucleotide sequences and inserted into the full-length cDNA of A36 that was amplified by PCR and subcloned into pEASY-Blunt Simple Cloning vector by BamHI/KpnI, and then the full-length cDNA of A36 with eGFP was cloned into pCAMBIA1300-ProA36:TerNOS by BamHI/KpnI. To generate pA39:SP:GFP:A39, the A39 promoter obtained as described above was cloned into the pCAMBIA1300-Pro-35S:TerNOS binary vector by PstI/XhoI, eGFP with XhoI not a stop codon was amplified by PCR from pCAMBIA1300-Pro-35S:GFPBS-2, and then the full-length cDNA of A39 with eGFP was cloned into pCAMBIA1300-ProA36:TerNOS by BamHI/KpnI.
Primers Used in This Study
Primers used in this study are listed in Supplemental Tables S1 and S2.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: A36 (At5g36260), A39 (At1g65240), APTG1 (At5g14850), COBL10 (At3g20580), and ACTIN7 (At5g098103).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Evolutionary relationships of A36 and A39 amino acid sequences from Drosophila melanogaster, Homo sapiens, Caenorhabditis elegans, S. cerevisiae, Chlamydomonas reinhardtii, Arabidopsis, Brachypodium distachyon, Zea mays, and Oryza sativa.
Supplemental Figure S2. Expression patterns of A36 and A39 by GUS staining.
Supplemental Figure S3. Coomassie Brilliant Blue staining of purified GST fusion proteins by SDS-PAGE and BSA standard curve.
Supplemental Figure S4. Phenotypes of the a36 and a39 single mutants.
Supplemental Figure S5. SEM observation and staining of pollen grains from a36 and a39 mutants.
Supplemental Figure S6. a36-2/+ displays slightly reduced pollen grain activity and pollen germination ratio in vitro.
Supplemental Figure S7. Pollen tube growth of a36 and a39 mutants is normal in wild-type transmitting tract.
Supplemental Figure S8. Ovules with a mature seven-celled embryo sac at FG7 observed by DIC and CLSM.
Supplemental Figure S9. Pollen germination ratio and pollen grain viability were recovered in the complemented a36-2 mutant.
Supplemental Figure S10. Early embryo development in the wild type and a36-2 a39.
Supplemental Figure S11. CLSM analysis of ovules at FG7 in wild-type, a36, a39, and trans-heterozygous plants.
Supplemental Figure S12. Crosses among wild-type, a36-2/− a39/+, and a36 a39 plants.
Supplemental Figure S13. Characterization of pollen grains in the a36 a39 mutant.
Supplemental Figure S14. Anther development of transverse sections in the wild type and the a36 a39 double mutant.
Supplemental Figure S15. a36-2/− a39/+ displays reduced pollen grain activity in vitro.
Supplemental Figure S16. TUNEL assay of mature pollen grains from the wild type, a36-2, a36-2/− a39/+, and a36-2 a39.
Supplemental Figure S17. Pollen tube growth of a36 a39 mutants is normal in wild-type transmitting tract.
Supplemental Figure S18. Compromised directional growth of a36 a39 pollen tubes in semi in vivo pollination assay.
Supplemental Figure S19. The fragment of A36 cDNA fused with GFP (pA36:SP:GFP:A36cDNA) and A39 cDNA fused with GFP (pA39:SP:GFP:A39cDNA) could rescue the phenotype of a36-2 a39.
Supplemental Figure S20. Localization of A36-GFP in pollen tubes of the aptg1/+ mutant and immunolabeling of cell wall polysaccharides in pollen tubes of a36-2 a39.
Supplemental Table S1. Primers for the identification of T-DNA insertion mutants.
Supplemental Table S2. Primers used for constructs.
Supplementary Material
Acknowledgments
We thank Hongju Li (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for the critical review for this article and suggestions; Xiansheng Zhang (Shandong Agricultural University) for generously providing the aptg1/+ seeds, Yan Zhang (Shandong Agricultural University) for the pCOBL10:SP:citrine:COBL10 seeds, and Yihua Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the JIM5 antibody; Wenqiang Tang and Dr. Xiaorui Yang (Hebei Normal University) for purifying the GST fusion proteins; and the ABRC for providing the seeds of Arabidopsis T-DNA insertion mutants.
Glossary
- PCD
programmed cell death
- ER
endoplasmic reticulum
- GPI
glycosylphosphatidylinositol
- RT
reverse transcription
- BSA
bovine serum albumin
- FITC
fluorescein isothiocyanate
- ABRC
Arabidopsis Biological Resource Center
- SEM
scanning electron microscopy
- DAPI
4′,6-diamidino-2-phenylindole
- FDA
fluorescein diacetate
- PI
propidium iodide
- FG7
female gametophyte stage 7
- DIC
differential interference contrast
- CLSM
confocal laser scanning microscopy
- TEM
transmission electron microscopy
- TUNEL
terminal-deoxynucleotidyl transferase-mediated nick end labeling
- ROS
reactive oxygen species
- HG
homogalacturonan
- RGI
rhamnogalacturonan I
- PBS
phosphate-buffered saline
Footnotes
This work was supported by the National Science Foundation of China (grant nos. 31270357 and 31540005), the Program for New Century Excellent Talents in University (grant no. NECT–12–0687), the Key Project of the Chinese Ministry of Education (grant no. 211019), the Natural Science Foundation of Hebei Province (grant no. C2013205160), the Foundation for High-Level Talents in Higher Education of Hebei, and the 100 Innovative Talents Program of Higher Education of Hebei (to Y.G.).
Articles can be viewed without a subscription.
References
- Ahmed SZ, Ferguson G, Glidewell C (1999) Keto-acid chloride and diketone intermediates in the acylation of ferrocene by arylene bis-acid chlorides. Crystal and molecular structure of 4-ferrocenylcarbonyl-4′-chlorocarbonylbiphenyl: intermolecular association by means of strong C-H···O hydrogen bonds and Π···Π stacking interactions. J Organomet Chem 585: 331–340 [Google Scholar]
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Asakura T, Matsumoto I, Funaki J, Arai S, Abe K (2000) The plant aspartic proteinase-specific polypeptide insert is not directly related to the activity of oryzasin 1. Eur J Biochem 267: 5115–5122 [DOI] [PubMed] [Google Scholar]
- Asakura T, Watanabe H, Abe K, Arai S (1995) Rice aspartic proteinase, oryzasin, expressed during seed ripening and germination, has a gene organization distinct from those of animal and microbial aspartic proteinases. Eur J Biochem 232: 77–83 [DOI] [PubMed] [Google Scholar]
- Bach AS, Derocq D, Laurent-Matha V, Montcourrier P, Sebti S, Orsetti B, Theillet C, Gongora C, Pattingre S, Ibing E, et al. (2015) Nuclear cathepsin D enhances TRPS1 transcriptional repressor function to regulate cell cycle progression and transformation in human breast cancer cells. Oncotarget 6: 28084–28103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G, Rasheed M, Taigwal R, Sahoo R, Kaur R (2014) GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata. Biochem J 458: 323–334 [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Rawlings ND, Woessner JF (1998) Handbook of Proteolytic Enzymes. CD-ROM. Academic Press, San Diego [Google Scholar]
- Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294: 1351–1362 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Bosch M, Hepler PK (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C (2002) Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. (2008) Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20: 1519–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A (2012) The cell wall of the Arabidopsis pollen tube: spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol 160: 1940–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Foolad MR (1997) Molecular organization of a gene in barley which encodes a protein similar to aspartic protease and its specific expression in nucellar cells during degeneration. Plant Mol Biol 35: 821–831 [DOI] [PubMed] [Google Scholar]
- Chen J, Ding J, Ouyang Y, Du H, Yang J, Cheng K, Zhao J, Qiu S, Zhang X, Yao J, et al. (2008) A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci USA 105: 11436–11441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Li C, Zou YJ, Wu HM (2014) Glycosylphosphatidylinositol anchoring: control through modification. Plant Physiol 166: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Conti L, Tonelli C, Galbiati M (2013) Challenges and perspectives to improve crop drought and salinity tolerance. N Biotechnol 30: 355–361 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Dai XR, Gao XQ, Chen GH, Tang LL, Wang H, Zhang XS (2014) ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyltransferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR BIOSYNTHESIS B in human, is required for Arabidopsis pollen tube micropylar guidance and embryo development. Plant Physiol 165: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F, Lehner A, Ramdani Y, Bardor M, Lerouge P, Driouich A, Mollet JC (2010) Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiol 153: 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardelle F, Le Mauff F, Lehner A, Loutelier-Bourhis C, Bardor M, Rihouey C, Causse M, Lerouge P, Driouich A, Mollet JC (2015) Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann Bot (Lond) 115: 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pinto MC, Locato V, De Gara L (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35: 234–244 [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM (2001) Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol 125: 2040–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas C, Mogensen HL (1993) Gametes and fertilization: maize as a model system for experimental embryogenesis in flowering plants. Plant Cell 5: 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Bork P, Eisenhaber F (1998) Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng 11: 1155–1161 [DOI] [PubMed] [Google Scholar]
- Faro C, Gal S (2005) Aspartic proteinase content of the Arabidopsis genome. Curr Protein Pept Sci 6: 493–500 [DOI] [PubMed] [Google Scholar]
- Ge W, Song Y, Zhang C, Zhang Y, Burlingame AL, Guo Y (2011) Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim Biophys Acta 1814: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Dietrich C, Matsuno M, Li G, Berg H, Xia Y (2005) An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep 6: 282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Gillmor CS, Lukowitz W, Brininstool G, Sedbrook JC, Hamann T, Poindexter P, Somerville C (2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J, Heslop-Harrison Y (1970) Evaluation of pollen viability by enzymatically induced fluorescence: intracellular hydrolysis of fluorescein diacetate. Stain Technol 45: 115–120 [DOI] [PubMed] [Google Scholar]
- Higashiyama T, Takeuchi H (2015) The mechanism and key molecules involved in pollen tube guidance. Annu Rev Plant Biol 66: 393–413 [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao X, Cheng K, Jiang Y, Ouyang Y, Xu C, Li X, Xiao J, Zhang Q (2013) OsAP65, a rice aspartic protease, is essential for male fertility and plays a role in pollen germination and pollen tube growth. J Exp Bot 64: 3351–3360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwa KY. (2001) Glycosyl phosphatidylinositol-linked glycoconjugates: structure, biosynthesis and function. Adv Exp Med Biol 491: 207–214 [DOI] [PubMed] [Google Scholar]
- Ingram VM. (1951) Autolysis products of pepsin. Nature 167: 83. [DOI] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K (2001) The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Yamamoto Y, Murakami S, Sato F (2005) Post-translational regulation of CND41 protease activity in senescent tobacco leaves. Planta 222: 643–651 [DOI] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP (2007) A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci USA 104: 7628–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervinen J, Tobin GJ, Costa J, Waugh DS, Wlodawer A, Zdanov A (1999) Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting. EMBO J 18: 3947–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS (2005) Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell 4: 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GH, Lilley KS, Dupree P, Grossniklaus U, Twell D (2004) SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. (2004) Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol 5: 305–315 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43: D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015) Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Xue Y, Jia DJ, Wang T, Hi DQ, Liu J, Cui F, Xie Q, Ye D, Yang WC (2011) POD1 regulates pollen tube guidance in response to micropylar female signaling and acts in early embryo patterning in Arabidopsis. Plant Cell 23: 3288–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ge FR, Xu M, Zhao XY, Huang GQ, Zhou LZ, Wang JG, Kombrink A, McCormick S, Zhang XS, et al. (2013) Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J 74: 486–497 [DOI] [PubMed] [Google Scholar]
- Li Y, Kabbage M, Liu W, Dickman MB (2016) Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28: 233–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhong S, Guo X, Hao L, Wei X, Huang Q, Hou Y, Shi J, Wang C, Gu H, et al. (2013) Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr Biol 23: 993–998 [DOI] [PubMed] [Google Scholar]
- Lodish HF. (2003) Molecular Cell Biology, Ed 5 WH Freeman, New York [Google Scholar]
- Maeda Y, Kinoshita T (2011) Structural remodeling, trafficking and functions of glycosylphosphatidylinositol-anchored proteins. Prog Lipid Res 50: 411–424 [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Tsukamoto T (2012) Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdiscip Rev Dev Biol 1: 96–113 [DOI] [PubMed] [Google Scholar]
- Palin R, Geitmann A (2012) The role of pectin in plant morphogenesis. Biosystems 109: 397–402 [DOI] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BD, Creissen G, Lamb C, Chattoo BB (2009) Overexpression of rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol Plant Microbe Interact 22: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Rosenzweig C, Elliott J, Deryng D, Ruane AC, Müller C, Arneth A, Boote KJ, Folberth C, Glotter M, Khabarov N, et al. (2014) Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc Natl Acad Sci USA 111: 3268–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Jones GH (1996) A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res 4: 507–516 [DOI] [PubMed] [Google Scholar]
- Rudd PM, Morgan BP, Wormald MR, Harvey DJ, van den Berg CW, Davis SJ, Ferguson MA, Dwek RA (1997) Roles for glycosylation in the anti-inflammatory molecule CD59. Biochem Soc Trans 25: 1177–1184 [DOI] [PubMed] [Google Scholar]
- Runeberg-Roos P, Saarma M (1998) Phytepsin, a barley vacuolar aspartic proteinase, is highly expressed during autolysis of developing tracheary elements and sieve cells. Plant J 15: 139–145 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Niwa H, Yokota N, Kubota K, Inoue H (2008) Widespread tissue expression of nepenthesin-like aspartic protease genes in Arabidopsis thaliana. Plant Physiol Biochem 46: 724–729 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, et al. (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286: 735–741 [DOI] [PubMed] [Google Scholar]
- Wang T, Liang L, Xue Y, Jia PF, Chen W, Zhang MX, Wang YC, Li HJ, Yang WC (2016) A receptor heteromer mediates the male perception of female attractants in plants. Nature 531: 241–244 [DOI] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23: 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT (2016) Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170: 234–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HT, Wan ZY, Li S, Zhang Y (2014) Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26: 2007–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao X, Cheng K, Du H, Ouyang Y, Chen J, Qiu S, Huang J, Jiang Y, Jiang L, et al. (2012) A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Xiong W, Ye T, Wu Y (2012) Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J Exp Bot 63: 2579–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.