In Arabidopsis, gamete fusion requires the C2H2 transcription factor SUF4, which regulates the expression of the EGG CELL1 gene family.
Abstract
The EGG CELL1 (EC1) gene family of Arabidopsis (Arabidopsis thaliana) comprises five members that are specifically expressed in the egg cell and redundantly control gamete fusion during double fertilization. We investigated the activity of all five EC1 promoters in promoter-deletion studies and identified SUF4 (SUPPRESSOR OF FRIGIDA4), a C2H2 transcription factor, as a direct regulator of the EC1 gene expression. In particular, we demonstrated that SUF4 binds to all five Arabidopsis EC1 promoters, thus regulating their expression. The down-regulation of SUF4 in homozygous suf4-1 ovules results in reduced EC1 expression and delayed sperm fusion, which can be rescued by expressing SUF4-β-glucuronidase under the control of the SUF4 promoter. To identify more gene products able to regulate EC1 expression together with SUF4, we performed coexpression studies that led to the identification of MOM1 (MORPHEUS’ MOLECULE1), a component of a silencing mechanism that is independent of DNA methylation marks. In mom1-3 ovules, both SUF4 and EC1 genes are down-regulated, and EC1 genes show higher levels of histone 3 lysine-9 acetylation, suggesting that MOM1 contributes to the regulation of SUF4 and EC1 gene expression.
The female gametophyte (FG) of flowering plants, also called the embryo sac, is the haploid generation that produces the two female gametes, the egg cell and the central cell. The development of the FG of Arabidopsis (Arabidopsis thaliana) is a morphologically well-described multistep process (from FG1 to FG7; Drews and Koltunow, 2011). The mature embryo sac of Arabidopsis consists of four different cell types that possess distinctive morphologies and hold defined positions within the FG: three antipodal cells are located at the chalazal pole of the FG (the proximal end of the ovule), a homodiploid central cell with a large vacuole occupies the center of the FG, while the egg cell and two adjacent synergid cells are located at the micropylar (distal) end of the FG (Schneitz et al., 1995; scheme in Fig. 1A). The entire FG is enclosed by the maternal tissues of the ovule.
Figure 1.
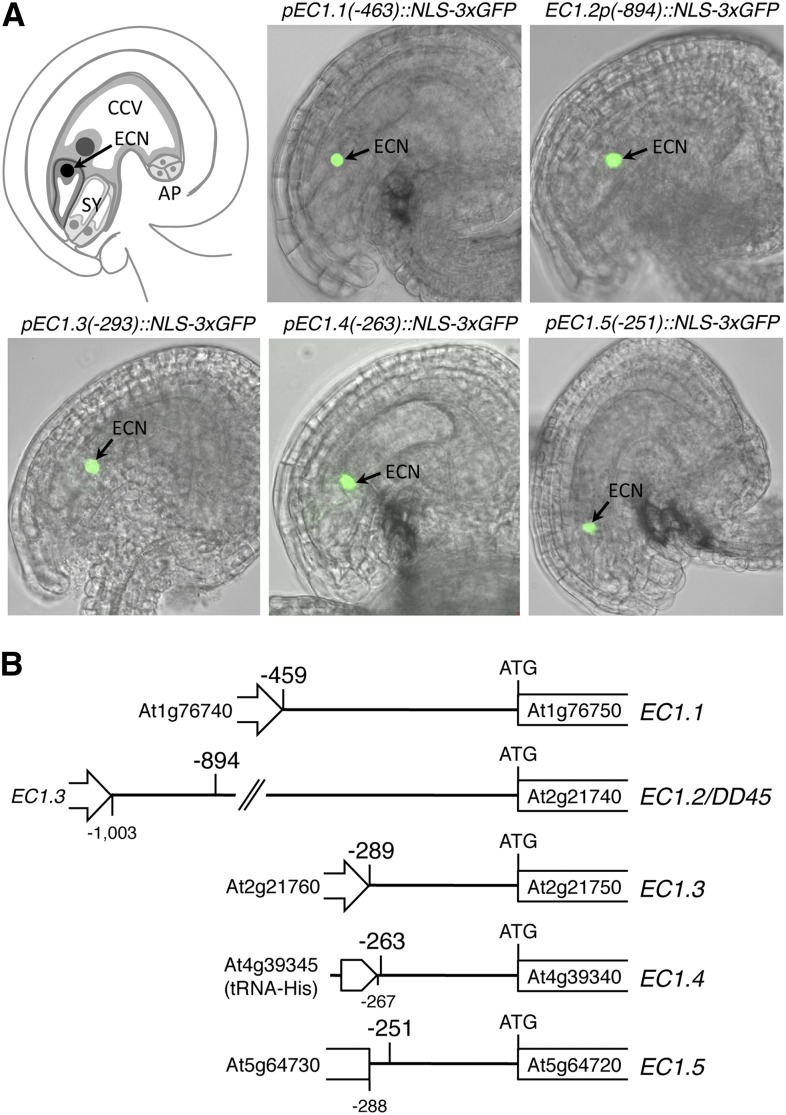
EC1 promoter regions drive egg cell-specific expression. A, Egg cell-specific reporter activity in mature ovules. Green fluorescent egg cell nuclei (arrows) indicate that all five promoters of the Arabidopsis EC1 gene family are functional and specifically active in the egg cell. AP, Antipodal cells; CCV, central cell vacuole; ECN, egg cell nucleus; SY, synergid cells. B, Schemes illustrating the genomic regions 5′ upstream of the sense strands of EC1 coding sequences. The position in a DNA sequence is designated relative to the predicted start codon (ATG) of the EC1 open reading frame. Arabidopsis Genome Initiative codes for EC1 genes and adjacent gene loci are given. Note that the promoter regions of EC1.3, EC1.4, and EC1.5 are short (−289 to −267 bp) but sufficient to drive egg cell-specific expression.
The molecular mechanisms regulating the establishment of cell identities within the FG are largely unknown, although several embryo sac-defective mutants have been isolated (Christensen et al., 1997; Pagnussat et al., 2005, 2007; Gross-Hardt et al., 2007; Matias-Hernandez et al., 2010; Masiero et al., 2011), and the impact of the phytohormones auxin and cytokinin on cell specification in the developing FG have become evident (Pagnussat et al., 2009; Yuan et al., 2016).
Besides genetic screens, a number of molecular approaches have been employed to clarify the mechanisms controlling embryo sac cell differentiation, such as differential gene expression analyses between the wild type and FG-defective mutants (Yu et al., 2005; Johnston et al., 2007; Jones-Rhoades et al., 2007; Steffen et al., 2007), microarray expression analysis of laser-dissected female gametophytic cells (Wuest et al., 2010), and exhaustive sequencing of ESTs from the cDNAs of manually isolated cells (Kumlehn et al., 2001; Lê et al., 2005; Márton et al., 2005; Yang et al., 2006; Koszegi et al., 2011). Isolation of egg cells and two-celled embryos from wheat (Triticum aestivum), by micromanipulation and subsequent EST analyses, resulted in the identification of the large, egg cell-specific transcript EST cluster termed EC1 (EGG CELL1; Sprunck et al., 2005). TaEC1 messengers encode small proteins having six conserved Cys residues and a predicted secretion signal sequence. Five EC1-related genes are present in the Arabidopsis genome, namely EC1.1, EC1.2, EC1.3, EC1.4, and EC1.5, all expressed exclusively in egg cells (Sprunck et al., 2012). Simultaneous silencing of all five EC1 genes prevents the fusion of the two male gametes with the egg cell and central cell during double fertilization. The observed sperm-activating effects of EC1 peptides suggest that EC1 proteins are secreted by the egg cell to promote sperm activation and, thereby, achieve rapid fusion with the female gametes (Sprunck et al., 2012; Rademacher and Sprunck, 2013).
To shed light on EC1 gene regulation, we investigated the promoter activities of all five EC1 genes in deletion studies and used the yeast one-hybrid approach to identify putative Arabidopsis EC1.1 transcriptional regulators. Among them, we identified the C2H2 zinc finger transcription factor SUPPRESSOR OF FRIGIDA4 (SUF4; Kim and Michaels, 2006). In vivo and in vitro evidence indicates that SUF4 is able to regulate all five EC1 genes; furthermore, suf4-1 mutants show a mild ec1 phenotype of delayed sperm fusion that can be rescued by the expression of pSUF4::SUF4-GUS. Bioinformatics approaches demonstrated that SUF4 is coexpressed with MOM1 (MORPHEUS’ MOLECULE1; Amedeo et al., 2000), and expression studies showed that SUF4 is down-regulated in mom1-3. Real-time reverse transcription (RT)-PCR analyses and genetic evidence indicate that MOM1 also controls EC1 expression by modulating the histone 3 Lys-9 acetylation (H3K9ac) of the EC1 loci.
RESULTS
EC1 Promoters Drive Egg Cell-Specific Expression of a Nucleus-Localized GFP Reporter
The promoter activities of EC1.1 and EC1.2/DD45 in the Arabidopsis egg cell have been reported previously (Steffen et al., 2007; Ingouff et al., 2009; Sprunck et al., 2012), while the upstream regulatory sequences of EC1.3, EC1.4, and EC1.5 have not been investigated to date. To compare the activity of all five EC1 promoters, we performed promoter-reporter studies using the nucleus-localized 3× GFP (NLS-3xGFP) as a reporter (Fig. 1A). Notably, all the EC1 promoters are able to drive a strong egg cell-specific expression of the reporter. Compared with the genomic regions 5′ upstream of the start codons of EC1.1 and EC1.2/DD45 (−459 and −1,003 bp, respectively), the 5′ upstream genomic regions of EC1.3, EC1.4, and EC1.5 are only 289 bp (EC1.3), 267 bp (EC1.4), and 287 bp (EC1.5) in length (Fig. 1B).
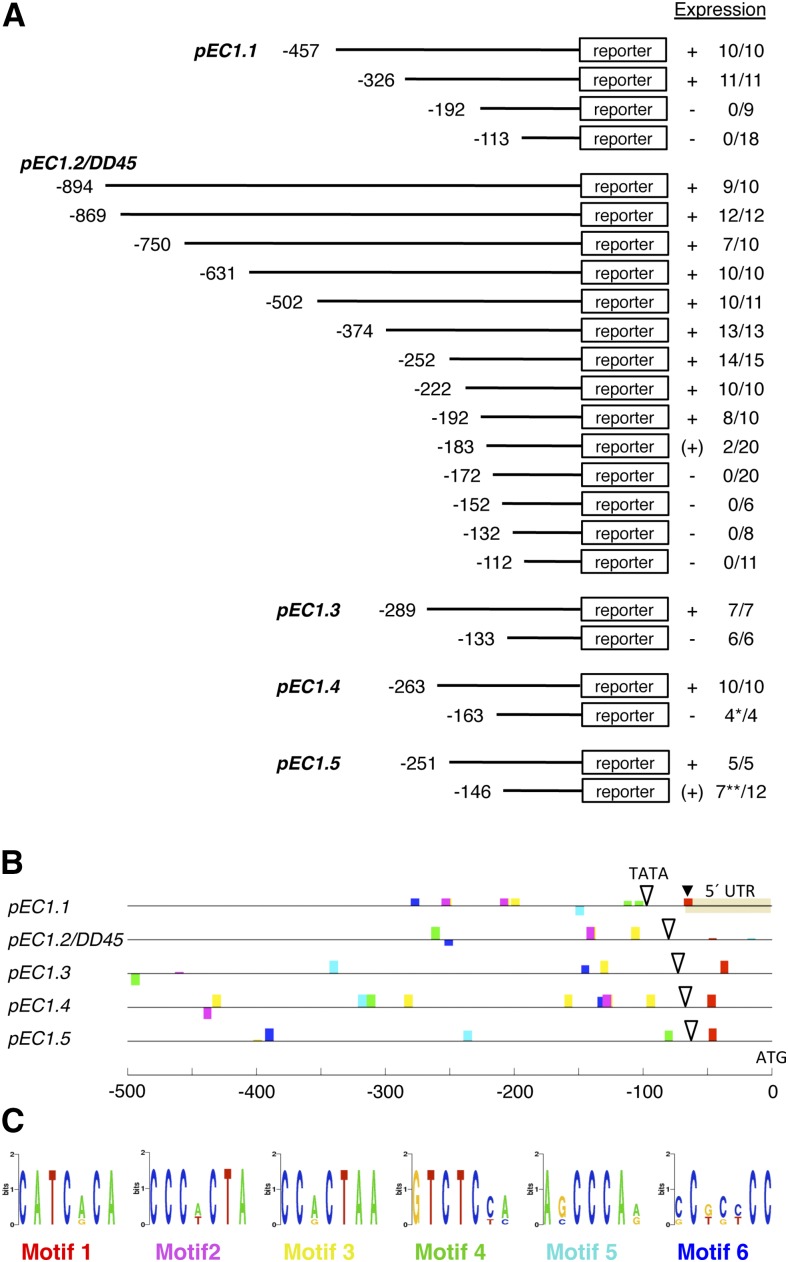
With the aim to narrow down the EC1 promoter regions sufficient to drive egg cell-specific gene expression, we generated a series of 5′ deletion constructs and investigated the ability of the deleted promoter fragments to drive reporter gene expression in vivo (Fig. 2A). Transgenic plants for the generated EC1.1 and EC1.2 promoter deletion constructs revealed that important cis-regulatory elements for egg cell-specific expression are located between −326 and −192 bp upstream of the translation start site of EC1.1 and between −192 and −172 bp upstream of the translation start site of EC1.2. Farther upstream, promoter deletions of EC1.1 and EC1.2 did not affect the reporter activity. The EC1.3(-133) and the EC1.4(-163) promoter deletions lost their ability to drive the expression of NLS-3xGFP in the egg cell. However, one of four independent pEC1.4(-163)::NLS-3xGFP lines exhibited ectopic fluorescence in the nuclei of sporophytic cells of the ovule. Seven of the 12 independent lines transgenic for pEC1.5(-146)::NLS-3xGFP showed expression of the reporter in the egg cell, but five of these lines revealed a very weak reporter activity, while two of the five lines showed ectopic expression of the NLS-3xGFP reporter in sporophytic cells of the ovule (Fig. 2A).
Figure 2.
EC1 promoter deletion studies and mapping of putative cis-regulatory motifs. A, Scheme summarizing the results from EC1 promoter deletion studies. A series of 5′ deletion constructs was tested for reporter activity in transgenic plants. Expression describes the observed reporter activity as present (+), weakly present [(+)], or absent (−) in the egg cell. Numbers indicate individual transgenic lines for a given deletion construct showing reporter activity compared with the total number of lines transgenic for this construct. *, One out of four lines showed misexpression of the reporter in sporophytic cells; **, five of seven lines showed only very weak reporter activity, and two of these five lines showed misexpression in sporophytic cells. B, Conserved sequence motifs (colored boxes) mapped in the −500-bp upstream regions of the five EC1 genes by Cistome (https://bar.utoronto.ca/cistome/cgi-bin/BAR_Cistome.cgi) using the prediction program MEME. White triangles mark the positions of TATA box motifs identified by AthMap (http://www.athamap.de/index.php). The transcription start site of EC1.1 is labeled with a black triangle. UTR, Untranslated region. C, Sequence logos of mapped sequence motifs shown in B. Motifs 2 and 3 show high sequence similarity.
Conserved Sequence Motifs in the EC1 Promoter Regions
To identify transcription factor-binding sites for TATA-binding proteins (TBPs), we used AthMap (http://www.athamap.de/index.php) and detected a putative TATA box in every EC1 promoter (Fig. 2B; Supplemental Table S1). The TATA box consensus sequence is TATAAA (EC1.1, EC1.2, EC1.3, and EC1.4) and TATATAT (EC1.5). The position of the predicted TATA box relative to the start codon (ATG) is nucleotide −99 for EC1.1, nucleotide −77 for EC1.2, nucleotide −73 for EC1.2, nucleotide −64 for EC1.4, and nucleotide −62 for EC1.5 (Fig. 2B). The distance from the annotated transcription start site for EC1.1 (black triangle in Fig. 2B) to the predicted TATA box is 31 nucleotides, matching the reported average distance of a TATA box to the transcription start site of 31.7 nucleotides (Molina and Grotewold, 2005).
To map conserved DNA motifs in the −500-bp upstream regions, relative to the start codons of the EC1 genes, we used the online tool Cistome (Austin et al., 2016; Bio-Analytic Resource at http://BAR.utoronto.ca). The comparison of all five EC1 promoters revealed that their overall sequence similarity is not very high. However, Cistome mapped a number of conserved DNA motifs in at least four out of five promoters (Fig. 2, B and C). Motif 1 [CATC(A/G)CA] (Fig. 2C) is present in all five EC1 promoters and locates to the core promoter region, downstream of the predicted TATA boxes (Fig. 2B). The spatial proximity of motif 1 to the predicted TATA boxes (12–33 nucleotides downstream of TATA) and the match of motif 1 with the annotated transcription start site for EC1.1 (Fig. 2B) suggest that this motif is close to, or part of, the initiator element, which is described as a loosely conserved element containing an A at the transcription start site and a C as the nucleotide preceding it, surrounded by a few pyrimidines (Smale and Kadonaga, 2003).
Motif 2 [CCC(A/T)CTA] and motif 3 [CC(A/G)CTAA] (Fig. 2C) share overlapping sequence identity and appear repeatedly in the −500-bp upstream regions of EC1.1, EC1.2, and EC1.4. However, the −500-bp upstream region of EC1.5 lacks both motifs, and just one motif 3 is detected in the EC1.3 promoter. Motif 5 [A(G/C)CCCA(A/G)] appears in the −500-bp upstream regions of all EC1 genes except EC1.2. Only motif 4 [GTCTC(C/T)(A/C)] and motif 6 [(C/G)C(G/T)(C/G)(C/T)CC] are detected in all five EC1 promoters. Nevertheless, our promoter-deletion studies (Fig. 2A) indicate that a major role for these motifs in mediating egg cell specificity is not very likely.
SUF4 Positively Regulates the Transcription of EC1 Genes
To dissect the molecular network controlling egg cell differentiation, we employed the EC1.1 promoter as bait in two yeast one-hybrid screens. The 463-bp EC1.1 upstream regulatory region was divided into two bait fragments (Fig. 3A) that were integrated into the MATα yeast strain Y187 and subsequently mated with yeast strain AH109 transformed previously with a normalized total plant Arabidopsis cDNA library (Costa et al., 2013; H. Sommer and S. Masiero, unpublished data). More than seven million diploid clones were analyzed in each single screening, and 31 positive clones matched a total of nine different proteins (Supplemental Table S2). All these clones were able to grow on medium lacking His and Leu and supplemented with 20 mm 3-amino-1,2,4-triazole (3-AT), a HIS3 competitive inhibitor. One of the transcription factors identified was the C2H2 zinc finger protein SUF4.
Figure 3.
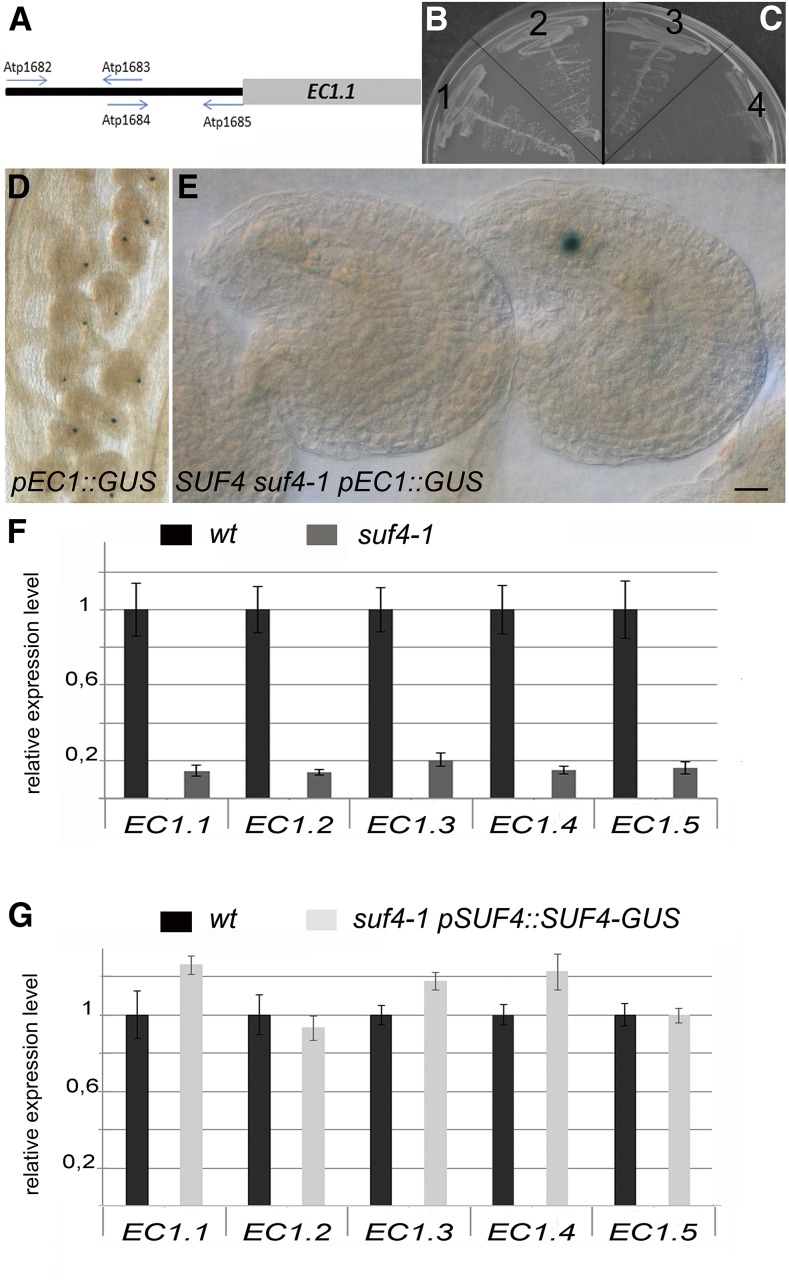
SUF4 regulates EC1.1 in yeast and in planta. A to C, Yeast one-hybrid analysis of interactions between SUF4 and pEC1.1. A, The EC1.1 promoter was divided into two bait fragments, and arrows indicate primers used for bait construction. B and C, Transformed yeast strains with the proximal fragment of the EC1.1 promoter were grown on either permissive –His–Leu medium (B) or selective –His–Leu with 5 mm 3-AT medium (C). Sections 1 and 4, pGADT7 without any insert (negative control); sections 2 and 3, pGAD-SUF4. D, GUS staining of homozygous pEC1.1(-457)::GUS plants. All egg cells show reporter activity. E, SUF4 is important for EC1.1 promoter activity in planta. suf4-1 mutants were crossed with homozygous pEC1.1(-457)::GUS plants. In the F1 carpels, only 25%, instead of the expected 50%, of egg cells were GUS positive; therefore, pEC1.1(-457)::GUS activation relies on SUF4. Bar = 20 µm. F, All five EC1 genes are down-regulated in suf4-1 mutant pistils, as indicated by real-time RT-PCR analyses. To normalize the expression level, we used UBIQUITIN10 or ACTIN8 (data not shown). The expression of each EC1 gene has been calibrated to 1 in wild-type pistils (wt). G, The normal EC1 gene expression is restored in suf4-1 suf4-1 pSUF4::SUF4-GUS pSUF4::SUF4-GUS pistils. The expression of each EC1 gene has been calibrated to 1 in wild-type pistils.
SUF4 binds the proximal fragment of the EC1.1 promoter (from −245 to −1 bp before the ATG; Fig. 3A). The full-length SUF4 cDNA was cloned into pGADT7 and reintroduced into the yeast strain containing the proximal region of the EC1.1 promoter. HIS3 reporter gene activation confirmed the ability of SUF4 to bind the EC1.1 promoter fragment (Fig. 3, B and C).
To confirm that SUF4 controls EC1.1 expression, transgenic plants homozygous for pEC1.1(-457)::GUS (Ingouff et al., 2009) and with 97.36% GUS-positive egg cells (n = 455 ovules; Fig. 1D) were crossed with homozygous suf4-1 plants. The F1 progeny plants were used to perform GUS assays on mature pistils collected 24 h after emasculation. The ratio expected for marker gene expression in the female gametes of heterozygous plants is 50% (Yadegari and Drews, 2004). If SUF4 positively regulates EC1.1, we would expect a reduction of GUS activity in egg cells from 50% to 25%. We analyzed 1,392 ovules and detected enzyme activity in only 356 egg cells (25.6%; Fig. 1E; Supplemental Table S3). We also analyzed the F2 segregating population and examined approximately 300 ovules produced by suf4-1 mutants homozygous for the pEC1.1(-457)::GUS T-DNA insertion (as suggested by the fact that all the progeny seedlings survived to BASTA application), and none showed GUS activity, although these plants were GUS positive in PCR analyses.
In addition, we also crossed homozygous pEC1.2(-893)::GUS plants (Sprunck et al., 2012) with suf4-1. In the F1 developing carpels, 301 FGs (24.6%) were GUS positive out of the 1,225 analyzed (Supplemental Table S3), suggesting that SUF4 also controls EC1.2 expressions.
Real-time RT-PCR analyses using cDNAs from suf4-1 pistils confirmed EC1.1 and EC1.2 down-regulation and provided evidence that SUF4 also regulates the other EC1 gene family members EC1.3, EC1.4, and EC1.5 (Fig. 3F). To confirm that SUF4 is a true regulator of the Arabidopsis EC1 genes, we analyzed EC1 expression in pSUF4::SUF4-GUS plants complementing suf4-1 (Kim and Michaels, 2006). Kim and Michaels (2006) introduced pSUF4::SUF4-GUS into the suf4-1 mutant background, demonstrating that the chimeric SUF4-GUS is biologically active, as these plants displayed a late-flowering phenotype. Real-time RT-PCR analyses using cDNAs from suf4-1 suf4-1 pSUF4::SUF4-GUS pSUF4::SUF4-GUS pistils showed that the expression of the five Arabidopsis EC1 genes also is fully restored (Fig. 3G).
SUF4 Is Expressed in the Developing FG
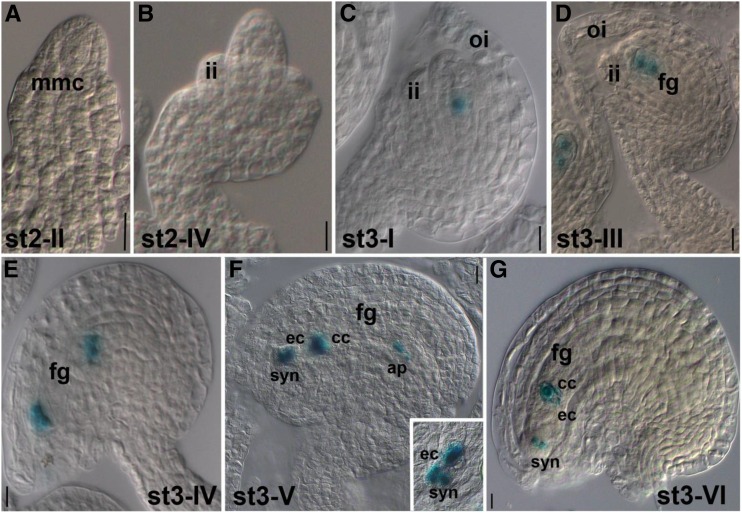
We also used the suf4-1 pSUF4::SUF4-GUS line (Kim and Michaels, 2006) to study SUF4 protein expression during embryo sac development. SUF4-GUS activity, driven by the genomic pSUF4::SUF4 locus, is detected neither in ovule primordia nor in the diploid megaspore mother cell or during meiosis (Fig. 4, A and B). SUF4-GUS becomes visible immediately after meiosis (Fig. 4C), when it localizes in the nucleus of the functional megaspore, and persists during megagametogenesis (Fig. 4, D–G). In the seven-celled embryo sac (FG stage 6 [FG6]) of stage 3-V ovules according to Schneitz et al. (1995), SUF4-GUS is detectable in all eight nuclei, including the two polar nuclei of the central cell and the egg cell nucleus (Fig. 4F). However, in the mature stage 3-VI ovule (FG7), SUF4-GUS is no longer detected in the egg cell nucleus (Fig. 4G). Such a peculiar expression pattern indicates that SUF4, detected during egg cell differentiation, is removed during egg cell maturation. This suggests a possible role for SUF4 in the developing egg cell and makes SUF4 a suitable marker to discriminate between immature egg cells, not yet competent for fertilization, and mature egg cells.
Figure 4.
SUF4 is expressed in developing FGs. A and B, pSUF4::SUF4-GUS activity is detected neither in the megaspore mother cell (mmc; A) nor in the tetrad of megaspores (B). C, SUF4-GUS is detected in developing ovules from stage 3-I on, initially in the nucleus of the functional megaspore forming the haploid FG. D and E, SUF4-GUS expression persists in the developing embryo sac. F, In the seven-celled embryo sac (stage 3-V), SUF4-GUS is detected in all seven nuclei. G, At stage 3-VI, SUF4-GUS is no longer expressed in the egg cell but only in the nuclei of the central cell and synergid cells. Ovule stages are according to Schneitz et al. (1995). ap, Antipodal cells; cc, central cell; ec, egg cell; fg, FG; ii, inner integument; oi, outer integument; syn, synergid cells. Bars = 20 µm
SUF4 Binds to EC1 Promoters
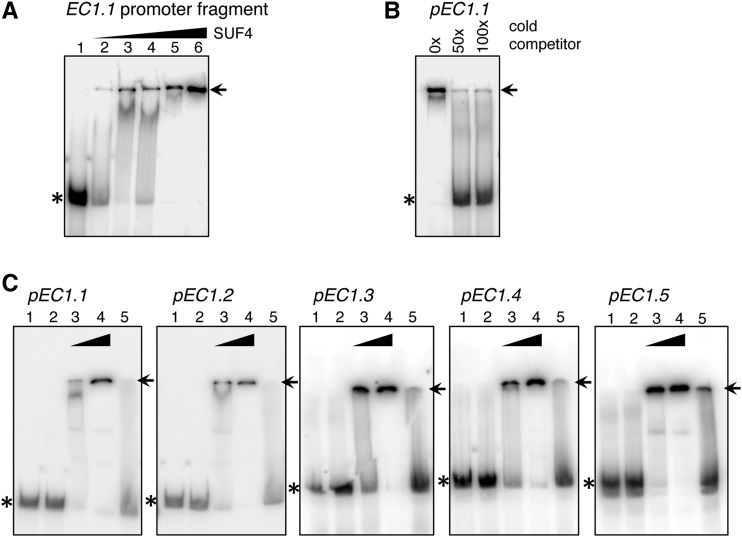
Recombinant SUF4, expressed either as a 6xHIS-SUF4-STREPII or as a 6xHIS-MBP-SUF4 fusion in Escherichia coli, was purified and used for in vitro DNA-binding assays. Electrophoretic mobility shift assays (EMSAs) were performed to confirm the interaction between SUF4 and the EC1.1 promoter as well as with all other Arabidopsis EC1 promoters (Fig. 5). A 108-bp EC1.1 promoter fragment, covering part of the proximal fragment that has been used in the yeast one-hybrid screening (Fig. 3A) and is known to be necessary for egg cell expression (Fig. 2A), was radioactively labeled with α-32P. This fragment showed significant binding to increasing amounts of purified 6xHIS-SUF4-STREPII (Fig. 5A). Competition experiments confirmed that SUF4 binding to the EC1.1 promoter fragment is displaced by the cold probe (Fig. 5B).
Figure 5.
SUF4 binds to all five EC1 promoters. A, Gel-shift assay without (lane 1) and with 10 (lane 2), 50 (lane 3), 100 (lane 4), 200 (lane 5), and 400 ng (lane 6) of recombinant 6xHIS-SUF4-STREPII added to a radioactively labeled 108-bp EC1.1 promoter fragment covering the DNA region used as bait in the yeast one-hybrid screen. B, Gel-shift assay with 50-fold (50×) and 100-fold (100×) excess of unlabeled EC1.1 promoter fragment as a cold competitor added to the reaction mix with 200 ng of 6xHIS-SUF4-STREPII. The control reaction is without cold competitor (0×). C, Fifty and 150 ng of recombinant 6xHIS-MBP-SUF4, and 150 ng of 6xHIS-MBP as a control, were mixed with 10 ng of radioactively labeled EC1 promoter fragments. Lane 1, Radioactively labeled promoter fragment only; lane 2, radioactively labeled promoter fragment with 150 ng of 6xHIS-MBP tag only; lane 3, radioactively labeled promoter fragment with 50 ng of 6xHIS-MBP-SUF4; lane 4, radioactively labeled promoter fragment with 150 ng of MBP-SUF4; lane 5, radioactively labeled promoter fragment with 150 ng of MBP-SUF4 and 100-fold excess of cold competitor (unlabeled promoter fragment). Asterisks mark free probes, arrows mark shifted bands of protein-DNA complexes.
We used 6xHIS-MBP-SUF4 and the fusion protein 6xHIS-MBP as a control to show that MBP-tagged SUF4 is able to specifically bind the radioactively labeled fragments of all five EC1 promoters (Fig. 5C).
In summary, the DNA-binding assays, together with the yeast data and the loss of GUS reporter activity of pEC1.1(-457)::GUS and pEC1.2(-893)::GUS in the suf4-1 mutant (Supplemental Table S3), clearly prove that SUF4 binds to and activates EC1 promoters. This is further supported by real-time RT-PCR analyses of EC1 gene expression in suf4-1 and in the complemented suf4-1 line (Fig. 3, F and G), suggesting that SUF4 binding to EC1 promoter sequences is necessary to promote EC1 gene activation.
suf4-1 Shows a Moderate ec1 Phenotype
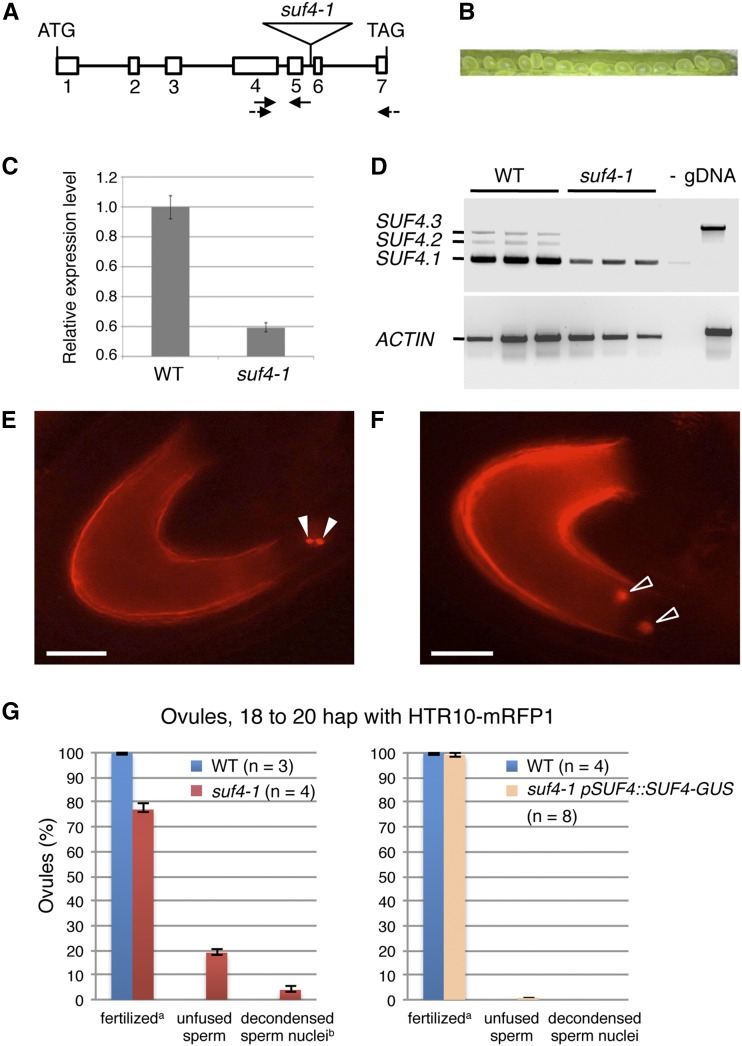
The simultaneous down-regulation of EC1.2 and EC1.3 by RNA interference in the homozygous triple mutant ec1.1/ec1.4/ec1.5 (termed ec1-RNAi) severely affects double fertilization (Sprunck et al., 2012). The sperm cells, delivered into ec1-RNAi mutant ovules, do not fuse with the two female gametes, causing polytubey, multiple sperm delivery, and reduced seed set (Sprunck et al., 2012). Therefore, we analyzed the siliques of homozygous suf4-1 plants, but no seed set defects were observed (Fig. 6B). However, the presence of functional SUF4.1 transcripts revealed that suf4-1 is not a null mutant (Fig. 6, C and D). This is likely the reason why suf4-1 is still able to accumulate lower EC1 transcript levels (Fig. 3F).
Figure 6.
suf4-1 ovules show a moderate ec1 phenotype. A, Genomic organization of SUF4, composed of seven exons and six introns. The T-DNA in suf4-1 is inserted in intron 5, 2,325 bp downstream of the predicted translation start site. B, Siliques of homozygous suf4-1 show normal seed set. C, Quantitative RT-PCR analyses revealed that residual SUF4 transcript is detectable in suf4-1. D, Three alternative splicing variants of SUF4 (SUF4.1, SUF4.2, and SUF4.3) are expressed in pistils of the wild type (WT). The functional splicing variant SUF4.1 (Kim and Michaels, 2006) also is detectable in pistils of homozygous suf4-1 plants. E and F, Phenotypes of suf4-1 pistils pollinated with the sperm cell marker line HTR10-mRFP1. Fluorescence microscopy 18 to 20 h after pollination revealed ovules with unfused sperm cells (arrowheads in E) or sperm cell nuclei with decondensed chromatin (arrowheads in F). At that time, gamete fusion in wild-type ovules has been accomplished (data not shown). Bars = 20 µm. G, Quantification of the suf4-1 ovule phenotypes shown in E and F. n, Number of pistils (Col-0, 167 ovules; suf4-1, 232 ovules). In the complemented line suf4-1 pSUF4::SUF4-GUS (at right), the suf4-1 phenotype of unfused or delayed-fusing sperm cells is not detectable. n, Number of pistils (Col-0, 178 ovules; suf4-1, 400 ovules). a, Fertilized ovules, no HTR10-mRFP1 fluorescence visible; b, includes two ovules with decondensed sperm chromatin and two additional unfused sperm cells. Error bars in C and G represent sem.
To investigate sperm cell behavior during double fertilization, we emasculated the pistils of wild-type and homozygous suf4-1 plants and pollinated them with the sperm cell marker line HTR10-mRFP1 (Ingouff et al., 2007). With this marker line, successful plasmogamy and ongoing karyogamy of male and female gametes are recognizable by the spatial separation of the two sperm nuclei and the decondensation of sperm chromatin, respectively.
When we prepared suf4-1 pistils 18 to 20 h after pollination, we detected a significant portion of suf4-1 ovules (23%; 53 of 232 ovules) exhibiting either nonfused sperm cells or sperm cells delayed in fusion (Fig. 6, E–G). These phenotypes were not observed in wild-type ovules (Fig. 6G), where gamete fusion is accomplished 6 to 9 h after pollination (Sprunck et al., 2012). Seed set is not affected in suf4-1 siliques, suggesting that unfused sperm cells do fuse later. Late-fusing sperm cells also have been described in individual ec1-RNAi lines (Rademacher and Sprunck, 2013) and are likely a result of variable EC1.2 and EC1.3 knockdown efficiencies in the triple ec1.1/ec1.4/ec1.5 mutant by the EC1.2/EC1.3 RNA interference construct.
Importantly, the delay in sperm fusion was reversed when pistils of the double homozygous line suf4-1 pSUF4::SUF4-GUS were pollinated with the sperm marker line HTR10-mRFP (Fig. 6G, right chart; 400 ovules analyzed), indicating that the complementation with pSUF4::SUF4-GUS is able to rescue the moderate ec1 phenotype in suf4-1.
Altogether, the observed delayed gamete fusion phenotype in suf4-1 ovules and the lack of undeveloped seeds in suf4-1 siliques suggest that the down-regulation of SUF4, and in turn the down-regulation of EC1 gene expression, impair rapid sperm fusion without abolishing it.
MOM1 Participates with SUF4 in Regulating the EC1 Genes
To better understand how SUF4 can regulate EC1 gene expression, we performed correlation analyses on around 1,700 microarray-based transcriptomic measurements (Menges et al., 2008). Gene coexpression often highlights a functional linkage between genes, and we observed that MOM1 shows a significant correlation value with SUF4 (Supplemental Table S4). We focused on MOM1, since it modulates epigenetic stress memory (Iwasaki and Paszkowski, 2014). MOM1 is a CHD3 chromatin-remodeling factor, which has nucleosome-remodeling and histone deacetylation activities (Tong et al., 1998).
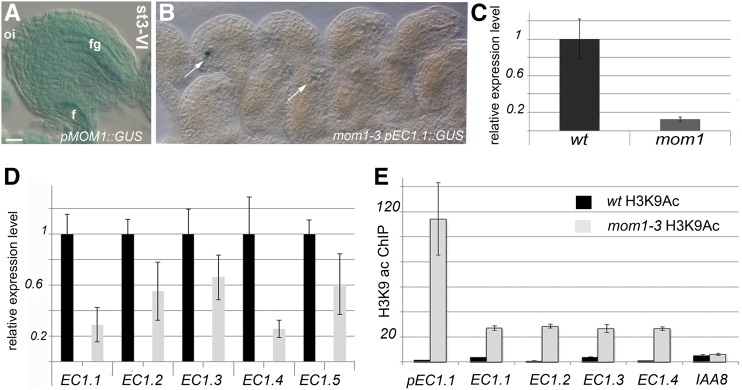
MOM1 messenger is detected in siliques (3–6 DAP), leaves, and inflorescences (Supplemental Fig. S1A). In transgenic pMOM1::GUS plants, GUS activity was found in the placenta tissue when ovule primordia arise (Supplemental Fig. S1B). In developing ovules, MOM1 is expressed from stage 2-III on (Supplemental Fig. S1D). In mature ovules (stage 3-VI), MOM1 promoter activity is detected in the sporophytic tissues of the ovule and in the mature FG, although the reporter gene activity is weak (Fig. 7A). Furthermore, MOM1 expression in the embryo sac is corroborated by transcriptome analyses (Yu et al., 2005; Johnston et al., 2007; Zhang et al., 2015).
Figure 7.
MOM1 is expressed in developing ovules and participates in SUF4 and EC1 expression. A, GUS activity driven by pMOM1::GUS is detected in the FG and in the sporophytic tissues of mature ovules. f, Funiculus; fg, FG; oi, outer integument. Bar = 20 µm. B, In mom1-3 mutants hemizygous for pEC1.1(-457)::GUS, enzymatic GUS activity is detected in 25% to 36% of the analyzed egg cells (arrows). A total of 589 ovules were analyzed. C, Quantitative RT-PCR analyses to monitor SUF4 expression in mom1-3 flowers. SUF4 expression is reduced compared with wild-type (wt) flowers. D, In mom1-3 mutant pistils, all five EC1 genes are down-regulated, as shown by quantitative RT-PCR analysis. E, ChIP using an anti-H3K9ac antibody. ChIP enrichment was evaluated by quantitative PCR analyses. EC1 genes are enriched in H3K9ac in mom1-3 inflorescence in comparison with wild-type ones. Immunoprecipitation efficiency was tested by quantifying H3K9ac marks in the IAA8 locus (Zhou et al., 2010). Cycle threshold values were used to calculate the immunoprecipitation/input signal. ChIP enrichments are presented as the percentage of bound/input signal.
To investigate the impact of MOM1 on SUF4 and EC1 gene expression, we performed real-time RT-PCR analyses and crossed homozygous pEC1.1(-457)::GUS plants with mom1-3. In the F2 segregating population, we looked for homozygous mom1-3 plants also homozygous for the pEC1.1(-457)::GUS insertion. In these plants, GUS enzymatic activity was detected in 68% to 73% of egg cells analyzed (four plants and three carpels per plant were analyzed; n = 589). Coherently, in mom1-3 mutants hemizygous for pEC1.1(-457)::GUS, the enzymatic activity was detected in a range from 25% to 37% of analyzed egg cells (Fig. 7B). Quantitative RT-PCR analyses with mom1-3 inflorescences showed that SUF4 expression is down-regulated (Fig. 7C). Although the members of the EC1 gene family also are down-regulated in mom1-3 (Fig. 7D), the reduction in EC1 expression is not as strong as that observed in suf4-1 (Fig. 3E).
In an attempt to clarify EC1 family gene regulation by MOM1, we explored their epigenetic landscape focusing on H3K9ac. Chromatin immunoprecipitation (ChIP) experiments revealed that, in mom1-3, especially the EC1.1 promoter region shows a higher level of H3K9ac (Fig. 7E) but also the tested EC1 gene loci displayed higher H3K9ac levels compared with the wild type, while the IAA8 gene locus was not affected in mom1-3 (Fig. 7E). Altogether, our data indicate that histone modifications also participate in EC1 regulation, as we show that, in mom1-3 flowers, the epigenetic landscape of these loci changes toward a state that favors the transcription, thus counteracting the SUF4 reduction recorded in mom1-3 mutant plants.
DISCUSSION
The few-celled FG of flowering plants has become an attractive model system in which to study the mechanisms involved in pattern formation and the differentiation of distinct cell types (Sprunck and Gross-Hardt, 2011). Considerable progress has been made in the past decade toward the identification of genes involved in the differentiation of FG cells (Evans, 2007; Gross-Hardt et al., 2007; Pagnussat et al., 2007, 2009; Moll et al., 2008; Krohn et al., 2012; Yuan et al., 2016). Nevertheless, not much is known about the transcriptional regulatory network involved in egg cell specification. One exception is the RKD subfamily of plant-specific RWP-RK transcription factors, which provoke an egg cell-like transcriptional profile when ectopically expressed in Arabidopsis seedlings (Koszegi et al., 2011) and act in egg and sperm cell differentiation in the liverwort Marchantia polymorpha (Koi et al., 2016; Rövekamp et al., 2016).
In this work, we used the egg cell-specific EC1.1 promoter as a tool to identify transcription factors participating in egg cell differentiation. We show that all five Arabidopsis EC1 promoters drive egg cell-specific reporter gene expression and share some common DNA sequence motifs. In 5′ deletion studies, we observed that relatively short proximal promoter regions are sufficient to drive egg cell-specific expression, indicating that important cis-regulatory elements for egg cell specificity are present in these regions. Using the yeast one-hybrid technique, we aimed to identify transcription factors binding to the EC1.1 promoter, which has been used as a developmental marker for the egg cell (Ingouff et al., 2009; Völz et al., 2012; Denninger et al., 2014; Kong et al., 2015; Mendes et al., 2016). The yeast one-hybrid system detects protein-DNA interactions in vivo, as prey proteins can acquire their native configuration (Lopato et al., 2006). The yeast one-hybrid technique is a simple, rapid, and sensitive tool (Reece-Hoyes et al., 2011) that nevertheless suffers certain limitations, such as its inability to identify transcription factors that bind the target DNA only if posttranslationally modified or those that are members of higher order complexes (Deplancke et al., 2006).
Unequivocal evidence for cis-regulatory motifs involved in egg cell-specific promoter activity is not yet given. Therefore, we split the 463-bp 5′ upstream region of the EC1.1 promoter into two bait fragments. This facilitates the interaction of transcription factors with the EC1.1 regulatory sequences even without detailed knowledge of the key cis-regulatory elements. Quite large promoter fragments have already been used successfully as bait in yeast one-hybrid screenings (Roccaro et al., 2005; Brady et al., 2011), although it is common to perform yeast one-hybrid screenings using multiple copies of small bait elements, such as cis-regulatory motifs (Tran et al., 2004; Lopato et al., 2006). One potential difficulty in using larger promoter fragments is the presence of several cis-regulatory elements, which might be bound by yeast DNA-binding proteins activating the transcription of the reporter gene even without any prey GAL4AD chimeric protein. Nevertheless, we did not experience self-activation for either of the two EC1.1 bait fragments.
Our in vitro and in vivo data indicate that SUF4 exerts a direct positive regulation on the EC1 gene family. SUF4 is a C2H2 protein already identified in secondary genetic screenings performed to isolate loci able to suppress the Columbia-0 (Col-0) FRIGIDA late-flowering phenotype. SUF4 binds the FLOWERING LOCUS C (FLC) promoter and subsequently recruits FRIGIDA and FRIGIDA-LIKE1 (Choi et al., 2011). FRIGIDA acts as a scaffold protein, forming a transcription activator complex that recruits, among others, chromatin modifiers to regulate FLC. Repression of FLC causes early flowering, and it is accompanied by covalent histone modification, like histone 3 Lys-9 and histone 3 Lys-14 deacetylation and histone 3 Lys-9 and histone 3 Lys-27 methylation (Sung and Amasino, 2004).
SUF4 binds the FLC promoter through the A/T-rich consensus sequence 5′-CCAAATTTTAAGTTT-3′ (Choi et al., 2011). Although we have not been able to recognize this consensus sequence in the EC1 promoters, it is well accepted that interacting proteins may modulate a transcription factor-binding specificity. Indeed, SUF4 interacts with several proteins, like MEDIATOR18 (Lai et al., 2014), members of the SPINDLE ASSEMBLY CHECKPOINT complex (Bao et al., 2014), and LUMINIDEPENDS (Kim et al., 2006). SUF4 also contains a BED-finger domain with DNA-binding ability, named after the Drosophila melanogaster proteins BEAF and DREF (Aravind, 2000). Interestingly, the human zinc BED proteins (ZBED1–ZBED6; Mokhonov et al., 2012) act as transcriptional regulators by modifying the local chromatin structure upon binding to GC-rich sequences.
In eukaryotic organisms, transcription factors regulate gene expression through binding to cis-regulatory-specific sequences in the promoters of their target genes. Nevertheless, also the chromatin structure actively participates in gene regulation, favoring or not the access of the DNA-binding proteins to their regulatory sites. Indeed, the chromatin structure is modulated in a highly cell-specific manner, as reported extensively for flowering time regulation (He, 2009) and flower development (Gan et al., 2013).
Our data on the SUF4-dependent EC1 expression in egg cells and on the strong down-regulation of SUF4 in mom1-3 mutant ovules (accompanied by an enrichment of H3K9ac in EC1 loci) suggest a complex regulation of EC1 gene expression involving chromatin remodeling. We provide evidence that SUF4 is involved in regulating EC1 gene expression in the developing egg cell, while in the mature egg cell, SUF4 is not detectable anymore. Therefore, it is possible that SUF4 participates in the recruitment of chromatin modifiers in the developing egg cell to promote EC1 gene expression.
We were able to show that histone modifications participate in EC1 gene regulation, at least in mom1-3 flowers. MOM1, which is coexpressed with SUF4, was identified during a genetic screen set up to monitor the release of transcriptional gene silencing of a cluster of transgenes (Amedeo et al., 2000). Remnants of the gypsy‐like retrotransposon Athila also are transcriptionally activated in mom1-3 mutants (Habu et al., 2006). The C-terminal region of MOM1 is similar to the C terminus of eukaryotic enhancer of polycomb proteins, which have roles in heterochromatin formation. However, the mechanism by which MOM1 contributes to chromatin changes is still quite elusive, as mom1-3 mutants display none or poor alterations of the epigenetic landscape of the released loci (Vaillant et al., 2006). Nevertheless, Numa et al. (2010) demonstrated that MOM1 targets also map in euchromatic regions. By ChIP experiments, they showed that the promoter of SDC (SUPPRESSOR OF drm1 drm2 cmt39), a MOM1 target, is enriched in histone 3 Lys-9 dimethylation. SDC is activated in mom1-3, and ChIP experiments revealed that the level of dimethylated histone 3 Lys-9 in tandem repeats of the SDC promoter is reduced.
The EC1 loci in mom1-3 flowers are enriched in H3K9ac, and both SUF4 and EC1 genes are differentially expressed in mom1-3 ovules, suggesting that MOM1 also participates in remodeling the chromatin organization of SUF4 and, thus, regulates its transcriptional activity. However, whether the chromatin status of SUF4 is changed in mom1-3, or whether SUF4 and MOM1 interact directly to regulate EC1 gene expression in the developing egg cell, remains to be investigated.
The observed enrichment of H3K9ac in EC1 loci of mom1-3 flowers indicates that MOM1 affects the modification of histones in EC1 genomic loci. Histone tail acetylation results in chromatin decondensation, and thus in remodeling the chromatin organization into transcriptionally active chromatin, as Lys acetylation removes the positive charge of this amino acid, favoring chromatin relaxation and access to transcription factors and other transcriptional coactivators. In mom1-3, therefore, the epigenetic landscape of EC1 loci changes toward a state that favors transcription. Our studies revealed, however, that SUF4 binding to the EC1 promoter sequences is necessary to promote EC1 gene activation but that SUF4 is strongly down-regulated in mom1-3. Although EC1 expression is lower in mom1-3 compared with the wild type, it is not as reduced as in suf4-1, suggesting that the SUF4 reduction and the resulting down-regulation of EC1 genes are partially counteracted in mom1-3 FGs. In addition to H3K9ac, other altered epigenetic events, such as histone methylation, histone phosphorylation, and DNA methylation, also may be involved in the regulation of EC1 gene expression.
The identification of egg cell-specific genes, the analyses of their promoter activities, and the characterization of transcriptional regulatory networks acting during egg cell differentiation are essential to improve our understanding of how this important cell becomes specified and how it acquires its unique features and functions in sexual reproduction. The discovery of SUF4 and MOM1 as regulators of the egg cell-specific EC1 gene family of Arabidopsis is an important step toward the identification of the egg cell transcriptional regulatory network. Nevertheless, we are only just beginning to understand how the complex expressional control of the EC1 genes is achieved.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) suf4-1 mutants and suf4-1 pSUF4:SUF4::GUS seeds were donated by S.D. Michaels, and mom1-3 mutants were donated by J. Paszkowski. Plants were grown under long-day conditions (14 h of light/10 h of dark) at 22°C. Gentotyping was done using gene-specific primers, specific T-DNA primers, and primers able to anneal to the GUS gene. All primers are listed in Supplemental Table S5.
Constructs for Promoter-Reporter Studies
All five EC1 upstream regulatory sequences were cloned as PCR fragments extending in the 5′ direction from the −1 position (referring to the respective start codon) toward the previous gene (Fig. 1A). EC1 promoters were amplified from genomic DNA of Arabidopsis (accession Col-0) using Phusion High-Fidelity DNA Polymerase (New England Biolabs) and the primer pairs EC1.1p(-463bp)_fw/EC1.1p_rev, EC1.2p(-894)_fw/EC1.2p_rev, EC1.3p(-289)_fw/EC1.3p_rev, EC1.4p(-263)_fw/EC1.4p_rev, and EC1.5p(-251)_fw/EC1.5p_rev (primer sequences are available in Supplemental Table S5) The PCR products were cloned into the Gateway entry vector pENTR/D-TOPO (Thermo Fisher Scientific). Subsequently, the promoter fragments were transferred into a Gateway-compatible version of the pGreenII-based vector NLS:3GFP:NOSt (Takada and Jürgens, 2007) termed pGII_GW:NLS:3GFP:NOSt (Zheng et al., 2011) by LR reaction using Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific). For deletion studies with NLS-3xGFP as a reporter, 5′ truncated promoter fragments were amplified using genomic DNA of Arabidopsis (Col-0) as a template and the primer combinations EC1.3p(-133)_fw/EC1.3p_rev, EC1.4p(-163)_fw/EC1.4p_rev, and EC1.5p(-156)_fw/EC1.5p_rev (Supplemental Table S5). EC1.3, EC1.4, and EC1.5 promoter deletions were cloned into pENTR/D-TOPO and recombined into pGII_GW:NLS:3GFP:NOSt. For studies with GFP as a reporter EC1.2 promoter, deletion fragments were generated by PCR using primers introducing unique restriction enzyme sites (PstI and BamHI; Supplemental Table S5). The PCR fragments were digested and ligated with pBI101.GFP (Yadegari et al., 2000). The binary vectors pEC1.1(-457)::GUS and pEC1.2(-893)::GUS have been described previously (Ingouff et al., 2009; Sprunck et al., 2012). pEC1.1(-457)::GUS served as a template to generate the deletion constructs pEC1.1(-326)::GUS and pEC1.1(-192)::GUS, applying the forward primers EC1.1p(-326)_fw and EC1.1p(-192)_fw (Supplemental Table S5). The deletion construct pEC1.1(-113)::GUS was generated by digesting pEC1.1(-457)::GUS with PmeI and HpaI, followed by religation. All constructs were sequence verified.
T-DNA constructs with pEC1.2 in pBI101.GFP were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Arabidopsis plants (Col-0) were transformed using a modified floral dip procedure (Clough and Bent, 1998). Transformed progeny were selected by germinating surface-sterilized T1 seeds on growth medium containing antibiotics (30 µg mL−1 kanamycin sulfate) supplemented with 15 µg mL−1 cefotaxime. Resistant seedlings were transplanted to soil 10 d after germination. The pEC1::NLS3xGFP expression vectors were delivered into A. tumefaciens strain GV3101 pSOUP, and pEC1::GUS expression vectors were delivered into strain GV3101 pMP90RK. Arabidopsis plants (Col-0) were transformed by floral dip. T1 seeds were collected, sown on soil, and vernalized for 3 d at 4°C in the dark. Starting 3 d after germination, BASTA-resistant seedlings were selected by spraying three times with 200 mg L−1 BASTA (Bayer Crop Science) supplemented with 0.1% (v/v) Tween. Transgene identity was verified by PCR.
Cloning of pMOM1:GUS
For the pMOM1:GUS construct, a 1.1-kb genomic region upstream of the MOM1 ATG start codon was amplified by Phusion High-Fidelity DNA Polymerase (Finnzymes; Supplemental Table S5). The product was cloned in the pBGWFS7 vector (Karimi et al., 2002) using the Gateway system (Thermo Fisher Scientific). The construct was verified by sequencing and used to transform Arabidopsis Col-0 plants (Clough and Bent, 1998). GUS assays were done according to Colombo et al. (2008).
Yeast Experiments and Cloning
The EC1.1 upstream regulatory region of 463 bp was amplified as two distinct fragments using primer pairs pAtEC1.11 plus EcoRI_fw/pAtEC1.11 plus XbaI_rev and pAtEC1.12 plus EcoRI_fw/pAtEC1.12 plus XbaI_rev (Supplemental Table S5), digested, and ligated into the EcoRI/XbaI-digested pHISi vector (Clontech). The two bait plasmids were linearized with XhoI and used to transform the Saccharomyces cerevisiae yeast strain Y187. A whole normalized total plant cDNA library (H. Sommer and S. Masiero, unpublished data) was cloned in pGADT7-rec and introduced into yeast strain the Saccharomyces cerevisiae AH109. The yeast containing the expression library was mated with modified Y187 strains (containing the EC1.1 regulatory regions) as described in the Clontech user manual PT4085-1. Diploids were selected on medium lacking Leu and His and supplemented with 20 mm 3-AT (Sigma-Aldrich). Plasmids were extracted from positive colonies and retransformed into Y187 to discard the false positives.
Purification of Recombinant SUF4 and EMSAs
Expression vectors for recombinant protein expression in Escherichia coli were cloned using the Gateway system (Invitrogen). The coding sequence of SUF4 was amplified by PCR from inflorescence cDNA (Supplemental Table S5) and cloned into pENTR/D-TOPO. LR-Clonase reactions were performed using the SUF4 entry vector and the destination vectors pET-53-DEST (Novagen) and pDEST-HisMBP (Nallamsetty et al., 2005). The resulting expression vectors were used to express a 6xHis-SUF4-StrepII fusion protein and a 6xHIS-MBP-SUF4 fusion protein. After expressing 6xHis-SUF4-StrepII in E. coli RosettaTM(DE3) (Novagen), the soluble fraction of the crude cell extract was purified by immobilized metal ion affinity chromatography under native conditions using nickel-nitrilotriacetic acid agarose (Qiagen) and gravity flow columns, following the manufacturer´s instructions. The 6xHis-MBP and 6xHis-MBP-SUF4 recombinant proteins were expressed in E. coli BL21-Codon Plus(DE3)-RIPL cells (Stratagene) and purified under native conditions using TALON Metal Affinity Resin (Clontech).
The EC1 promoter fragments were amplified with terminal XbaI restriction sites via PCR using Taq polymerase (Fermentas), resulting in fragments for EC1.1 (108 bp), EC1.2 (115 bp), EC1.3 (167 bp), EC1.4 (199 bp), and EC1.5 (189 bp; primer sequences are available in Supplemental Table S5). The purified promoter fragments were digested with XbaI and radioactively labeled using Klenow enzyme (Fermentas) and [α-32P]dATP. Unincorporated [α-32P]dATP was removed by spin-column chromatography (Illustra ProbeQuant G-50 Micro columns; GE Healthcare).
For the EMSAs, the radioactively labeled promoter fragments (10 or 18 ng) were incubated with different amounts of SUF4 (10 to 400 ng) in 1× EMSA buffer (10 mm Tris-HCl, pH 7.5, 100 mm KCl, 1 mm EDTA, 0.1 mg mL−1 bovine serum albumin, 100 μm ZnCl2, 6% glycerol, and 1 mm dithiothreitol) in 20-μL reaction volumes for 1 h at 4°C. Afterward, the reactions were separated on a 5% polyacrylamide gel in TAE buffer (40 mm Tris and 2.5 mm EDTA, pH 7.8) at 10 V cm−1 gel length for 1 h. For the competitor assays, the respective unlabeled probe was added in excess (50× and 100×) to the binding mixture. Gel images were obtained using autoradiography (Cyclone Phosphoimager A431201; Packard).
Comparative Promoter Studies
For motif discovery, we used the online tool Cistome (https://bar.utoronto.ca/cistome/cgi-bin/BAR_Cistome.cgi) to map conserved sequence motifs in the −500-bp upstream regions of EC1 genes relative to their translation start sites. Cistome predicts cis-elements in the promoters of sets of coexpressed genes. The cis-element prediction program MEME (Bailey et al., 2009) was selected, with the following parameters: width, 7; number of motifs, 6; mode: oops. Transcription factor-binding sites for TBPs were mapped using AthMap (http://www.athamap.de/index.php).
Correlation Analysis
Calculation of the Pearson correlation coefficient and the microarray data set employed were as described previously (Menges et al., 2008; Berri et al., 2009).
ChIP and Quantitative PCR Analyses
For ChIP experiments, chromatin was extracted from Arabidopsis Col-0 and mom1-3 mutant flowers (before fertilization occurred). ChIP experiments were done as described previously (Mizzotti et al., 2014). Real-time PCR analyses were performed on input and immunoprecipitated samples, and percentage of input was calculated. IAA8 (At2g22670) was used as a reference as it carries the H3K9ac mark (Mizzotti et al., 2014). Quantitative expression analyses were performed using the iQ5 multicolor real-time PCR detection system (Bio-Rad). Primers used for ChIP experiments are listed in Supplemental Table S5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. MOM1 and IAA8 expression pattern.
Supplemental Table S1. Predicted transcription factor-binding sites for TBP.
Supplemental Table S2. Proteins able to bind the EC1.1 promoter in yeast.
Supplemental Table S3. SUF4 affects the activity of EC1.1 and EC1.2 promoters.
Supplemental Table S4. Genes coexpressed with SUF4.
Supplemental Table S5. Primers used in this work.
Supplementary Material
Acknowledgments
We thank Scott D. Michaels for suf4-1 and suf4-1 pSUF4::SUF4-GUS seeds, J. Paszkowski for mom1-3 seeds, M. Ron for making the Gateway-compatible pGreenII-based vector GW::NLS:3GFP:NOSt available to us, M. Kammerer for cloning and plant care, and M. Grasser for support in the gel-shift assays.
Glossary
- FG
female gametophyte
- RT
reverse transcription
- 3-AT
3-amino-1,2,4-triazole
- EMSA
electrophoretic mobility shift assay
- H3K9ac
histone 3 Lys-9 acetylation
- ChIP
chromatin immunoprecipitation
- Col-0
Columbia-0
Footnotes
This work was supported by the German Research Council (grant nos. SP 686/1–2 and SFB924 to S.S.), by the German Academic Exchange Service (PPP Vigoni 2009 award to T.D. and L.C.), by the Università degli Studi di Milano (Ph.D. fellowship to F.R.), and by the CARIPLO Foundation (grant no. 2011–2257 to S.M.).
References
- Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J (2000) Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405: 203–206 [DOI] [PubMed] [Google Scholar]
- Aravind L. (2000) The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci 25: 421–423 [DOI] [PubMed] [Google Scholar]
- Austin RS, Hiu S, Waese J, Ierullo M, Pasha A, Wang TT, Fan J, Foong C, Breit R, Desveaux D, Moses A, Provart NJ (2016) New BAR tools for mining expression data and exploring Cis-elements in Arabidopsis thaliana. Plant J 88: 490–504 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Zhang N, Hua J (2014) Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nat Commun 5: 5628. [DOI] [PubMed] [Google Scholar]
- Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro ACM, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè ME, et al. (2009) Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol 9: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. (2011) A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Costa M, Nobre MS, Becker JD, Masiero S, Amorim MI, Pereira LG, Coimbra S (2013) Expression-based and co-localization detection of arabinogalactan protein 6 and arabinogalactan protein 11 interactors in Arabidopsis pollen and pollen tubes. BMC Plant Biol 13: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denninger P, Bleckmann A, Lausser A, Vogler F, Ott T, Ehrhardt DW, Frommer WB, Sprunck S, Dresselhaus T, Grossmann G (2014) Male-female communication triggers calcium signatures during fertilization in Arabidopsis. Nat Commun 5: 4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJ (2006) Gateway-compatible yeast one-hybrid screens. CSH Protoc 2006: pdb.prot4590. [DOI] [PubMed] [Google Scholar]
- Drews GN, Koltunow AMG (2011) The female gametophyte. The Arabidopsis Book 9: e0155 doi/10.1199/tab.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MMS. (2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan ES, Huang J, Ito T (2013) Functional roles of histone modification, chromatin remodeling and microRNAs in Arabidopsis flower development. Int Rev Cell Mol Biol 305: 115–161 [DOI] [PubMed] [Google Scholar]
- Gross-Hardt R, Kägi C, Baumann N, Moore JM, Baskar R, Gagliano WB, Jürgens G, Grossniklaus U (2007) LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol 5: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu Y, Mathieu O, Tariq M, Probst AV, Smathajitt C, Zhu T, Paszkowski J (2006) Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep 7: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. (2009) Control of the transition to flowering by chromatin modifications. Mol Plant 2: 554–564 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F (2007) Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Ingouff M, Sakata T, Li J, Sprunck S, Dresselhaus T, Berger F (2009) The two male gametes share equal ability to fertilize the egg cell in Arabidopsis thaliana. Curr Biol 19: R19–R20 [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Paszkowski J (2014) Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc Natl Acad Sci USA 111: 8547–8552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Meier P, Gheyselinck J, Wuest SEJ, Federer M, Schlagenhauf E, Becker JD, Grossniklaus U (2007) Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol 8: R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D (2007) Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet 3: 1848–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I (2006) SUPPRESSOR OF FRIGIDA4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18: 2985–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD (2006) SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133: 4699–4707 [DOI] [PubMed] [Google Scholar]
- Koi S, Hisanaga T, Sato K, Shimamura M, Yamato KT, Ishizaki K, Kohchi T, Nakajima K (2016) An evolutionarily conserved plant RKD factor controls germ cell differentiation. Curr Biol 26: 1775–1781 [DOI] [PubMed] [Google Scholar]
- Kong J, Lau S, Jurgens G (2015) Twin plants from supernumerary egg cells in Arabidopsis. Curr Biol 25: 225–230 [DOI] [PubMed] [Google Scholar]
- Koszegi D, Johnston AJ, Rutten T, Czihal A, Altschmied L, Kumlehn J, Wüst SEJ, Kirioukhova O, Gheyselinck J, Grossniklaus U, et al. (2011) Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J 67: 280–291 [DOI] [PubMed] [Google Scholar]
- Krohn NG, Lausser A, Juranić M, Dresselhaus T (2012) Egg cell signaling by the secreted peptide ZmEAL1 controls antipodal cell fate. Dev Cell 23: 219–225 [DOI] [PubMed] [Google Scholar]
- Kumlehn J, Kirik V, Czihal A, Altschmied L, Matzk F, Lörz H, Bäumlein H (2001) Parthenogenetic egg cells of wheat: cellular and molecular studies. Sex Plant Reprod 14: 239–243 [DOI] [PubMed] [Google Scholar]
- Lai Z, Schluttenhofer CM, Bhide K, Shreve J, Thimmapuram J, Lee SY, Yun DJ, Mengiste T (2014) MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat Commun 5: 3064. [DOI] [PubMed] [Google Scholar]
- Lê Q, Gutièrrez-Marcos JF, Costa LM, Meyer S, Dickinson HG, Lörz H, Kranz E, Scholten S (2005) Construction and screening of subtracted cDNA libraries from limited populations of plant cells: a comparative analysis of gene expression between maize egg cells and central cells. Plant J 44: 167–178 [DOI] [PubMed] [Google Scholar]
- Lopato S, Bazanova N, Morran S, Milligan AS, Shirley N, Langridge P (2006) Isolation of plant transcription factors using a modified yeast one-hybrid system. Plant Methods 2: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márton ML, Cordts S, Broadhvest J, Dresselhaus T (2005) Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576 [DOI] [PubMed] [Google Scholar]
- Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2011) The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, Kater MM, Colombo L (2010) VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22: 1702–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes MA, Guerra RF, Castelnovo B, Silva-Velazquez Y, Morandini P, Manrique S, Baumann N, Groß-Hardt R, Dickinson H, Colombo L (2016) Live and let die: a REM complex promotes fertilization through synergid cell death in Arabidopsis. Development 143: 2780–2790 [DOI] [PubMed] [Google Scholar]
- Menges M, Dóczi R, Okrész L, Morandini P, Mizzi L, Soloviev M, Murray JAH, Bögre L (2008) Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol 179: 643–662 [DOI] [PubMed] [Google Scholar]
- Mizzotti C, Ezquer I, Paolo D, Rueda-Romero P, Guerra RF, Battaglia R, Rogachev I, Aharoni A, Kater MM, Caporali E, et al. (2014) SEEDSTICK is a master regulator of development and metabolism in the Arabidopsis seed coat. PLoS Genet 10: e1004856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhonov VV, Theendakara VP, Gribanova YE, Ahmedli NB, Farber DB (2012) Sequence-specific binding of recombinant Zbed4 to DNA: insights into Zbed4 participation in gene transcription and its association with other proteins. PLoS ONE 7: e35317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C, Grotewold E (2005) Genome wide analysis of Arabidopsis core promoters. BMC Genomics 6: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll C, von Lyncker L, Zimmermann S, Kägi C, Baumann N, Twell D, Grossniklaus U, Gross-Hardt R (2008) CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J 56: 913–921 [DOI] [PubMed] [Google Scholar]
- Nallamsetty S, Austin BP, Penrose KJ, Waugh DS (2005) Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci 14: 2964–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numa H, Kim JM, Matsui A, Kurihara Y, Morosawa T, Ishida J, Mochizuki Y, Kimura H, Shinozaki K, Toyoda T, et al. (2010) Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana. EMBO J 29: 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V (2009) Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Sundaresan V (2007) Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19: 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher S, Sprunck S (2013) Downregulation of egg cell-secreted EC1 is accompanied with delayed gamete fusion and polytubey. Plant Signal Behav 8: e27377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Barutcu AR, McCord RP, Jeong JS, Jiang L, MacWilliams A, Yang X, Salehi-Ashtiani K, Hill DE, Blackshaw S, et al. (2011) Yeast one-hybrid assays for gene-centered human gene regulatory network mapping. Nat Methods 8: 1050–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccaro M, Li Y, Masiero S, Saedler H, Sommer H (2005) ROSINA (RSI), a novel protein with DNA-binding capacity, acts during floral organ development in Antirrhinum majus. Plant J 43: 238–250 [DOI] [PubMed] [Google Scholar]
- Rövekamp M, Bowman JL, Grossniklaus U (2016) Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr Biol 26: 1782–1789 [DOI] [PubMed] [Google Scholar]
- Schneitz K, Hulskamp M, Pruitt RE (1995) Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J 7: 731–749 [Google Scholar]
- Smale ST, Kadonaga JT (2003) The RNA polymerase II core promoter. Annu Rev Biochem 72: 449–479 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T (2005) The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J 41: 660–672 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Gross-Hardt R (2011) Nuclear behavior, cell polarity, and cell specification in the female gametophyte. Sex Plant Reprod 24: 123–136 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T (2012) Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN (2007) Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Takada S, Jürgens G (2007) Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134: 1141–1150 [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395: 917–921 [DOI] [PubMed] [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Schubert I, Tourmente S, Mathieu O (2006) MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep 7: 1273–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völz R, von Lyncker L, Baumann N, Dresselhaus T, Sprunck S, Gross-Hardt R (2012) LACHESIS-dependent egg-cell signaling regulates the development of female gametophytic cells. Development 139: 498–502 [DOI] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, Weiss M, Gheyselinck J, Lohr M, Wellmer F, Rahnenführer J, von Mering C, Grossniklaus U (2010) Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr Biol 20: 506–512 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell (Suppl) 16: S133–S141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Kinoshita T, Lotan O, Cohen G, Katz A, Choi Y, Nakashima K, Harada JJ, Goldberg RB, Fischer RL, et al. (2000) Mutations in the FIE and MEA genes that encode interacting Polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12: 2367–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kaur N, Kiriakopolos S, McCormick S (2006) EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta 224: 1004–1014 [DOI] [PubMed] [Google Scholar]
- Yu HJ, Hogan P, Sundaresan V (2005) Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol 139: 1853–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Liu Z, Song X, Johnson C, Yu X, Sundaresan V (2016) The CKI1 histidine kinase specifies the female gametic precursor of the endosperm. Dev Cell 37: 34–46 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang L, Yang Y, Cui J, Chang F, Wang Y, Ma H (2015) Analysis of Arabidopsis floral transcriptome: detection of new florally expressed genes and expansion of Brassicaceae-specific gene families. Front Plant Sci 5: 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Chen X, McCormick S (2011) The anaphase-promoting complex is a dual integrator that regulates both microRNA-mediated transcriptional regulation of cyclin B1 and degradation of cyclin B1 during Arabidopsis male gametophyte development. Plant Cell 23: 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, He K, Charron JB, Elling AA, Deng XW (2010) Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol 72: 585–595 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.