The IPE1 gene, encoding a putative glucose-methanol-choline oxidoreductase targeted to the endoplasmic reticulum, plays an essential role in anther cuticle and pollen exine development in maize.
Abstract
Anther cuticle and pollen exine are protective barriers for pollen development and fertilization. Despite that several regulators have been identified for anther cuticle and pollen exine development in rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), few genes have been characterized in maize (Zea mays) and the underlying regulatory mechanism remains elusive. Here, we report a novel male-sterile mutant in maize, irregular pollen exine1 (ipe1), which exhibited a glossy outer anther surface, abnormal Ubisch bodies, and defective pollen exine. Using map-based cloning, the IPE1 gene was isolated as a putative glucose-methanol-choline oxidoreductase targeted to the endoplasmic reticulum. Transcripts of IPE1 were preferentially accumulated in the tapetum during the tetrad and early uninucleate microspore stage. A biochemical assay indicated that ipe1 anthers had altered constituents of wax and a significant reduction of cutin monomers and fatty acids. RNA sequencing data revealed that genes implicated in wax and flavonoid metabolism, fatty acid synthesis, and elongation were differentially expressed in ipe1 mutant anthers. In addition, the analysis of transfer DNA insertional lines of the orthologous gene in Arabidopsis suggested that IPE1 and their orthologs have a partially conserved function in male organ development. Our results showed that IPE1 participates in the putative oxidative pathway of C16/C18 ω-hydroxy fatty acids and controls anther cuticle and pollen exine development together with MALE STERILITY26 and MALE STERILITY45 in maize.
Male sterility is a common biological phenomenon in plants and widely used in the production of hybrid seeds, which can reduce costs and enhance seed purity (Tester and Langridge, 2010). According to inheritance or origin, male sterility includes three types: genic male sterility, cytoplasmic male sterility, and cytoplasmic-genic male sterility (Rhee et al., 2015). The generation of mature pollen grains relies on anther development. The start of anther formation occurs in differentiated flower tissues (floral meristem), which consist of three histogenic layers: L1, L2, and L3. After continuous cell division and differentiation, L1 forms the epidermis and the L3 layer develops into the stomium and vascular bundles. L2 is the most important layer; it undergoes a series of periclinal and anticlinal divisions and eventually grows into the endothecium, the middle layer, the tapetum, and the pollen mother cells. When anther morphogenesis is completed, the anther has centrally localized pollen mother cells enclosed by four somatic layers, which are, from the surface to the interior, the epidermis, endothecium, middle layer, and tapetum. Then, the pollen mother cells undergo meiosis and mitosis, resulting in trinucleate pollen grains, and the endothecium, middle layer, and tapetum are gradually degraded (Goldberg et al., 1993, 1995; Ma, 2005).
The anther cuticle and pollen exine are two barriers for anther development and fertilization. However, their chemical nature is obscure because of the limitation of purifying and obtaining a sufficient quantity of materials for analysis. Previous results indicate that the anther cuticle, covering the outer surface of anthers, contains cutin and wax. The polymerization of hydroxylated and epoxy C16 and C18 fatty acids produces cutin (Pollard et al., 2008; Beisson et al., 2012), and a mixture of very-long-chain fatty acids, alkanes, alkenes, and fatty alcohols forms wax (Samuels et al., 2008). Pollen exine, occurring on the outer surface of pollen, consists of two sublayers, the sexine and nexine. Sexine is further divided into bacula and tectum. The major component of pollen exine is the very insoluble sporopollenin, which may be made of aliphatic derivatives such as fatty acids and phenolic compounds (Ariizumi and Toriyama, 2011). The innermost of the anther wall, the tapetum, has been considered the place where nutrients and enzymes are synthesized (Stieglitz, 1977; Ariizumi and Toriyama, 2011). These materials can be transferred to the surface of anthers and pollen with the help of transporters and the specialized structures called Ubish bodies (Wang et al., 2003). Until now, several genes have been discovered in rice (Oryza sativa) that are involved in the development of the anther cuticle and pollen wall, including WAX-DEFICIENT ANTHER1 (WDA1; Jung et al., 2006), CYP704B2 (Li et al., 2010), DEFECTIVE POLLEN WALL (DPW; Shi et al., 2011), CYP703A3 (Yang et al., 2014), POSTMEIOTIC DEFICIENT ANTHER1 (Hu et al., 2010; Zhu et al., 2013), OsABCG15 (Niu et al., 2013; Qin et al., 2013), OsABCG26 (Zhao et al., 2015), and TAPETUM DEGENERATION RETARDATION (TDR; Li et al., 2006). Nevertheless, their regulatory networks still need to be explored.
With advances of forward and reverse genetics, some male-sterile genes have been isolated in maize, such as MALE STERILE CONVERTED ANTHER1 (Chaubal et al., 2003), MULTIPLE ARCHESPORIAL CELLS1 (Wang et al., 2012), MALE STERILITY32 (MS32; Moon et al., 2013), MS8 (Wang et al., 2013), AMEIOTIC1 (Pawlowski et al., 2009), ABSENCE OF FIRST DIVISION1 (Golubovskaya et al., 2006), RECOMBINATION PROTEIN51 (Li et al., 2007), MS26 (Albertsen et al., 2006), and MS45 (Albertsen et al., 1993). Only MS26 and MS45 are required for pollen wall development, but their detailed functions have not been confirmed. MS45 may encode a strictosidine synthase, which is localized in the tapetum during the early vacuolization stage of microspore development (Cigan et al., 2001; Skibbe and Schnable, 2005). In the ms26 mutant, tapetal cells degenerate abnormally. Microspores show early vacuolation and are aborted after the tetrad stage. MS26 probably encodes a cytochrome P450 monooxygenase (Djukanovic et al., 2013) that is homologous to CYP704B1 in Arabidopsis (Arabidopsis thaliana), CYP704B2 in rice, and BnMS1 and BnMS2 in Brassica napus, the mutation of which results in defective pollen exine. CYP704B1 and CYP704B2 have a similar function in the ω-hydroxylation of C16 and C18 fatty acids (Dobritsa et al., 2009; Li et al., 2010; Yi et al., 2010). The mechanism of anther cuticle and pollen wall development remains largely unknown in maize.
In this study, we obtained a complete maize male-sterile mutant, irregular pollen exine1 (ipe1), which had a smooth outer anther surface, defective Ubisch bodies, abnormal pollen exine, and no mature pollen grains. Through position cloning and transgenic experiments, we cloned the IPE1 gene as a putative Glc-methanol-choline (GMC) oxidoreductase that was expressed mainly in the tapetum. The expression of IPE1 fused with GFP indicated that IPE1 was localized in the endoplasmic reticulum by the N-terminal signal peptide. A remarkably altered composition of cutin, wax, and fatty acids was observed in the ipe1 mutant. Moreover, the expression of wax and flavonoid metabolism genes, and fatty acid synthesis and elongation genes, was changed in ipe1 anthers. Our data indicated that the putative oxidative pathway of ω-hydroxy fatty acids depending on IPE1 plays an important role in anther cuticle and pollen exine formation in maize.
RESULTS
Isolation and Phenotypic Analysis of the ipe1 Mutant
To identify more male-sterile genes in maize, we screened a MuDR library (Dong et al., 2013) and obtained the complete male-sterile mutant ipe1. When crossed with the wild type, all F1 progeny were fertile, with the F2 population segregating at an approximate ratio of 3:1 (fertility:sterility = 208:58, χ2 < χ2(0.05, 1) = 3.84), indicating that the mutant phenotype was controlled by a single recessive gene. Compared with the wild type, there were no obvious differences in ipe1 during vegetative growth (Fig. 1, A and B). However, the mutant anthers could not be spread from glumes (Fig. 1, C and D) and became smaller, wilted, and brown with no mature pollen grains (Fig. 1, E–H). ipe1 plants could normally set seeds when pollinated with wild-type pollen, implying that female fertility was unaffected.
Figure 1.
Morphological comparisons between the wild type and ipe1. A and B, No significant difference was observed between the wild type (A) and ipe1 (B) at the tasseling stage. C and D, Mutant anthers (D) are hidden in the glume compared with wild-type anthers (C) after flowering. E and F, In the wild type, six anthers are filled and dark yellow (E), whereas ipe1 anthers are wilted, smaller, and brown (F). G and H, At the mature pollen grain stage, by staining with 1% odine-potassium iodide (I2-KI) solution, abundant mature pollen grains are present in wild-type anthers (G) but completely absent in ipe1 mutant anthers (H). Bars = 50 μm.
Anther Development Was Defective in the ipe1 Mutant
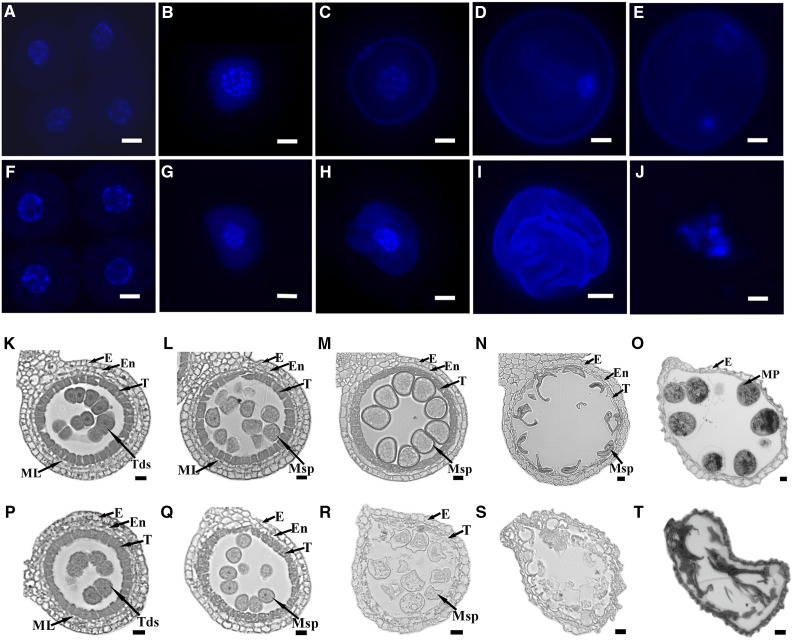
To check the defect of ipe1 during anther development, we first analyzed microspore development in the wild type and ipe1 by 4′,6-diamidino-2-phenylindole (DAPI) staining. Normal meiosis and tetrads were observed in the ipe1 mutant (Fig. 2, A and F; Supplemental Fig. S1). At the early uninucleate microspore stage, similar to that in the wild type, ipe1 microspores were normally released from the tetrad and the nucleus was located in the middle (Fig. 2, B and G). At the following stages, in the wild type, the size of microspores increased gradually and the pollen wall was formed; the nucleus moved to the peripheral region and then underwent mitosis to generate two nuclei: a generative nucleus and a vegetative nucleus (Fig. 2, C–E). However, the pollen wall of ipe1 was undeveloped, which resulted in microspores slowly collapsing and aborting (Fig. 2, H and I). Eventually, only some remnant material was left at the binucleate microspore stage (Fig. 2J), implying that defects of the ipe1 microspore start with pollen wall formation after the tetrad stage.
Figure 2.
Phenotypic analysis of ipe1 anthers at various developmental stages. A to J, DAPI staining of wild-type (A–E) and ipe1 (F–J) microspores. A and F, At the tetrad stage, the normal tetrad is shown in the wild type (A) and mutant (F). B and G, At the early uninucleate stage, normal microspores of the wild type (B) and mutant (G) are free from the tetrad. C and H, At the middle uninucleate stage, the pollen wall of wild-type microspores is distinct (C), while the shape of the microspore is irregular and the pollen wall is not well formed in the mutant (H). D and I, At the late uninucleate stage, compared with the wild-type microspore (D), the ipe1 microspore is severely disorganized (I). E and J, At the binucleate stage, in the wild type (E), the larger microspore has a vegetative nucleus and a generative nucleus, while the ipe1 microspore is completely degenerated (J). Bars = 10 μm. K to T, Transverse sections of wild-type and ipe1 anthers. A single locule is shown from cross sections of the wild type (K–O) and the ipe1 mutant (P–T). K and P, At the tetrad stage, both the wild-type (K) and mutant (P) anthers with four somatic layers and tetrads are shown. L and Q, At the early uninucleate stage, the tapetum is condensed and deeply stained in the wild type (L). While the ipe1 middle layer is missing, the tapetum is lightly stained with more vacuoles present (Q). M and R, At the late uninucleate stage, the ipe1 tapetum is almost completely degraded and microspores degenerated without the dark pollen exine (R) compared with the wild type (M). N and S, At the binucleate stage, in the mutant (S), collapsed anther layers and microspores are observed compared with the wild type (N). O and T, At the mature pollen grain stage, no mature pollen grains are found in the mutant (T) compared with the wild type (O). E, Epidermis; En, endothecium; ML, middle layer; MP, mature pollen; Msp, microspore; T, tapetum; Tds, tetrads. Bars = 20 μm.
Furthermore, a transverse section was made for more detailed characterization of the ipe1 defect. At the tetrad stage, like the wild-type anther, four anther wall layers persisted, surrounding the tetrad in ipe1 (Fig. 2, K and P). At the early uninucleate microspore stage, in wild-type anthers, there was the thin middle layer and the tapetum was degraded and appeared more condensed and deeply stained (Fig. 2L). However, the ipe1 middle layer could not be seen, and tapetal cells were lightly stained. More vacuoles appeared in the tapetum than in the wild type, suggesting more severe degradation of the tapetum. Microspores could be released normally from the tetrad with a natural shape (Fig. 2Q). During the late uninucleate microspore stage, the middle layer of wild-type anthers disappeared and the tapetum continued to be degraded. Microspores became vacuolated and gradually enlarged, and the pollen exine was clearly visible and became darker, suggesting that more sporopollenin precursors had been deposited on the surface of microspores (Fig. 2M). By contrast, in ipe1 anthers, the tapetum was almost completely degraded and less cellular material was left. Microspores appeared to be degraded and exhibited an irregular appearance, and the pollen exine could not be observed distinctly (Fig. 2R). At the binucleate microspore stage, the wild-type tapetum displayed a strip shape and vacuolated microspores showed the falcate form (Fig. 2N). Unlike the wild type, the ipe1 microspores and anther wall had collapsed (Fig. 2S). At the mature pollen grain stage, many pollen grains were observed in the anther locule of the wild type (Fig. 2O). On the contrary, in ipe1 anthers, no pollen grains existed and only some debris remained (Fig. 2T). These phenotypes confirmed that IPE1 is involved in the degeneration of the tapetum in anther development.
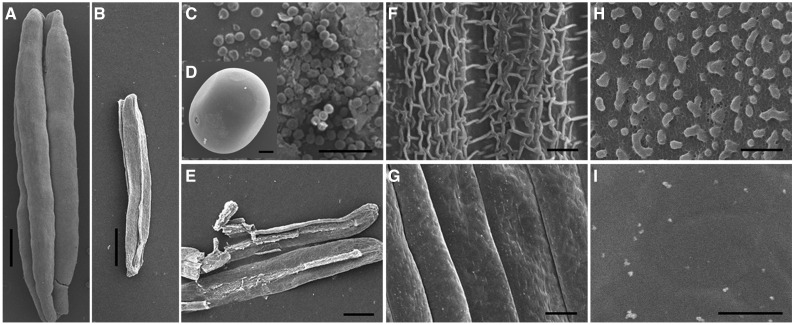
Scanning electron microscopy (SEM) was used to investigate anther development at the mature pollen grain stage. Consistent with the above morphological results, ipe1 anthers produced smaller, wilted anthers with no pollen grains in the anther locule (Fig. 3, A–E). Moreover, reticulate cuticle coated the outermost surface of anthers, and abundant Ubisch bodies were distributed on the outside of tapetal cells in wild-type anthers (Fig. 3, F and H). However, the ipe1 anther surface was smooth (Fig. 3G), indicating that anther cuticle formation was disrupted in the mutant. In addition, Ubisch bodies could not be seen on the inner surface of mutant anthers (Fig. 3I).
Figure 3.
SEM analysis of wild-type and ipe1 anthers at the mature pollen grain stage. A and B, The ipe1 anther (B) is smaller and wilted than the normal anther (A). C to E, Abundant mature pollen grains are observed in the wild type (C) but have vanished in the mutant (E). A wild-type pollen grain is shown in D. F to I, The outer (F) and inner (H) surfaces of wild-type anther compared with the outer (G) and inner (I) surfaces of ipe1 anther. The ipe1 anther outer surface is glossy with no Ubisch bodies on the inner anther surface. Bars = 600 μm in A and B, 400 μm in C and E, 15 μm in D, 6 μm in F and G, and 2 μm in H and I.
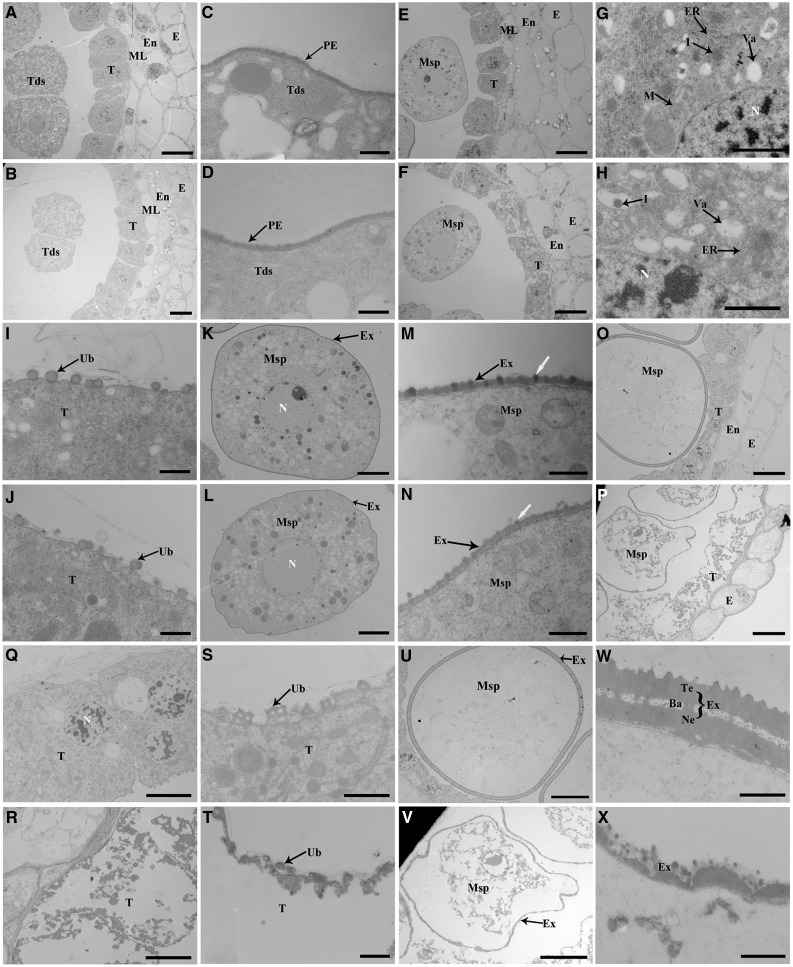
To further examine the ipe1 defects, transmission electron microscopy (TEM) was employed. According to DAPI staining and transverse sectioning, no apparent differences were detected at the tetrad stage between the wild type and ipe1 (Fig. 4, A–D). At the early uninucleate microspore stage, tapetal cells of the wild type contained numerous subcellular organelles (Fig. 4, E and G). In the ipe1 tapetal cells, the Ubisch bodies were normally obvious, as in the wild type (Fig. 4, I and J). However, fewer subcellular organelles and lipid bodies but more vacuoles were observed, which indicated that lipid metabolism was influenced (Fig. 4, F and H). At this stage, microspores of the wild type had begun to form the exine, consisting of bacula and nexine (Fig. 4, K and M). By contrast, ipe1 microspores did not develop the dark bacula structure (Fig. 4, L and N). During the late uninucleate microspore stage, the wild-type tapetum degenerated but parts of subcellular organelles existed (Fig. 4, O and Q). Ubisch bodies accumulated enough sporopollenin precursors (Fig. 4S). However, the ipe1 tapetum had been degraded completely and barely functional Ubisch bodies could be observed (Fig. 4, P, R, and T). Meanwhile, wild-type microspores formed a thicker pollen exine that consisted of tectum, bacula, and nexine (Fig. 4, U and W). In contrast, sporopollenin precursors were distributed randomly and the pollen exine did not have the typical three-layer structure in ipe1 (Fig. 4, V and X). These observations illustrated that IPE1 functions in the formation of the anther cuticle, Ubisch bodies, and pollen exine.
Figure 4.
TEM of anthers in the wild type and the ipe1 mutant. A and B, Normal anthers of the wild type (A) and the mutant (B) are shown at the tetrad stage. C and D, On the surface of the microspore, the primexine is normally developed in the wild type (C) and the mutant (D) at the tetrad stage. E and F, Anthers of the wild type (E) and the mutant (F) show the defective ipe1 tapetum with light staining at the early uninucleate microspore stage. G, Higher magnification of the wild-type tapetum in E shows abundant subcellular organelles. H, Higher magnification of the ipe1 tapetum in F shows fewer subcellular organelles. I and J, Normal Ubisch bodies of the wild type (I) and the mutant (J) are shown at the early uninucleate microspore stage. K and L, The shape of microspores is normal in the wild type (K) and the mutant (L) at the early uninucleate microspore stage. M and N, The pollen exine of the wild type (M) and the mutant (N) shows the primary structure of the pollen exine; the dark bacula (white arrows) are not clearly visible in the ipe1 microspore at the early uninucleate microspore stage. O and P, Anthers of the wild type (O) and the mutant (P) show the defects of ipe1 anthers with abnormal tapetum and microspores at the late uninucleate microspore stage. Q, Higher magnification of the wild-type tapetum in O shows that a portion of subcellular organelles are present in the degenerated tapetum. R, Higher magnification of the mutant tapetum in P shows that the ipe1 tapetum is almost completely degraded with only remnants remaining. S and T, Ubisch bodies have accumulated abundant electron-dense sporopollenin precursors in the wild type (S), while ipe1 Ubisch bodies are severely degenerated and have almost disappeared (T) at the late uninucleate microspore stage. U and V, Microspores of the wild type (U) and the mutant (V) show the disordered microspore of ipe1 at the late uninucleate microspore stage. W and X, The wild-type pollen exine contains tectum, bacula, and nexine (W), while in the ipe1 mutant, the structure of pollen exine is not intact (X) at the late uninucleate microspore stage. Ba, Bacula; E, epidermis; En, endothecium; ER, endoplasmic reticulum; Ex, exine; l, lipid body; M, mitochondria; ML, middle layer; Msp, microspore; N, nucleus; Ne, nexine; PE, primexine; T, tapetum; Tds, tetrads; Te, tectum; Ub, Ubisch body; Va, vacuole. Bars = 10 μm in A, B, E, F, O, P, U, and V, 5 μm in K, J, Q, and R, 1 μm in G and H, 1 μm in S, T, W, and X, and 0.5 μm in C, D, I, J, M, and N.
Alteration of Wax, Cutin, and Fatty Acids in ipe1 Anther
The phenotypic defects of the anther cuticle, pollen wall, and Ubisch bodies in ipe1 revealed that abnormalities may occur in the synthesis or transport of aliphatic compounds. To test this hypothesis, we used gas chromatography-mass spectrometry (GC-MS) to determine the constituents of chloroform-extractable cuticular waxes, cutin, and fatty acids in wild-type and ipe1 anthers. We calculated the surface areas of randomly chosen anthers and then plotted them against the weight of each sample to determine the surface areas of corresponding samples (Li et al., 2010; Shi et al., 2011; Supplemental Fig. S2).
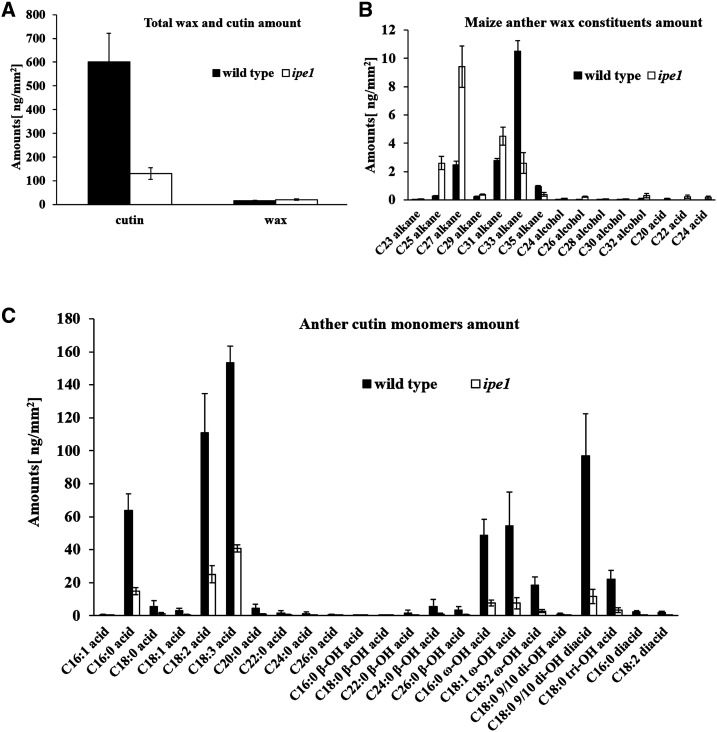
Our results showed that there was no significant variation in the total wax between ipe1 anthers (21.17 ng mm−2) and wild-type anthers (17.37 ng mm−2; P > 0.05). By contrast, the total cutin of ipe1 anthers (130.91 ng mm−2) decreased by 78.25% compared with the wild type (601.79 ng mm−2; P < 0.01; Fig. 5A). For the wax constituents, C23, C25, C27, C29, and C31 alkanes were increased significantly in ipe1 (P < 0.01) while C33 and C35 alkanes were obviously decreased (P < 0.01; Fig. 5B). In addition, significant increases in the levels of C24, C26, C28, C30, and C32 alcohols (P < 0.05) were detected in mutant anthers (Fig. 5B). In the wild-type anther, the major aliphatic cutin monomers were C16:0, C18:2, C18:3, C16:0 ω-OH, C18:1 ω-OH, C18:2 ω-OH, C18:0 9/10 di-OH, and C18:0 tri-OH acids (Fig. 5C). Compared with the wild type, all aliphatic cutin monomers observed in the ipe1 anthers were reduced significantly (Fig. 5C; Supplemental Table S1). Fatty alcohols, alkanes, and other aliphatic molecules are derivatives of fatty acids (Zhang et al., 2008). To understand the alteration of the levels of fatty acids, the total fatty acids of anthers were extracted for analysis. The level of total fatty acids of wild-type anthers with carbon length from C14 to C28 was 61.51 μg mg−1, whereas that of ipe1 anthers was only 32.15 μg mg−1 (Table I). Chemical analysis indicated that IPE1 participates in lipidic metabolism or transport necessary for anther development.
Figure 5.
Analysis of anther wax and cutin in the wild type and the ipe1 mutant. A, Total cutin and wax amounts per unit of surface area (ng mm−2) in wild-type (black bars) and ipe1 (white bars) anthers. Error bars indicate sd (n = 5). B, Wax constituent amounts per unit of surface area (ng mm−2) in wild-type (black bars) and ipe1 (white bars) anthers. Error bars indicate sd (n = 5). C, Cutin constituent amounts per unit of surface area (ng mm−2) in wild-type (black bars) and ipe1 (white bars) anthers. Error bars indicate sd (n = 5). Compound names are abbreviated as follows: C16:1 acid, 7-hexadecenoic acid; C16:0 acid, hexadecanoic acid; C18:0 acid, octadecanoic acid; C18:1 acid, oleic acid; C18:2 acid, linoleic acid; C18:3 acid, linolenic acid; C20:0 acid, eicosanoic acid; C22:0 acid, docosanoic acid; C24:0 acid, tetracosanoic acid; C26:0 acid, hexacosanoic acid; C16:0 β-OH acid, 2-hydroxy-hexadecanoic acid; C18:0 β-OH acid, 2-hydroxy-octadecanoic acid; C22:0 β-OH acid, 2-hydroxy-docosanoic acid; C24:0 β-OH acid, 2-hydroxy-tetracosanoic acid; C26:0 β-OH acid, 2-hydroxy-hexacosanoic acid; C16:0 ω-OH acid, 16-hydroxy-hexadecanoic acid; C18:1 ω-OH acid, 18-hydroxy-oleic acid; C18:2 ω-OH acid, 18-hydroxy-linoleic acid; C18:0 9/10 di-OH acid, 9,10-dihydroxy-octadecanoic acid; C18:0 9/10 di-OH diacid, 9,10-dihydroxy-octadecanoic-1,18-dioic acid; C18:0 tri-OH acid, 9,10,18-trihydroxy-octadecanoic acid; C16:0 diacid, hexadecanoic-1,16-dioic acid; and C18:2 diacid, linoleic-1,18-dioic acid.
Table I. Total fatty acids of wild-type and ipe1 anthers.
Fatty acid values shown are means ± sd.
| Lipids | Wild Type | ipe1 | Up |
|---|---|---|---|
| μg mg−1 dry weight | |||
| C14:0 acid | 0.066 ± 0.001 | 0.061 ± 0.009 | −7.62% |
| C16:1 acid | 0.041 ± 0.005 | 0.137 ± 0.029 | 234.37% |
| C16:0 acid | 10.173 ± 1.060 | 4.285 ± 0.790 | −57.88% |
| C17:0 acid | 8.240 ± 0.043 | 8.268 ± 0.054 | 0.33% |
| C18:2 acid | 15.055 ± 1.477 | 12.666 ± 2.573 | −15.87% |
| C18:3 acid | 26.167 ± 4.709 | 3.715 ± 0.487 | −85.80% |
| C18:1 acid | 0.257 ± 0.017 | 0.463 ± 0.104 | 80.59% |
| C18:0 acid | 0.623 ± 0.066 | 0.890 ± 0.117 | 42.77% |
| C20:0 acid | 0.457 ± 0.042 | 0.727 ± 0.067 | 59.27% |
| C22:0 acid | 0.158 ± 0.008 | 0.398 ± 0.030 | 151.65% |
| C24:0 acid | 0.099 ± 0.008 | 0.292 ± 0.039 | 195.92% |
| C26:0 acid | 0.038 ± 0.003 | 0.092 ± 0.008 | 141.41% |
| C28:0 acid | 0.140 ± 0.018 | 0.158 ± 0.048 | 12.83% |
| Total acids | 61.513 ± 7.456 | 32.150 ± 4.355 | −47.73% |
Map-Based Cloning of IPE1
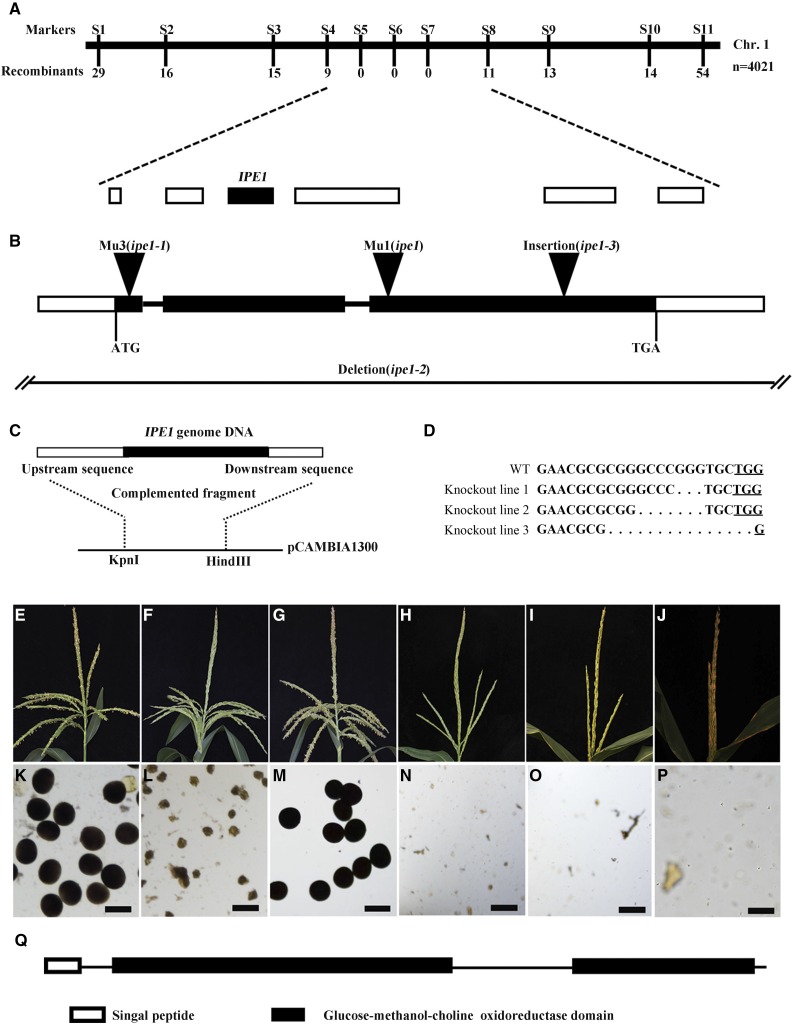
We used the map-based cloning approach to isolate the responsible gene for the ipe1 mutant. The IPE1 gene was initially mapped on the chromosome 1 short arm between simple sequence repeat markers S1 and S11 (Fig. 6A). We then developed nine pairs of insertion-deletion (Indel) markers for fine mapping (Supplemental Table S2). Using 4,021 mutant individuals from the F2 population, the target genomic region was ultimately narrowed to the approximately 290-kb interval between Indel markers S4 and S8, with nine and 11 recombination individuals, respectively (Fig. 6A). Six predicted genes were located in this interval (Fig. 6A), and one of these genes, GRMZM2G434500, was identified to have a Mutator1 transposon (Mu1) insertion at the 64th base pair from the beginning of the third exon by sequencing the ipe1 genomic DNA (Fig. 6B).
Figure 6.
Positional cloning and confirmation of the IPE1 gene. A, Fine-mapping of the IPE1 gene. The IPE1 locus was mapped to a 290-kb region between genetic markers S4 and S8 on the chromosome 1 short arm, containing six genes. Three Indel markers (S5, S6, and S7) cosegregated with the male-sterile phenotype. B, Structure and mutation sites of the IPE1 gene. Black boxes indicate exons, intervening lines indicate introns, and white boxes indicate untranslated regions. The triangles represent the locations of the insertions. In the ipe1-2 mutant, there is a deletion containing the IPE1 gene. C, The complemented fragment contains a 1,644-bp upstream sequence, the 2,585-bp IPE1 genomic DNA, and a 969-bp downstream sequence after the stop codon. D, Mutation sites of three knockout lines generated by CRISPR-Cas9. Compared with the wild-type sequence (WT), the deletions of three nucleotides, seven nucleotides, and 15 nucleotides were detected in knockout lines 1, 2, and 3, respectively. Sequences in boldface, Target sites; dots, deleted nucleotides; underlined TGG, protospacer-adjacent motif sequences. E to P, Phenotypes of tassels (E–J) and pollen grains stained with 1% I2-KI solution (K–P) at the mature pollen grain stage. E and K, Wild-type tassel and mature pollen grains. F and L, The ipe1-3 anthers could not be exserted from glumes, and the pollen grains are abnormal. G and M, The complemented line of ipe1-3 was restored to the wild-type phenotype. H to J and N to P, In knockout lines 1 (H and N), 2 (I and O), and 3 (J and P), the anthers could not be spread from glumes, and no pollen grains remain in the anther loculus. Bars = 100 μm. Q, The IPE1 protein contains a signal peptide at the N terminus and two Glc-methanol-choline oxidoreductase domains.
In addition, we obtained three other male-sterile lines, named ipe1-1 (mu1019442::Mu), ipe1-2, and ipe1-3 (ms*-6044). They all exhibited a smooth anther surface, which was very similar to the ipe1 phenotype (Supplemental Fig. S3). Allelism tests confirmed that ipe1-1, ipe1-2, and ipe1-3 were all allelic to ipe1 (Supplemental Table S3). Sequencing of the GRMZM2G434500 gene in these mutants revealed the mutated sites: there was a Mu3 transposon insertion at the 49th base pair from the starting nucleotide of translation in ipe1-1, a deletion containing the entire GRMZM2G434500 in ipe1-2, and a 1,331-bp insertion at the 691st base pair from the beginning of the third exon in ipe1-3 (Fig. 6B), verifying that GRMZM2G434500 was responsible for the male-sterile phenotype in ipe1. The IPE1 gene was further confirmed by functional complementation of ipe1-3 homozygous plants and targeted gene knockouts of IPE1 by the CRISPR-Cas9 system (Fig. 6, C–P).
Sequence analysis of cDNA confirmed that the IPE1 gene consisted of three exons and two introns (Fig. 6B) and encoded a 582-amino acid protein. Structure prediction using SMART (http://smart.embl-heidelberg.de/) indicated that IPE1 included two domains of the GMC oxidoreductase at the N terminus (50–323) and C terminus (424–571; Fig. 6Q). Further analysis by National Center for Biotechnology Information BLAST revealed that IPE1 was a putative GMC oxidoreductase.
Partially Functional Conservation of the IPE1 Orthologous Gene in Arabidopsis
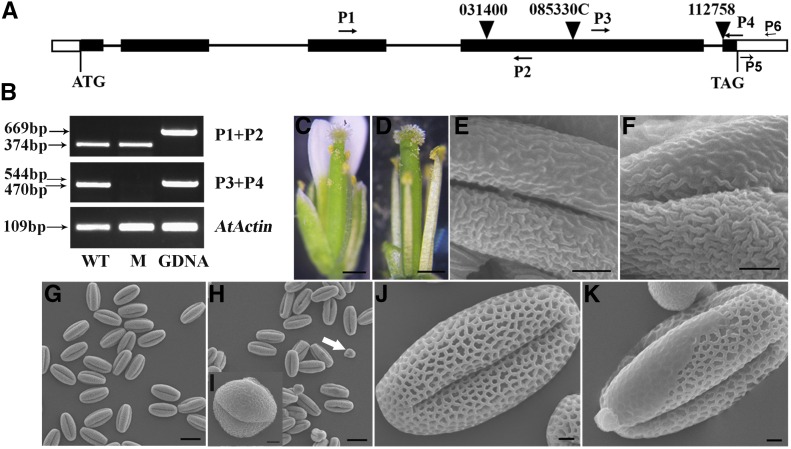
To demonstrate whether the function of IPE1 in anther cuticle and pollen exine development is conserved, we constructed a phylogenetic tree based on IPE1 and 17 of the most similar protein sequences from maize, rice, and Arabidopsis (Supplemental Fig. S4). Most of these genes have a common GMC domain and are highly expressed in the male reproductive organs (Supplemental Fig. S5; Supplemental Table S4), indicating that they may have a conserved function in the formation of the male reproductive organs. The phylogenetic tree suggested that the AT1G12570 gene was the most closely orthologous gene for IPE1 in Arabidopsis. To address the function of AT1G12570, we obtained a T-DNA line (SALK-085330C) with an insertion in the fourth exon (Fig. 7A), resulting in premature transcription termination (Fig. 7B). Compared with the wild type, the T-DNA line had the normal pollen-grain extrusion and anther cuticle development (Fig. 7, C–F). However, pollen grains that were smaller than wild-type ones were detected and the percentage of mutant pollen grains was about 23% (293 of 1,272; Fig. 7, G–I). Moreover, the surface of some pollen grains with normal size was smooth to varying degrees (Fig. 7, J and K). Two allelic mutants (SALK-112758 and SALK-031400) of AT1G12570 showed similar phenotypes to the mutant (Supplemental Fig. S6), indicating that AT1G12570 is involved in pollen grain maturation and pollen exine development but not in anther cuticle development. These data suggested that the Arabidopsis ortholog of IPE1 plays a partially conserved role in anther development.
Figure 7.
Phenotypic analysis of a T-DNA insertional line of the IPE1 orthologous gene in Arabidopsis. A, Gene structure and the position of the T-DNA insertion in AT1G12570. B, Reverse transcription (RT)-PCR analysis with the primers shown in A. Amplification occurred with primers P1+P2 in the T-DNA insertional line but failed with primers P3+P4. AtACTIN was used as a positive control. WT, Wild type; M, T-DNA insertional line; GDNA, genomic DNA. C and D, The pollen grains of the wild type (C) and the T-DNA insertional line (D) are observed. E to K, SEM analysis of the wild type and the T-DNA insertional line. E and F, The normal anther surfaces of the wild type (E) and the T-DNA insertional line (F) are shown. G and H, Smaller pollen grains appeared in the T-DNA insertional line (arrow; H) compared with the wild type (G). I, Magnification of a smaller pollen grain. J and K, A few limited areas without a visible exine network were detected in the T-DNA insertional line (K) compared with the wild type (J). Bars = 500 μm in C and D, 2 μm in E, F, and I to K, and 20 μm in G and H.
IPE1 Is Expressed Predominantly in the Tapetum
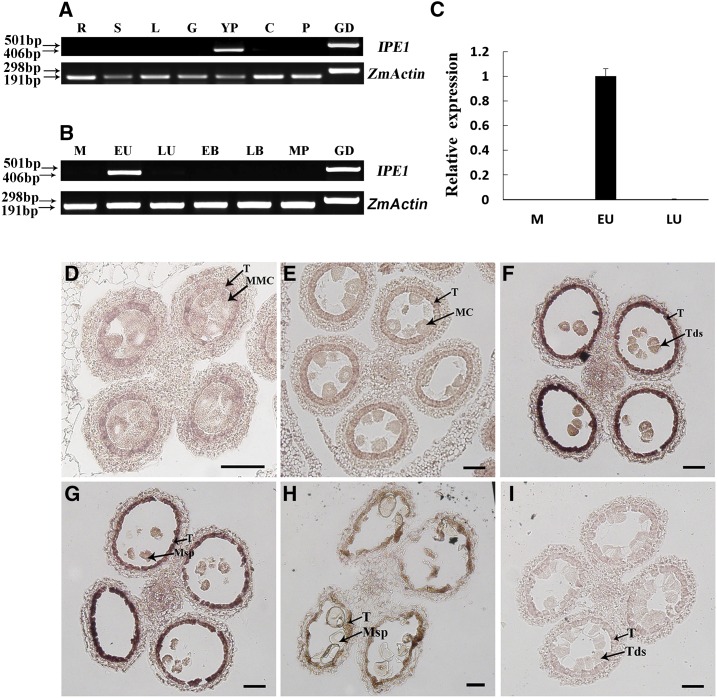
According to phenotypic analysis, the formation of anther cuticle and pollen exine was abnormal in ipe1 anthers, while there was no obvious mutant phenotype in vegetative growth. To examine whether the expression and function of IPE1 are consistent, the expression pattern of IPE1 was evaluated. Total RNA extracted from vegetative and reproductive organs was used to perform RT-PCR. The IPE1 transcript was detected in young spikelets with no apparent detection in roots, stems, leaves, glumes, cobs, and pistils, which agreed with the mutant phenotype (Fig. 8A). Anthers at different developmental stages were collected to further determine the expression pattern. IPE1 was expressed specifically at the early uninucleate microspore stage (Fig. 8B), which was confirmed by quantitative reverse transcription (qRT)-PCR (Fig. 8C). However, no evident expression was detected at the meiosis stage, late uninucleate microspore stage, binucleate microspore stage, and mature pollen grain stage (Fig. 8B).
Figure 8.
Expression analysis of the IPE1 gene. A, Detection of IPE1 transcript in wild-type selected tissues by RT-PCR, showing that IPE1 was expressed only in the young spikelets. ZmACTIN was used as a positive control. R, Roots; S, stems; L, leaves; G, glumes; YP, young spikelets; C, cobs; P, pistils; GD, genomic DNA. B, Detection of IPE1 transcript in wild-type anthers at different developmental stages by RT-PCR, showing that IPE1 was restricted temporally to the anthers during the early uninucleate microspore stage. M, Meiosis stage; EU, early uninucleate microspore stage; LU, late uninucleate microspore stage; EB, early binucleate microspore stage; LB, late binucleate microspore stage; MP, mature pollen grain stage. C, qRT-PCR analysis of IPE1 expression in wild-type anthers. Each data point is the average of three biological replicates. Error bars indicate sd (n = 3). D to I, RNA in situ hybridization of wild-type anthers using an IPE1-specific antisense probe (D–H) and a negative control sense probe (I). A strong hybridization signal in the tapetum was detected at the tetrad stage (F) and early uninucleate microspore stage (G). The signal in the tapetum disappeared at the early meiosis stage (D), meiosis stage (E), and late uninucleate microspore stage (H). Hybridization with the sense probe produced no signal at the tetrad stage (I). MC, Meiotic cell; MMC, meiotic mother cell; Msp, microspore; T, tapetum; Tds, tetrads. Bars = 50 μm.
To accurately determine the spatial and temporal expression of IPE1, we employed RNA in situ hybridization with wild-type anthers at various stages. Consistent with the RT-PCR results, IPE1 was strongly and predominantly expressed in the tapetum at the tetrad stage and early uninucleate stage (Fig. 8, F and G), and there was no expression in other stages (Fig. 8, D, E, and H). No signal was detected in the negative control using sense IPE1 transcript as a probe (Fig. 8I). This spatiotemporal specificity in IPE1 expression is consistent with its role in modulating anther cuticle and pollen exine formation via the synthesis of aliphatic materials in the tapetum.
IPE1 Was Localized Mainly in the Endoplasmic Reticulum
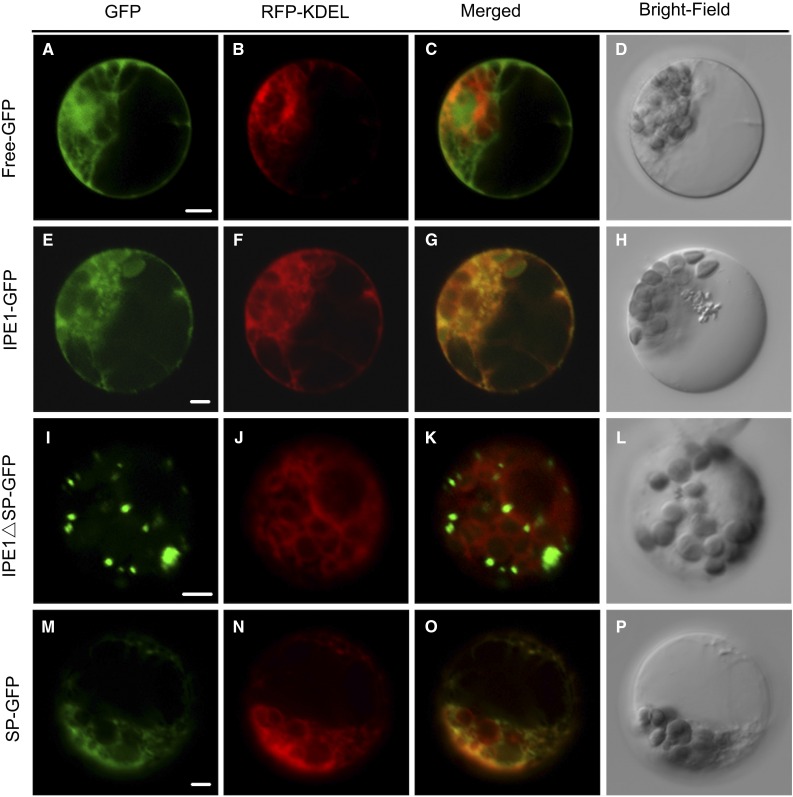
There is a predicted secretory pathway signal peptide (SP) with 27 amino acids at the N terminus of the IPE1 protein according to the TargetP1.1 server (http://www.cbs.dtu.dk/services/TargetP/; Fig. 6Q). To investigate the cellular localization of the IPE1 protein, we generated three constructs: pUbi::IPE1-GFP, pUbi::IPE1ΔSP-GFP, and pUbi::SP-GFP. pUbi::IPE1-GFP contained the full-length IPE1 coding sequence fused to the 5′ terminus of GFP. In pUbi::IPE1ΔSP-GFP, the putative N-terminal signal peptide of the coding sequence was deleted and fused to GFP. pUbi::SP-GFP represented the DNA fragment of the putative N-terminal signal peptide fused with GFP. These constructs including pUbi::GFP were cointroduced with the endoplasmic reticulum marker red fluorescent protein (RFP)-KDEL (Liu et al., 2012) into the protoplast isolated from the maize seeding. Observations by confocal laser scanning microscopy showed that the free GFP signal was distributed widely in the cytoplasm (Fig. 9, A–D). IPE1-GFP fluorescence coincided with the fluorescence of the endoplasmic reticulum (Fig. 9, E–H). In contrast, IPE1ΔSP-GFP fluorescence did not clearly colocalize with the fluorescence of the endoplasmic reticulum (Fig. 9, I–L). Furthermore, the putative signal peptide of IPE1 could transfer GFP to the endoplasmic reticulum (Fig. 9, M–P). Therefore, the IPE1 protein was located in the ER.
Figure 9.
Subcellular localization of the IPE1 protein in maize protoplasts. A to D, A maize protoplast showing green fluorescent signals by expressing pUbi::GFP (A), red fluorescent signals by expressing the endoplasmic reticulum marker RFP-KDEL (B), merged signals (C) of A and B, and bright field (D). E to H, A maize protoplast showing green fluorescent signals by expressing pUbi::IPE1-GFP (E), red fluorescent signals by expressing the endoplasmic reticulum marker RFP-KDEL (F), merged signals (G) of E and F, and bright field (H). I to L, A maize protoplast showing green fluorescent signals by expressing pUbi::IPE1ΔSP-GFP (I), red fluorescent signals by expressing the endoplasmic reticulum marker RFP-KDEL (J), merged signals (K) of I and J, and bright field (L). M to P, A maize protoplast showing green fluorescent signals by expressing pUbi::SP-GFP (M), red fluorescent signals by expressing the endoplasmic reticulum marker RFP-KDEL (N), merged signals (O) of M and N, and bright field (P). Bars = 5 μm.
Independent Roles of IPE1, MS26, and MS45 in Anther Development
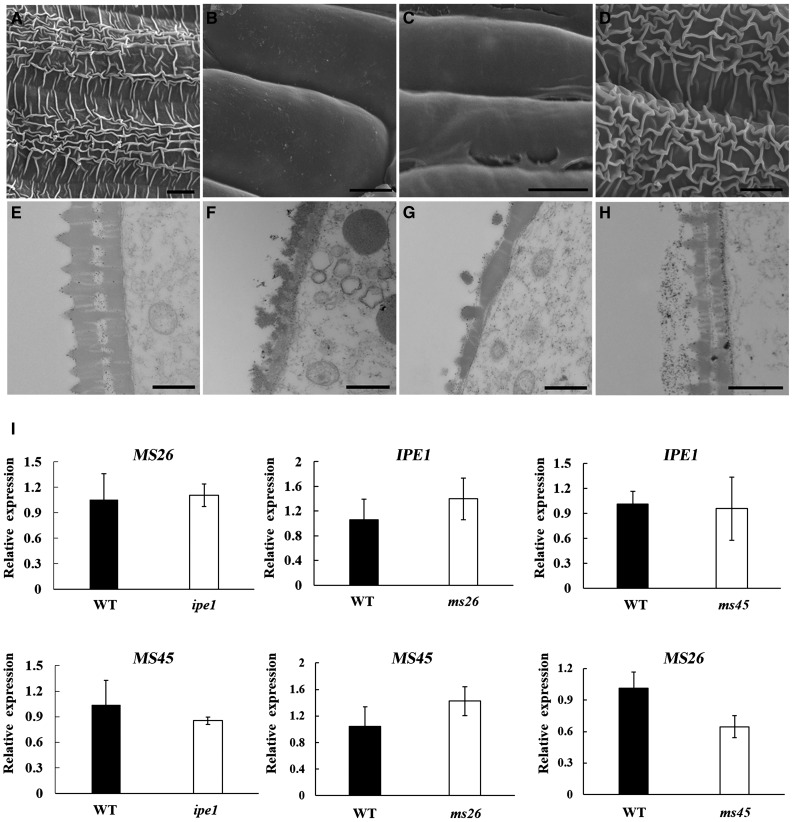
According to reports in the Maize Genetics and Genomics Database, two cloned genes, MS26 and MS45, had been reported to participate in microspore development. To elucidate in detail the roles of IPE1, MS26, and MS45 in regulating anther cuticle and pollen exine development, phenotypes of these mutants were analyzed. Available studies indicate that both anther cuticle and pollen exine development can be divided mainly into two steps: the formation of anther cuticle followed by maturation, and the formation of the three-layer structure of pollen exine followed by thickening (Li et al., 2010; Qin et al., 2013). Similar to the ipe1 phenotype, the outer surface of ms26 anthers was smooth at the mature pollen grain stage, and pollen exine did not form the typical three-layer structure at the late uninucleate microspore stage (Fig. 10, B and F). Compared with ipe1 and ms26, the ms45 anther epidermis was not glossy but lacked the normal reticular pattern (Fig. 10D). The typical three-layer structure of the pollen exine was well organized but thinner than that of the wild type (Fig. 10H). These results demonstrated that the similar functions of IPE1 and MS26 differed from that of MS45: IPE1 and MS26 are involved in the formation of the anther cuticle and pollen exine, whereas MS45 serves in anther cuticle maturation and pollen exine thickening.
Figure 10.
Phenotypic and expression analyses of IPE1, MS26, and MS45. A to D, Scanning electron micrographs of the anther outer surface at the mature pollen grain stage. The mature reticular anther cuticle was observed in the wild type (A), while its formation was disturbed in ms26 (B) and ipe1 (C), and ms45 anther cuticle did not mature normally (D). Bars = 6 μm. E to H, Transmission electron micrographs of the pollen wall at the late uninucleate microspore stage. The wild-type pollen exine had three thick layers composed of the tectum, bacula, and nexine (E), whereas the pollen exine of ms26 (F) and ipe1 (G) did not form the three-layer structure, and the thickening of pollen exine was affected in the ms45 mutant (H). Bars = 0.5 μm. I, Transcript levels of IPE1, MS26, and MS45 in reciprocal mutants, showing that their expression was not apparently altered by reciprocal mutations. WT, Wild type. Values are means ± sd of three replicates.
Expression analysis showed that MS26 and MS45 expression was observed only in young spikelets and was higher in anthers at the early uninucleate microspore stage (Supplemental Fig. S7). Subsequently, we investigated transcript levels of MS26 and MS45 in wild-type and ipe1 anthers at the early uninucleate microspore stage, and no distinct difference was detected. Similarly, the expression of IPE1 and MS45 in ms26 and the expression of IPE1 and MS26 in ms45 did not differ (Fig. 10I). These results suggested that these three genes act independently in controlling anther development.
Transcriptome Analysis of Wild-Type and ipe1 Anthers
To further dissect the regulatory network of IPE1 during anther cuticle and pollen wall development, we performed transcript profiling of ipe1 anthers at the early uninucleate microspore stage by RNA sequencing (RNA-Seq) technology. Using a cutoff (false discovery rate < 0.05 and fold change > 2), we identified 3,085 differentially expressed genes (DEGs), including 1,218 up-regulated genes and 1,867 down-regulated genes, in ipe1 anthers. To validate the results, we randomly selected nine DEGs for qRT-PCR (Supplemental Fig. S8). The correlation of fold change values between RNA-Seq and qRT-PCR was 0.98, indicating that our RNA-Seq data were reliable. The analysis of Gene Ontology (GO) term enrichment indicated that the DEGs were involved in many biology processes, some of which were related to anther cuticle and microspore development, such as lipid biosynthetic processes and carbohydrate metabolic processes (Supplemental Fig. S9).
Fatty acid synthesis and elongation are essential for the formation of anther cuticle and pollen exine, which involve many enzymatic conversions. In plants, heteromeric acetyl-CoA carboxylase and fatty acid synthase are involved in de novo fatty acid biosynthesis, occurring in plastids and resulting in acyl-ACP moieties that are 16 or 18 carbons long (O’Hara et al., 2001). In ipe1 anthers, the expression of two genes (GRMZM2G124335 and GRMZM2G072205) involved in fatty acid biosynthesis was decreased (Table II). Very-long-chain fatty acids can be synthesized by a fatty acid elongase, 3-ketoacyl-CoA synthase, in vegetative tissues (Todd et al., 1999). Our results showed that transcription levels of 11 genes encoding β-ketoacyl-CoA synthase were altered in ipe1 (Table II). Alkanes are the main component of wax. In Arabidopsis, the protein ECERIFERUM1 (CER1) is associated with long-chain alkane synthesis and pollen exine development (Aarts et al., 1995). In our study, GRMZM2G066578, a homolog of CER1, was down-regulated in ipe1 anthers. However, another wax synthesis gene, GRMZM2G114642, was up-regulated (Table II).
Table II. A selection of DEGs putatively involved in anther cuticle and pollen exine development.
| Gene Identifier | Gene Description | Log2 (ipe1/Wild Type) |
|---|---|---|
| GRMZM2G066578 | Wax synthesis | −4.28 |
| GRMZM2G114642 | Wax synthesis | 4.67 |
| GRMZM2G422750 | Chalcone synthase | −3.56 |
| GRMZM2G151227 | Chalcone synthase | −6.66 |
| GRMZM2G346095 | Chalcone synthase | 2.38 |
| GRMZM2G175812 | Chalcone synthase | 4.93 |
| GRMZM2G114471 | Chalcone synthase | 2.69 |
| GRMZM2G124335 | Ketoacyl-ACP synthase | −3.55 |
| GRMZM2G072205 | Ketoacyl-ACP synthase | −4.67 |
| GRMZM2G022558 | β-Ketoacyl-CoA synthase | −2.58 |
| GRMZM2G162508 | β-Ketoacyl-CoA synthase | −2.67 |
| GRMZM2G168304 | β-Ketoacyl-CoA synthase | −2.14 |
| GRMZM2G445602 | β-Ketoacyl-CoA synthase | −3.34 |
| GRMZM2G164974 | β-Ketoacyl-CoA synthase | −1.73 |
| GRMZM2G003501 | β-Ketoacyl-CoA synthase | −2.42 |
| GRMZM2G063024 | β-Ketoacyl-CoA synthase | −3.59 |
| GRMZM2G097469 | β-Ketoacyl-CoA synthase | 3.19 |
| GRMZM2G149636 | β-Ketoacyl-CoA synthase | 2.07 |
| GRMZM2G075140 | β-Ketoacyl-CoA synthase | 2.41 |
| GRMZM2G003138 | β-Ketoacyl-CoA synthase | 3.22 |
Flavonoids also were considered constituents of the pollen wall, and several genes participating in flavonoid metabolism were identified. The expression of five genes involved in the synthesis of chalcones was altered, including two down-regulated genes (GRMZM2G422750 and GRMZM2G151227) and three up-regulated genes (GRMZM2G346095, GRMZM2G175812, and GRMZM2G114471; Table II).
DISCUSSION
IPE1 Is Crucial for the Development of the Anther Cuticle and Pollen Exine in Maize
In this study, we showed that ipe1 anthers exhibited a smooth outer surface of the epidermis (Fig. 3G; Supplemental Fig. S3). However, there were no apparent differences in the cuticular structures of stems, leaves, and glumes between the wild type and ipe1 (Supplemental Fig. S10), which was consistent with the expression of the IPE1 gene being detected only in the anther (Fig. 8A). The outer surface phenotype of ipe1 anthers was very similar to that of mutants defective in the formation of the anther cuticle, such as wda1 (Jung et al., 2006), dpw (Shi et al., 2011), cyp704b2 (Li et al., 2010), cyp703a3 (Yang et al., 2014), osabcg15 (Niu et al., 2013; Qin et al., 2013), osabcg26 (Zhao et al., 2015), and tdr (Li et al., 2006), in rice. Therefore, IPE1 plays a vital role in the development of the anther cuticle in maize.
The anther cuticle is made of cuticular wax and cutin synthesized in the tapetum. In this study, we detected 17.37 ng mm−2 total wax and 601.79 ng mm−2 total cutin in wild-type anthers, suggesting that cutin is the major component of maize anther epidermis compared with the wax. Furthermore, the levels of total cutin and all cutin monomers in the ipe1 mutant were decreased significantly while that of total wax was not obviously affected, indicating that the function of IPE1 is involved in the synthesis of cutin. However, in the Arabidopsis hothead-12 (hth-12) mutant, a homolog of IPE1, most of the cutin monomer remains unchanged, except for a reduction in the amount of C16 and C18 α-,ω-dicarboxylic acids and an increase in that of ω-hydroxy acids (Kurdyukov et al., 2006). This disagreement between the metabolic changes in maize and Arabidopsis may result from the fact that the functions of IPE1 in maize and HTH in Arabidopsis are diverse. IPE1 is involved in anther cuticle and pollen wall formation, and HTH is involved in the fusions of floral buds. Moreover, IPE1 in maize may have less redundancy than in Arabidopsis. Surprisingly, a significant change of ipe1 anther wax constituents was detected (Fig. 5). Therefore, we assumed that IPE1 controls wax metabolism by feedback regulation. Consistent with this assumption, our RNA-Seq data showed that the expression of two wax synthesis genes was altered in the mutant (Table II). It should be noted that fatty acids are important precursors of anther wax and cutin (Shi et al., 2015). Chemical analysis revealed that ipe1 anthers had a dramatic reduction in fatty acids (Table I), which was further confirmed by the fact that 11 genes involved in fatty acid synthesis and elongation were expressed differently in ipe1 (Table II). Taken together, these data suggested that the IPE1 mutation leads to the alteration of anther aliphatic molecules, eventually resulting in abnormal anther cuticle.
The tapetum is essential for sporopollenin deposition and pollen exine synthesis, which commence at the tetrad stage; the structure of the pollen exine is evident at the uninucleate microspore stage. Afterward, the size increases gradually (Blackmore et al., 2007). Our TEM observations revealed that sporopollenin deposition and the morphology of ipe1 pollen exine were defective, due to the bacula being undeveloped at the early uninucleate microspore stage (Fig. 4, N and X). Similarly, the development of bacula structure and pollen exine is disrupted in rice tdr (Zhang et al., 2008) and cyp704b2 (Li et al., 2010) and Arabidopsis acyl-coa synthetase5 (de Azevedo Souza et al., 2009). These data indicated that the bacula is indispensable for the sporopollenin deposition required for pollen exine synthesis. In addition, the abnormal pollen exine resulted from the dysfunction of Ubisch bodies in transferring sporopollenin precursors (Fig. 4T). Furthermore, IPE1 was expressed specifically in anthers during the tetrad and uninucleate microspore stages (Fig. 8, F and G), when the primexine and bacula were developing. Strikingly, IPE1 was expressed mainly in the tapetum, where sporopollenin precursors were synthesized. Collectively, our data indicated that IPE1 is crucial for pollen exine development.
IPE1 Mediates the Lipid Metabolic Pathway in the Tapetum
Hydroxyacids and dicarboxylic acids are the important constituents of the protective cuticle in anthers (Li et al., 2010; Shi et al., 2011). However, the mechanism underlying the conversion from hydroxyacids to dicarboxylic acids in the reproductive organs is little known. It is believed that modifications of fatty acids from the plastid occur in the endoplasmic reticulum, which contributes to anther cuticle and pollen wall development (Kunst and Samuels, 2003; Lallemand et al., 2013). In our work, we cloned a novel maize male-sterile gene, IPE1, putatively encoding a member of the GMC oxidoreductase superfamily. Further analysis revealed that the IPE1 protein was targeted to the endoplasmic reticulum (Fig. 9). These results supported the hypothesis that IPE1 regulates fatty acid metabolism.
GMC oxidoreductases have a wide variety of substrates and can catalyze the oxidation of an alcohol to the corresponding aldehyde (Dreveny et al., 2001; Wongnate and Chaiyen, 2013). In Arabidopsis, the ADHESION OF CALYX EDGES (ACE)/HTH protein was found to be a single-domain protein related to GMC oxidoreductase domain-containing proteins and could oxidize long-chain ω-hydroxy fatty acids produced in the ω-oxidation pathway of cytochrome P450 fatty acid ω-hydroxylases. Supportively, the content of α-,ω-dicarboxylic fatty acids has been found to decrease and that of ω-hydroxy fatty acids to increase in the ace/hth mutant, resulting in defects in cuticle development (Krolikowski et al., 2003; Kurdyukov et al., 2006). Meanwhile, the biochemical activity of ω-hydroxyacid dehydrogenase and ω-oxoacid dehydrogenase has been proven in epidermis of young Vicia faba leaves and potato (Solanum tuberosum) discs (Kolattukudy et al., 1975; Agrawal and Kolattukudy, 1977). Consistently, an abnormal anther cuticle and the significant reduction of C16:0 and C18:2 diacids were observed in the ipe1 mutant. However, in contrast to the ace/hth mutant, the amount of ω-hydroxy fatty acids detected in ipe1 anthers and C16 and C18 fatty acids was reduced dramatically. The discrepancy could be explained by the following reasons. (1) In lipid metabolism, some other genes are involved in the metabolism of C16 and C18 fatty acids and C16/C18 ω-hydroxy fatty acids, such as POLYKETIDE SYNTHASE A and POLYKETIDE SYNTHASE B (Kim et al., 2010). IPE1 may regulate their metabolism by the feedback pathway. (2) Mutation of the IPE1 gene leads to wilted anthers, which may cause the degradation of some fatty acids. In addition, MS26 may hydroxylate C16 and C18 fatty acids at the ω-carbon position (Dobritsa et al., 2009; Li et al., 2010; Djukanovic et al., 2013). To sum up, we speculate that the major function of IPE1 in anther development is as follows: ω-hydroxy C16/C18 fatty acids formed by MS26 can be converted into C16/C18 diacids by IPE1 in the tapetum and long-chain ω-aldehyde dehydrogenases, which will further enrich the lipid metabolic pathway in anthers, especially the formation of dicarboxylic acids. Surprisingly, C16/C18 diacids only account for less than 1% of the total amount of cutin in our study. How do the diacids play the vital role in anther development? This may be caused by the following reasons. (1) The diacids are not the main constituent but are a crucial constituent for anther cuticle and pollen exine development. (2) The altered conversion from hydroxy C16/C18 fatty acids to C16/C18 diacids will result in the disruption of other metabolic pathways involved in cutin and wax synthesis. Compared with previous reports (Li et al., 2010; Shi et al., 2011), we detected that the C18:0 9/10 di-OH diacid is a cutin monomer and that the level is reduced significantly in the mutant, indicating that IPE1 participates in the metabolism of the C18:0 9/10 di-OH diacid indirectly.
Interestingly, our phenotypic analysis found that the tapetum was degraded early in the mutant (Figs. 2R and 4R), consistent with the preferential expression patterns of IPE1 in tapetum, suggesting that IPE1 can control tapetal development. Transcription factors are the master regulator of the development of tapetal cells and pollen exine. In Arabidopsis and rice, many genes encoding enzymes needed for wax, cutin, and sporopollenin biosynthesis can be modulated by transcription factors (Shi et al., 2015). For example, CYP703A3, encoding a cytochrome P450 fatty acid hydroxylase, can be regulated by GAMYB and TDR and can be involved in tapetal degradation (Yang et al., 2014). In Arabidopsis, DYSFUNCTIONAL TAPETUM1 (Gu et al., 2014), ABORTED MICROSPORES (Xu et al., 2010, 2014), and MS1 (Wilson et al., 2001; Ito et al., 2007; Yang et al., 2007) can regulate multiple genes involved in pollen wall development. Thus, we hypothesized that IPE1 indirectly regulates tapetal degeneration via transcription factors.
IPE1, MS26, and MS45 Cooperatively Regulate Anther Cuticle and Pollen Exine Development in an Independent Manner
Aliphatic materials containing wax, cutin, and sporopollenin share one common catalytic pathway in the tapetum, and they will be transported to the surface of anthers and microspores (Zhao et al., 2015). In maize, knowledge about the development of the anther cuticle and pollen exine is limited (Cigan et al., 2001; Skibbe and Schnable, 2005; Djukanovic et al., 2013). In this study, we compared three male-sterile mutants of maize (ipe1, ms26, and ms45) in which the phenotypes of pollen exine and anther cuticle were all defective (Fig. 10, A–H). In accordance with that, the three genes were expressed specifically in the anthers (Fig. 8A; Supplemental Fig. S7). Furthermore, IPE1 and MS45 were expressed mainly in the tapetum (Cigan et al., 2001; Fig. 8, D–I). In addition, CYP704B2, the rice homolog of MS26, also was expressed in the tapetum (Li et al., 2010). The phenotypes of ipe1, ms26, and ms45, coupled with their expression patterns, suggested that IPE1, MS26, and MS45 are all required for anther cuticle and pollen exine development. However, their phenotypes were different: the anther outer surface was smooth in ipe1 and ms26, while in ms45, it was not smooth but lacked the regular reticular structure, implying that IPE1 and MS26 play a role in the formation of the anther cuticle whereas MS45 functions in its maturation. The three layers of pollen exine did not form in ipe1 and ms26 and appeared but did not thicken in ms45, indicating that IPE1 and MS26 initiate pollen exine formation and MS45 enhances its thickness. Furthermore, expression pattern analysis of the three genes suggested that they function independently in anther development (Fig. 10I).
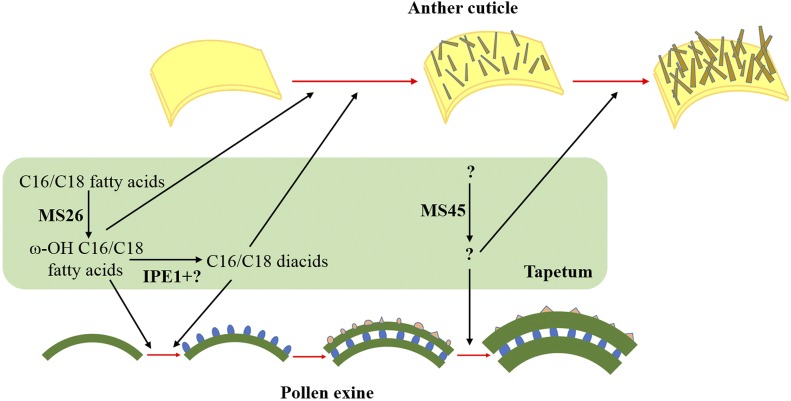
Taken together, we propose an initial working model of how IPE1 regulates anther cuticle and pollen wall development in maize (Fig. 11). In the tapetum, MS26 functions as a cytochrome P450 monooxygenase to generate ω-hydroxy C16/C18 fatty acids, which can be oxidized to C16/C18 dioic acids by IPE1 and corresponding aldehyde dehydrogenases. ω-Hydroxy C16/C18 fatty acids and dioic acids can be transferred to the surface of anthers and microspores to form the anther cuticle and the three layers of pollen exine. Afterward, the MS45 product participates in the maturation of the anther cuticle and thickening of the pollen exine.
Figure 11.
Model for the role of IPE1 during anther cuticle and pollen exine development. C16/C18 fatty acids synthesized in the plastid are transported to the endoplasmic reticulum and hydroxylated to ω-hydroxy C16/C18 fatty acids by MS26. After IPE1 and an aldehyde dehydrogenase reaction, ω-hydroxy C16/C18 fatty acids are converted to C16/C18 diacids. ω-Hydroxy C16/C18 fatty acids and C16/C18 diacids are required for anther cuticle and pollen exine formation. The product formed by MS45 is involved in the maturation of the anther cuticle and thickening of the pollen exine.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Maize (Zea mays) mu1019442::Mu (ipe1-1), ms*-6044(ipe1-3), ms26, and ms45-6006 (ms45) were obtained from the Maize Genetics Cooperation Stock Center. ipe1-2 was a radiation-treated mutant. The Arabidopsis (Arabidopsis thaliana) T-DNA insertional lines were from The Arabidopsis Information Resource. All maize materials with the exception of ms*-6044(ipe1-3) were grown in experimental stations of the China Agricultural University in Beijing and San Ya. ms*-6044(ipe1-3) was grown in experimental stations of the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences in Beijing and San Ya. The maize transgenic plants and population were grown in a greenhouse of the Institute of Genetics and Developmental Biology. ipe1 was backcrossed to inbred line zheng58 five times for morphological comparisons. Arabidopsis lines were grown in a greenhouse under 16 h of light/8 h of dark at 22°C.
Characterization of Phenotypes
A Nikon D40 digital camera or Olympus SZX10 dissecting microscope was used to take photographs for plants and flowers.
For SEM, anthers at different stages were prefixed in 2.5% glutaraldehyde overnight at 4°C and then rinsed three times using phosphate-buffered saline (pH 7.2). Samples were postfixed for 1 h in 1% osmium tetraoxide and rinsed three times using phosphate-buffered saline. The samples were dehydrated using a graded ethanol series (30%–100%) and then rinsed three times with isoamyl acetate. The samples were critical point dried and gold coated. The observation and recording of images were performed using a Hitachi S-3400N scanning electron microscope.
For TEM analysis, anthers were vacuum infiltrated and prefixed in 2.5% glutaraldehyde followed by rinsing with 0.1 m phosphate buffer. The samples were fixed in 1% osmium tetraoxide and rinsed with 0.1 m phosphate buffer. The fixed samples were dehydrated using an acetone series from 30% to 100% and embedded in epoxy resin. Ultra-thin sections were prepared using a Leica UC6 I ultramicrotome and were stained with uranyl acetate and double stained with lead citrate. Images were obtained with a JEM-1230 transmission electron microscope.
For meiosis analysis, young panicles from the wild type and ipe1 were harvested and fixed in Carnoy’s solution (ethanol:glacial acetic acid, 3:1, v/v). Anthers undergoing meiosis were crushed with forceps in an acetocarmine solution on glass slides and then covered with a cover slip. First, slides were examined with a light microscope, and then cover slips of ideal slides were removed in liquid nitrogen. Chromosomes were stained with DAPI, and images were captured with an Olympus BX61 fluorescence microscope.
Analysis of Anther Wax, Cutin, and Fatty Acids
To describe amounts per unit of surface area, we determined a ratio of anther weight to surface area (Supplemental Fig. S2). The area was calculated according to the length and width of anthers in microscope images, assuming a cylindrical body for maize anthers. To extract waxes, 200 mg of fresh anther sample corresponding to 1,521 to 2,292 mm2 of surface area was submersed in 3 mL of chloroform for 1 min. The resulting chloroform extracts were spiked with 30 µg of docosanol and 10 µg of heptadecanoic acid as internal standards and transferred to a new vial. The solvents were evaporated under a gentle stream of nitrogen gas. The residue was derivatized with 100 µL of N-methyl-N-(trimethylsilyl) trifluoroacetamide and incubated for 1 h at 50°C. To extract anther cutin, the remaining anthers were submersed in 3 mL of chloroform:methanol (1:1, v/v). They were first incubated at 50°C for 30 min and then 72 h with constant shaking at room temperature. The anthers were freeze dried and submersed in 1 mL of 1 n methanolic HCl for 2 h at 80°C with 10 µg of heptadecanoic acid as an internal standard. The hydrophobic monomers were subsequently extracted three times with 1 mL of hexane after the addition of 2 mL of saturated NaCl solution. The solvent evaporated, and the remaining samples were derivatized as described above. These derivatized samples were then analyzed by GC-MS (Agilent gas chromatograph coupled to an Agilent 5975C quadrupole mass selective detector). The results of anther wax and cutin analysis were related to unit surface area.
To extract total fatty acids, 6 mg of freeze-dried anther material was trans-esterified in 1 mL of 1 n methanolic HCl (containing 50 µg of heptadecanoic acid as an internal standard) for 4 h at 50°C. After the addition of 1.5 mL of 0.9% (w/v) NaCl, the hydrophobic monomers were subsequently extracted with 1.5 mL of hexane. The organic phases were evaporated under a gentle stream of nitrogen gas. The residue was dissolved in 100 µL of hexane and then analyzed by GC-MS (Agilent gas chromatograph coupled to an Agilent 5975C quadrupole mass selective detector). The results of the total fatty acid analysis were related to unit dry weights of anthers.
Molecular Cloning and Allelism Test
ipe1 was crossed with inbred line zheng58, and the F1 plants were self-pollinated to generate an F2 population; individuals with mutations were chosen for the mapping population. Simple sequence repeat markers distributed on 10 chromosomes were used for initial mapping. For fine-mapping, several Indel markers were designed with Oligo7 software. Primers used for fine-mapping are listed in Supplemental Table S2. The genotype was confirmed with 3% agarose gels. For the allelism test, six genetic crosses were produced among ipe1, ipe1-1, ipe1-2, and ipe1-3, whose progeny were analyzed for the segregating ratio between fertility and sterility.
Complementation of the Mutant
For functional complementation of the maize ipe1-3 mutant, the DNA fragment was obtained with primers 1F (5′-GAAGAAAGGTTGCGTCAG-3′) and 1R (5′-ATTGCGATGGGAGCGTAT-3′) and digested with HindIII and KpnI. The digested fragment was cloned into the binary vector pCAMBIA1300 to generate the p1300:IPE1 plasmids. These plasmids contained a 5,198-bp genomic fragment with the entire 2,585-bp IPE1 gene region, a 1,644-bp upstream sequence region, and a 969-bp downstream region and were transformed into the maize hybrid Hi-II with Agrobacterium tumefaciens AGL1. We backcrossed the positive transgenic individuals to ipe1-3 continuously twice to obtain positive transgenic plants with the ipe1-3 background and assayed its mature pollen grains with 1% I2-KI staining solution.
Constructs for CRISPR-Cas9 and Transformation
The vector pBUN411 from Genovo Biotech was used, and a 23-bp fragment from the second exon of IPE1 (sequence as shown in Fig. 6D) was introduced into the vector and then used for A. tumefaciens-mediated transformation into maize (Hi-II). Transgenic plants were identified by the enzyme digestion reaction with XmaI/SmaI and PCR amplification using the primers 2F (5′-CGCCCCTGGTGTCGCAGTACA-3′) and 2R (5′-CGCCGACAGGATCACCTCGTTC-3′). Mature pollen grains of transgenic lines were observed by I2-KI staining.
Phylogenetic Analysis
The full-length amino acid sequence of IPE1 and 17 of the most similar sequences identified via a BLAST search were aligned with the ClustalW tool using default parameters. A phylogenetic tree was constructed with the alignment of IPE1-like protein sequence using MEGA 5 with the following parameters: Poisson model, complete deletion, and 1,000 bootstrap replicates.
RNA Extraction, RT-PCR, and qRT-PCR
Total RNA was isolated with TRIzol reagent (Invitrogen) from maize roots, stems, glumes, young spikelets, cobs, pistils, and anthers and Arabidopsis flowers. Total RNA (1.5 µg) was treated with DNase I (Promega), and RT was conducted using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT)20 primers. RT product (1 µL) was used as the template for PCR. qRT-PCR was performed on an ABI 7500 real-time PCR system with SYBR Green PCR Master Mix (Takara). ZmACTIN and AtACTIN were used as the internal normalization controls. Every sample contained three biological replications and three technological replications. Relative expression was calculated using the 2−ΔΔCt method. The sd was calculated with three biological replications. All primers used for RT-PCR and qRT-PCR are listed in Supplemental Table S2.
In Situ Hybridization
The anthers of inbred line zheng58 at the different developmental stages were fixed overnight in 3.7% formalin-acetic acid-alcohol (3.7% formaldehyde, 5% glacial acetic acid, and 50% ethanol), dehydrated in a graded ethanol series (50%, 70%, 85%, 95%, and 100%), and then embedded in paraffin and sliced into 8-μm sections. The construct of the IPE1 probe was generated with the gene-specific primer (primer sequence is given in Supplemental Table S2). Probe synthesis and RNA in situ hybridization were performed according to previously described protocols (Ding et al., 2015).
Subcellular Localization of IPE1
For the analysis of subcellular localization, three fragments (the 1,746-bp coding sequence without the stop codon [IPE1], the 1,653-bp coding sequence without the 93-bp fragment encoding the putative signal peptide and stop codon [IPE1ΔSP], and the N-terminal signal peptide-encoding sequence [SP]) were amplified with the primers GFP-F/R, ΔSP-GFP-F/GFP-R, and GFP-F/SP-GFP-R and cloned into pJIT163-GFP to obtain C-terminal fusions with GFP. Maize protoplasts were isolated as described (Yoo et al., 2007), and the endoplasmic reticulum marker RFP-KDEL (Liu et al., 2012) was cotransformed with the plasmids pUbi::IPE1-GFP, pUbi::IPE1ΔSP-GFP, pUbi::SP-GFP, and pUbi::GFP individually using the polyethylene glycol-mediated method. Transformed protoplasts were incubated on six-well plates in the dark at 23°C overnight. Fluorescence was determined with a fluorescence confocal microscope (Zeiss LSM 710).
RNA-Seq Analysis
Anthers were collected from the wild type and ipe1 at the early uninucleate microspore stage, with two biological replicates for each genotype. Total RNA was extracted with TRIzol reagent (Invitrogen). Library construction was conducted according to Illumina TruSeq mRNA construction and sequenced on a HiSeq 2500 sequencer. Raw reads were preprocessed to remove low-quality bases and cut adapter sequences by employing the SolexQA (Cox et al., 2010) and Cutadapt (Martin. 2011) tools. Clean reads were then mapped to the maize genome sequence (AGPv3; MaizeSequence.org) using TopHat (Trapnell et al., 2009) with default parameters. Read counts of annotated genes were obtained separately by HTSeq-count (Anders et al., 2015). Lowly expressed genes were removed, and only genes with an expression level of at least 1 count per million in at least two samples were retained for further analysis. The R package edgeR was then employed to identify the DEGs (Robinson et al., 2010). Genes with more than 2-fold change in expression level and a false discovery rate less than 0.05 were considered DEGs. GO terms for each maize gene were obtained at http://www.gramene.org/. The GO enrichment analysis for DEGs was done with the R package TopGO (Alexa et al., 2006). Adrian Alexi’s improved weighted scoring algorithm and Fisher’s exact test were used to determine the significance of GO term enrichment. A GO term with P < 0.01 was thought to be a significant enrichment GO term.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MS26, NM_001137176.1; MS45, XM_008662089.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. DAPI staining of chromosomes of the wild type and ipe1 in meiosis.
Supplemental Figure S2. Weight-surface area ratio of wild-type and ipe1 anthers.
Supplemental Figure S3. SEM observation of the anther surface.
Supplemental Figure S4. Phylogenetic tree of IPE1-related proteins.
Supplemental Figure S5. Sequence alignment of IPE1 and 17 IPE1-related proteins.
Supplemental Figure S6. Phenotypes of two additional T-DNA insertional lines of AT1G12570.
Supplemental Figure S7. Spatial and temporal expression of MS26 and MS45 by RT-PCR.
Supplemental Figure S8. qRT-PCR analysis of nine randomly selected DEGs in ipe1.
Supplemental Figure S9. Significantly enriched GO terms of the up-regulated genes and down-regulated genes between the wild type and ipe1.
Supplemental Figure S10. SEM analysis of the surfaces of glume, leaf, and stem in the wild type and ipe1.
Supplemental Table S1. Detailed cutin compositions of wild-type and ipe1 anthers.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Fertile and sterile plant analysis of F1 plants from genetic crosses among ipe1, ipe1-1, ipe1-2, and ipe1-3.
Supplemental Table S4. Putative expression patterns of IPEI homologous genes.
Supplementary Material
Acknowledgments
We thank Dr. Xiaolan Zhang (China Agricultural University) and Chunju An (China Agricultural University) for valuable comments in the preparation of this article; Caixia Gao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the pUbi::GFP construct and for maize transformation; Fengxia Zhang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for wax, cutin, and fatty acid assays and primary analysis; and Yanbao Tian (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the subcellular localization of IPE1.
Glossary
- DAPI
4′,6-diamidino-2-phenylindole
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
- GC-MS
gas chromatography-mass spectrometry
- Indel
insertion-deletion
- RT
reverse transcription
- qRT
quantitative reverse transcription
- RNA-Seq
RNA sequencing
- DEGs
differentially expressed genes
- GO
Gene Ontology
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31471499, 31271729, 91535206, and 31421005), the National Key Research and Development Program of China (grant no. 2016YFD0101201), and the Ministry of Agriculture of China (grant no. 2016ZX08009003–003).
Articles can be viewed without a subscription.
References
- Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7: 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal VP, Kolattukudy PE (1977) Biochemistry of suberization: ω-hydroxyacid oxidation in enzyme preparations from suberizing potato tuber disks. Plant Physiol 59: 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen MC, Fox TW, Huffmann G, Trimnell M August 29, 2006. Nucleotide sequences affecting plant male fertility and methods of using same. United States Patent Application No. 7098388B2
- Albertsen MC, Fox TW, Trinmell MR (1993) Cloning and utilizing a maize nuclear male sterility gene. Proc Annu Corn Sorghum Ind Res Conf 48: 224–233 [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M (2012) Solving the puzzles of cutin and suberin polymer biosynthesis. Curr Opin Plant Biol 15: 329–337 [DOI] [PubMed] [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR (2007) Pollen wall development in flowering plants. New Phytol 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Chaubal R, Anderson JR, Trimnell MR, Fox TW, Albertsen MC, Bedinger P (2003) The transformation of anthers in the msca1 mutant of maize. Planta 216: 778–788 [DOI] [PubMed] [Google Scholar]
- Cigan AM, Unger E, Xu R, Kendall T, Fox TW (2001) Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex Plant Reprod 14: 135–142 [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ (2009) A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Yan S, Jiang L, Zhao W, Ning K, Zhao J, Liu X, Zhang J, Wang Q, Zhang X (2015) HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet 11: e1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukanovic V, Smith J, Lowe K, Yang M, Gao H, Jones S, Nicholson MG, West A, Lape J, Bidney D, et al. (2013) Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J 76: 888–899 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D (2009) CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Jiang C, Chen X, Zhang T, Ding L, Song W, Luo H, Lai J, Chen H, Liu R, et al. (2013) Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol 163: 1306–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreveny I, Gruber K, Glieder A, Thompson A, Kratky C (2001) The hydroxynitrile lyase from almond: a lyase that looks like an oxidoreductase. Structure 9: 803–815 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Sanders PM, Beals TP (1995) A novel cell-ablation strategy for studying plant development. Philos Trans R Soc Lond B Biol Sci 350: 5–17 [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJ, Braun D, Meeley R, Cande WZ (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119: 3306–3315 [DOI] [PubMed] [Google Scholar]
- Gu JN, Zhu J, Yu Y, Teng XD, Lou Y, Xu XF, Liu JL, Yang ZN (2014) DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J 80: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Hu L, Tan H, Liang W, Zhang D (2010) The Post-meiotic Deficicent Anther1 (PDA1) gene is required for post-meiotic anther development in rice. J Genet Genomics 37: 37–46 [DOI] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19: 3549–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, Lee YS, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim YW, et al. (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza CdeA, Geoffroy P, Heintz D, Krahn D, Kaiser M, et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy PE, Croteau R, Walton TJ (1975) Biosynthesis of cutin: enzymatic conversion of ω-hydroxy fatty acids to dicarboxylic acids by cell-free extracts of Vicia faba epidermis. Plant Physiol 55: 875–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE (2003) Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J 35: 501–511 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bär S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A (2006) Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224: 315–329 [DOI] [PubMed] [Google Scholar]
- Lallemand B, Erhardt M, Heitz T, Legrand M (2013) Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol 162: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Harper LC, Golubovskaya I, Wang CR, Weber D, Meeley RB, McElver J, Bowen B, Cande WZ, Schnable PS (2007) Functional analysis of maize RAD51 in meiosis and double-strand break repair. Genetics 176: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Yin ZJ, Xiao L, Xu YN, Qu Q (2012) Identification and evaluation of ω-3 fatty acid desaturase genes for hyperfortifying α-linolenic acid in transgenic rice seed. J Exp Bot 63: 3279–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Moon J, Skibbe D, Timofejeva L, Wang CJ, Kelliher T, Kremling K, Walbot V, Cande WZ (2013) Regulation of cell divisions and differentiation by MALE STERILITY32 is required for anther development in maize. Plant J 76: 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu BX, He FR, He M, Ren D, Chen LT, Liu YG (2013) The ATP-binding cassette transporter OsABCG15 is required for anther development and pollen fertility in rice. J Integr Plant Biol 55: 710–720 [DOI] [PubMed] [Google Scholar]
- O’Hara P, Slabas AR, Fawcett T (2001) Fatty acid synthesis in developing leaves of Brassica napus in relation to leaf growth and changes in activity of 3-oxoacyl-ACP reductase. FEBS Lett 488: 18–22 [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Wang CJ, Golubovskaya IN, Szymaniak JM, Shi L, Hamant O, Zhu T, Harper L, Sheridan WF, Cande WZ (2009) Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc Natl Acad Sci USA 106: 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Qin P, Tu B, Wang Y, Deng L, Quilichini TD, Li T, Wang H, Ma B, Li S (2013) ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol 54: 138–154 [DOI] [PubMed] [Google Scholar]
- Rhee SJ, Seo M, Jang YJ, Cho S, Lee GP (2015) Transcriptome profiling of differentially expressed genes in floral buds and flowers of male sterile and fertile lines in watermelon. BMC Genomics 16: 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R (2008) Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Shi J, Tan H, Yu XH, Liu Y, Liang W, Ranathunge K, Franke RB, Schreiber L, Wang Y, Kai G, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe DS, Schnable PS (2005) Male sterility in maize. Maydica 50: 367–376 [Google Scholar]
- Stieglitz H. (1977) Role of β-1,3-glucanase in postmeiotic microspore release. Dev Biol 57: 87–97 [DOI] [PubMed] [Google Scholar]
- Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327: 818–822 [DOI] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17: 119–130 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Datla R, Selvaraj G (2003) The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc Natl Acad Sci USA 100: 14487–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ (2012) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139: 2594–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Skibbe DS, Walbot V (2013) Maize Male sterile 8 (Ms8), a putative β-1,3-galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod 26: 329–338 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wongnate T, Chaiyen P (2013) The substrate oxidation mechanism of pyranose 2-oxidase and other related enzymes in the glucose-methanol-choline superfamily. FEBS J 280: 3009–3027 [DOI] [PubMed] [Google Scholar]
- Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang D (2014) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19: 3530–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, et al. (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56: 979–994 [DOI] [PubMed] [Google Scholar]
- Yi B, Zeng F, Lei S, Chen Y, Yao X, Zhu Y, Wen J, Shen J, Ma C, Tu J, et al. (2010) Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J 63: 925–938 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zhao G, Shi J, Liang W, Xue F, Luo Q, Zhu L, Qu G, Chen M, Schreiber L, Zhang D (2015) Two ATP binding cassette G transporters, rice ATP Binding Cassette G26 and ATP Binding Cassette G15, collaboratively regulate rice male reproduction. Plant Physiol 169: 2064–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Shi J, Zhao G, Zhang D, Liang W (2013) Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J Plant Biol 56: 59–68 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.