A regulatory module by which ethylene and cytokinin interaction affects rose flower senescence.
Abstract
In many plant species, including rose (Rosa hybrida), flower senescence is promoted by the gaseous hormone ethylene and inhibited by the cytokinin (CTK) class of hormones. However, the molecular mechanisms underlying these antagonistic effects are not well understood. In this study, we characterized the association between a pathogenesis-related PR-10 family gene from rose (RhPR10.1) and the hormonal regulation of flower senescence. Quantitative reverse transcription PCR analysis showed that RhPR10.1 was expressed at high levels during senescence in different floral organs, including petal, sepal, receptacle, stamen, and pistil, and that expression was induced by ethylene treatment. Silencing of RhPR10.1 expression in rose plants by virus-induced gene silencing accelerated flower senescence, which was accompanied by a higher ion leakage rate in the petals, as well as increased expression of the senescence marker gene RhSAG12. CTK content and the expression of three CTK signaling pathway genes were reduced in RhPR10.1-silenced plants, and the accelerated rate of petal senescence that was apparent in the RhPR10.1-silenced plants was restored to normal levels by CTK treatment. Finally, RhHB6, a homeodomain-Leu zipper I transcription factor, was observed to bind to the RhPR10.1 promoter, and silencing of its expression also promoted flower senescence. Our results reveal an ethylene-induced RhHB6-RhPR10.1 regulatory module that functions as a brake of ethylene-promoted senescence through increasing the CTK content.
The terminal phase of flower development is senescence, which is generally characterized by time to petal wilting, or withering, or by the time to turgid petal abscission (van Doorn, 2001). Flower senescence is a highly regulated process that exhibits many of the structural, biochemical, and molecular changes that are hallmarks of programmed cell death. These include a loss of membrane permeability, increase in reactive oxygen species, and decreased levels of protective enzymes, followed by protein degradation, fatty acid breakdown, and degradation of nucleic acids (Wagstaff et al., 2002; Jones et al., 2005; Xu et al., 2006; Tripathi and Tuteja, 2007; Yamada et al., 2009; Shahri and Tahir, 2011; Rogers, 2013).
The initiation of flower senescence can be triggered by external or internal cues (Gan and Amasino, 1997; Rogers, 2013; Zhang and Zhou, 2013), and phytohormones have been shown to play a pivotal role in triggering and modulating the progression of senescence, often through combinatorial interactions (Beaudoin et al., 2000; van Doorn and Woltering, 2008; Zhang and Zhou, 2013). Among these hormones, ethylene is considered a major regulator of flower senescence (van Doorn, 2001), and in ethylene-sensitive species, flower senescence is associated with a burst of ethylene production, and the coordinated expression of ethylene-responsive genes, including those with regulatory functions (Ichimura et al., 2009; Lerslerwong et al., 2009). For example, in Arabidopsis (Arabidopsis thaliana), the expression of microRNA164 was shown to be repressed by ethylene, thereby promoting senescence (Li et al., 2013), while in rose (Rosa hybrida), a reduction in transcript abundance of a homeodomain-Leu zipper (HD-Zip) I transcription factor gene, RhHB1, was observed to delay senescence (Lü et al., 2014). In contrast, the expression of another member of the HD-Zip I transcription factor family, HaHB-4, was reported to be up-regulated in response to ethylene in sunflower (Helianthus annuus), consequently delaying the senescence process (Manavella et al., 2006), suggesting that antagonistic mechanisms influence ethylene-induced senescence.
Members of another class of hormones, cytokinins (CTKs), have been proposed to act as antisenescence factors in flowers (Mayak and Halevy, 1970; Eisinger, 1977). Accordingly, it has been reported that rose varieties with long flower longevity contain higher levels of CTKs than those with shorter lived flowers (Mayak and Halevy, 1970), and CTK application has been shown to delay flower senescence in several plant species, including carnation (Dianthus caryophyllus), petunia (Petunia hybrida), and rose (Mayak and Halevy, 1970; Mor et al., 1983; Taverner et al., 1999). Conversely, an increase in transcript abundance of two genes encoding CTK oxidase/dehydrogenase during carnation petal senescence accelerated CTK degradation and promoted corolla senescence (Hoeberichts et al., 2007).
In addition to the individual effects of ethylene and CTK on flower senescence, these two hormones have been reported to interact in senescence. For example, in carnation, declining CTK levels act as a trigger for ethylene production (Eisinger, 1977), while treatment of carnation petals with CTK blocked the conversion of exogenously supplied 1-aminocyclopropane-1-carboxylic acid to ethylene (Mor et al., 1983; Taverner et al., 1999). In petunia, driving the overexpression of the CTK biosynthesis gene IPT with the Senescence-associated gene12 (SAG12) promoter resulted in a significant delay in flower senescence, accompanied by reduced ethylene sensitivity (Chang et al., 2003). In addition, it has been reported that the Arabidopsis KNOTTED-like homeodomain protein KNAT2 acts synergistically with CTKs and antagonistically with ethylene to delay senescence (Hamant et al., 2002). However, despite such observations exemplifying the antagonistic effects of ethylene and CTKs on flower senescence, the underlying molecular mechanisms and the many details of the genes in the associated regulatory networks are still largely unknown.
In this study of rose flower senescence, we identified a member of the pathogenesis-related (PR) PR-10 family as being involved in the cross talk between ethylene and CTK signaling. PR proteins, which function in a wide range of processes related to signal transduction and antimicrobial activity (Zubini et al., 2009), have been classified into 17 families based on structural features; of these, the PR-10 protein family is a large group containing more than 100 members (Somssich et al., 1988; Biesiadka et al., 2002; Hashimoto et al., 2004; Xu et al., 2014), which are typically small and localized in the cytosol (Fernandes et al., 2013). PR-10 proteins play multifunctional roles in defense mechanisms against abiotic and biotic stresses, and in developmental regulation via their RNase activity and/or interaction with ligands (Fernandes et al., 2013; Agarwal and Agarwal, 2014). In addition, CTKs can play a role in biological processes that are mediated by PR-10 proteins (Pasternak et al., 2006; Srivastava et al., 2006, 2007; Fernandes et al., 2008; Zubini et al., 2009). It has been proposed that when PR-10 proteins act as RNases, they may degrade certain types of tRNAs containing a CTK moiety, thereby contributing to the regulation of endogenous CTK content (Zubini et al., 2009). In support of this idea, overexpression of the pea (Pisum sativum) PR-10 gene ABR17 in Brassica napus and Arabidopsis enhanced abiotic stress tolerance through degradation of RNA to increase CTK levels (Srivastava et al., 2006, 2007). PR-10 proteins are also capable of binding CTKs; LIPR-10.2 from yellow lupine (Lupinus arboreus) has been shown to bind trans-zeatin, thereby acting as a reservoir of CTK molecules (Fernandes et al., 2008). However, the role of PR-10 proteins in modulating CTK content during flower senescence has not been characterized.
Roses are one of the most important ornamental crops worldwide, and their commodity value largely depends on the long vase life of the flowers. It is well known that ethylene promotes rose flower senescence (Ma et al., 2005), while CTKs delay the process (Mayak and Halevy, 1970; Lukaszewska et al., 1994). Here, we report that an ethylene-induced rose PR-10 family gene, RhPR10.1, inhibits ethylene-induced flower senescence via increasing the CTK content.
RESULTS
The Expression of RhPR10.1 Is Induced following Senescence and Exogenous Ethylene Treatment
To investigate the function of the PR-10 family genes during flower senescence, seven transcripts encoding putative PR-10 family proteins were identified in an in-house ethylene-treated rose (R. hybrida ‘Samantha’) petal transcriptome database (http://bioinfo.bti.cornell.edu/cgi-bin/rose_454/index.cgi.): RU05569, RU05592, RU06312, RU26673, RU20207, RU22712, and RU02982 (Supplemental Fig. S1). Expression analysis by quantitative reverse transcription PCR (qRT-PCR) showed that, of these genes, RU06312 exhibited a trend of increasing expression and the highest accumulation of transcripts during petal opening (Supplemental Fig. S2), and so we targeted this gene for further analysis.
We isolated a 766-bp RU06312 cDNA sequence with a 477-bp predicted open reading frame by rapid amplification of cDNA ends, encoding a deduced protein of 159 amino acids with a conserved P-loop motif and Bet v 1 motif (Lebel et al., 2010; Supplemental Fig. S3). Sequence alignment showed that RU06312 had a high degree of sequence homology to FaPR10-4 from Fragaria × ananassa (Supplemental Fig. S3), and the gene corresponding to RU06312 was named RhPR10.1. Phylogenetic analysis showed that RhPR10.1 belongs to the major pollen and food allergens subclass of PR10 protein family (Fernandes et al., 2013; Supplemental Fig. S3).
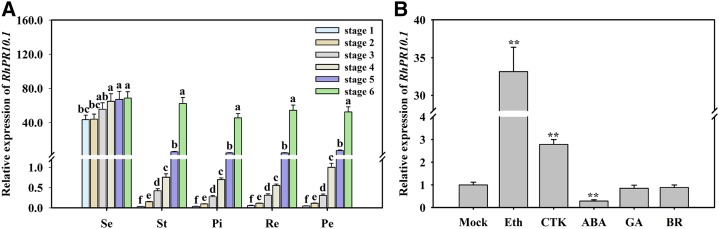
The expression of RhPR10.1 was evaluated in different organs during flower opening using qRT-PCR. The opening process of cut rose flowers can be divided into six stages (Ma et al., 2005). The expression patterns of RhPR10.1 in all the floral organs, including sepal, stamen, pistil, petal, and receptacle, showed similar trends, with transcript levels increasing progressively from the partially opened flower bud (stage 1) to the onset of petal wilting (stage 6; Fig. 1A). The expression was higher in sepals in all the flower opening stages (stages 1–6) than in the other floral organs (Fig. 1A). When the petals treated with hormones were tested, we observed that exogenous ethylene and CTK significantly induced RhPR10.1 expression, while an abscisic acid (ABA) treatment resulted in a decrease in expression. Gibberellic acid (GA) and brassinosteroid treatments did not alter RhPR10.1 transcript levels (Fig. 1B).
Figure 1.
Expression patterns of RhPR10.1 in different rose flower organs during various opening stages (A) and in rose petals in response to exogenous hormones (B). A, Se, sepal; St, stamen; Pi, pistil; Re, receptacle; Pe, petal. B, Eth, 10 µL L−1 ethylene; CTK, 100 µm 6-BA; ABA, 100 µm ABA; GA, 80 µm GA3; BR, 5 µm brassinosteroid. The petals at opening stage 2 were used as material. The results are the means of three biological replicates with standard deviations. Letters indicate significant differences according to Duncan’s multiple range test (P < 0.05), and asterisks indicate significant differences according to Student’s t test (*P < 0.05, **P < 0.01).
Silencing of RhPR10.1 Accelerates Rose Flower Senescence
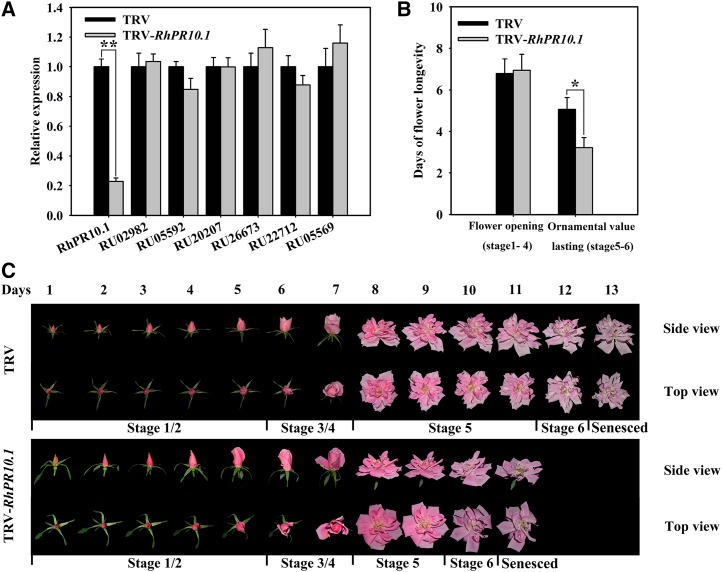
To investigate the potential role of RhPR10.1 in flower senescence, expression of the gene was silenced using virus-induced gene silencing (VIGS). We constructed a tobacco rattle virus vector (TRV-RhPR10.1) from the 3′ end region of RhPR10.1 (Supplemental Fig. S1) to specifically silence RhPR10.1 expression in rose plantlets and petal discs. As shown in Figure 2A, with the exception of RhPR10.1, none of the RhPR10 family genes exhibited reduced expression levels in RhPR10.1-silenced petals compared with the TRV control, thereby confirming the specificity of the gene silencing.
Figure 2.
Silencing of RhPR10.1 promotes flower senescence. A, Expression of the RhPR10 genes in RhPR10.1-silenced and TRV control flowers was analyzed by qRT-PCR. B, The time of flower opening (stages 1–4) and retention of ornamental value (stages 5 and 6) were recorded. C, Flower phenotypes were recorded and photographed every day. Stages of rose flowering were defined as follows: stage 1, bud with completely opened sepal; stage 2, bud with the outmost petal layer loosened; stage 3, flower with the outmost petal layer opened; stage 4, flower with the inner petal layer loosened; stage 5, fully opened flower; and stage 6, flower with fading color and loss of ornamental value. The results are the means of three biological replicates with standard deviations. Asterisks indicate statistically significant differences (Student’s t test, *P < 0.05, **P < 0.01).
The flower life span of rose plantlets can be divided into six stages. Stages 1 to 4 are defined as the flower opening phase, where the flowers have not fully opened, while stages 5 and 6 are defined as phases with ornamental value, starting from when the flower fully opens until color fading has occurred (Fig. 2C). The duration of the flower opening phase in the RhPR10.1-silenced plant was indistinguishable from that of the TRV control, with stages 1 to 4 lasting 6.8 ± 0.6 and 6.9 ± 0.8 d, respectively (Fig. 2, B and C). The flower sizes were also not statistically significant difference between TRV control and RhPR10.1-silenced plantlets. However, in the ornamental value phases, RhPR10.1 silencing caused an accelerated senescence phenotype, with stages 5 and 6 lasting only 3.7 ± 0.4 d compared with 5.1 ± 0.5 d in the TRV control (Fig. 2, B and C). In addition, we observed that the flower life spans of TRV control plantlets were indistinguishable from that of the wild type (Supplemental Fig. S4), indicating TRV control did not affect the flower opening and senescence.
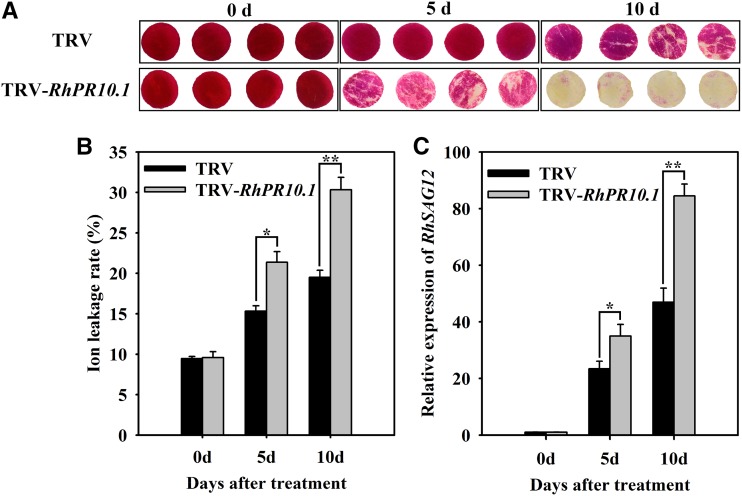
We further analyzed the effect of RhPR10.1 silencing on senescence of petal discs. In the TRV control, slight color fading occurred at 10 d (Fig. 3A). In contrast, RhPR10.1-silenced discs showed color fading at day 5, and almost all the petal discs were discolored at day 10 (Fig. 3A). In addition, the ion leakage rate and expression of the senescence marker gene RhSAG12 (Lü et al., 2014) were significantly higher in RhPR10.1-silenced petal discs than in the TRV control (Fig. 3, B and C).
Figure 3.
Silencing of RhPR10.1 promotes senescence in rose petal discs. A, The color-fading phenotypes of petal discs were recorded. B and C, Ion leakage rate (B) and expression of RhSAG12 (C) were analyzed in RhPR10.1-silenced and TRV control petals. The results are the means of three biological replicates with standard deviations. Asterisks indicate significant differences (Student’s t test, *P < 0.05, **P < 0.01).
RhPR10.1 Silencing-Induced Senescence Is Associated with the CTK Pathway
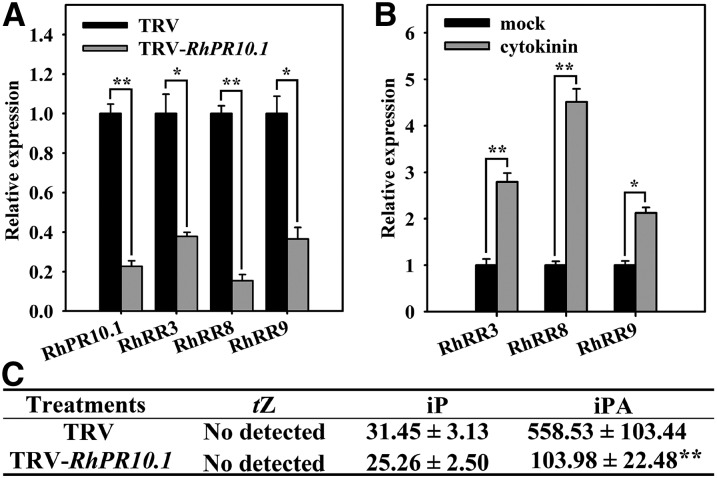
To investigate whether RhPR10.1 influences the CTK pathway during senescence, we measured the expression of three CTK signaling pathway genes, RhRR3, RhRR8, and RhRR9, in RhPR10.1-silenced rose petals. qRT-PCR analysis showed that the transcript abundance of all three genes, along with RhPR10.1, was reduced in petals of RhPR10.1-silenced flowers compared with TRV controls (Fig. 4A). In addition, the expression of the three genes increased in response to exogenous CTK, confirming their involvement in the CTK response pathway (Fig. 4B). We next examined the contents of the biologically active CTKs trans-zeatin, N6-(∆2-isopentenyl)-adenine (iP), and isopentenyl adenosine (iPA) in the petals of the RhPR10.1-silenced flowers. iPA levels were found to be significantly reduced in the silenced flowers, while trans-zeatin levels were undetectable in both the TRV control and the RhPR10.1-silenced petals (Fig. 4C), and no significant difference was observed in iP contents (Fig. 4C).
Figure 4.
RhPRh10.1 silencing reduces CTK signaling and levels in rose petals. A and B, Expression of genes related to the CTK signaling pathway in petals of RhPR10.1-silenced and TRV control flowers (A), and in rose wild-type petals treated with CTK (B). C, CTK contents in petals of RhPR10.1-silenced and TRV control flowers; tZ, trans-zeatin. Values are means (pg g−1 fresh weight) ± sd. The results are the means of three biological replicates with standard deviations. Asterisks indicate statistically significant differences (Student’s t test, *P < 0.05, **P < 0.01).
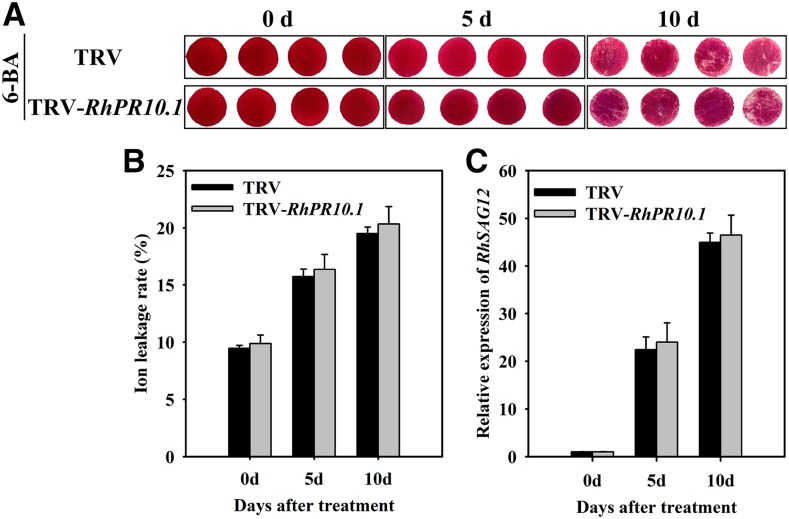
Since CTK response and content were altered in RhPR10.1-silenced flowers, we speculated that RhPR10.1-mediated flower senescence may function by regulating CTK pathway. We therefore examined the effect of treating RhPR10.1-silenced petals with 6-benzyl aminopurine (6-BA; a synthetic CTK), and found that its application resulted in a slight color fading after 10 d in both the TRV control and RhPR10.1-silenced petal discs (Fig. 5A). In addition, similar expression levels of RhSAG12 and ion leakage rates were detected in petal discs from the TRV control and the RhPR10.1-silenced plants (Fig. 5, B and C).
Figure 5.
The effect of exogenous CTK treatment on senescence of RhPR10.1-silenced rose petal discs. A, The phenotypes of petal discs treated with 100 µm 6-BA were recorded. B and C, The ion leakage rates (B) and transcript levels of RhSAG12 (C) in RhPR10.1-silenced and TRV control petal discs treated by 6-BA. Statistical differences were analyzed by Student’s t test (P < 0.05).
RhPR10.1 Is Directly Regulated by Transcription Factor RhHB6
To better understand the regulatory mechanism of RhPR10.1, a 1,378-bp promoter region upstream of the RhPR10.1 coding sequence was identified. Sequence analysis using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) suggested that the cis-elements in the promoter of RhPR10.1 included 13 homologous sequences of ARR1AT for CTK response regulator, five binding sites for MYB transcription factor, four binding sites for MYC, and three binding site for WRKY (Supplemental Fig. S5). In addition, a known binding site (CAATTATTA) for the HD-Zip subfamily I member ATHB6 (Himmelbach et al., 2002) was located between positions −706 and −715 (Supplemental Fig. S5). A search of an in-house-generated ethylene-treated rose petal transcriptome database (http://bioinfo.bti.cornell.edu/cgi-bin/rose_454/index.cgi) revealed three members of the HD-Zip I subfamily, RhHB1 (RU00522; Lü et al., 2014), RhHB6 (RU12591), and RhHB7 (RU00731), as well as a member of the HD-Zip II subfamily, RhHB2 (RU01217).
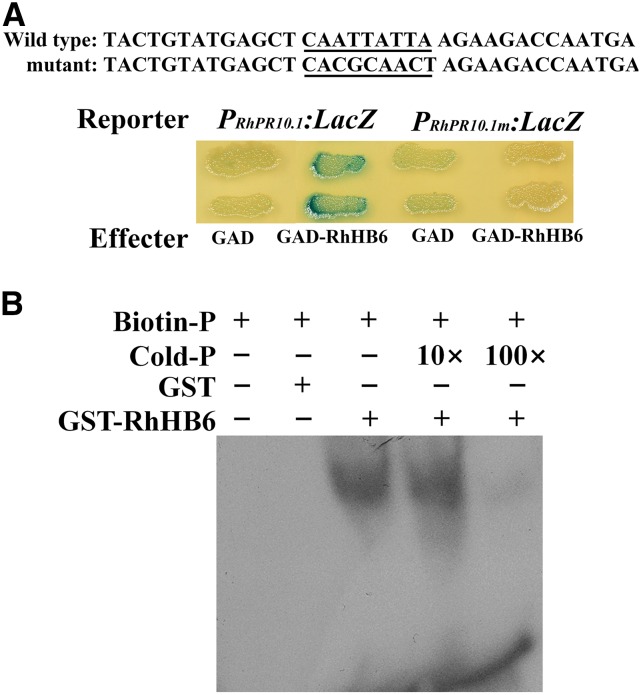
We conducted a yeast one-hybrid assay, using the cis-element in the RhPR10.1 promoter, CAATTATTA, as bait, to assess the interaction of the HD-Zip family members with the RhPR10.1 promoter (Supplemental Fig. S6). RhHB1, RhHB7, and RhHB2 did not bind to the RhPR10.1 promoter (Supplemental Fig. S6); however, RhHB6 bound and triggered expression of the LacZ reporter gene (Fig. 6A), but did not bind a mutated RhPR10.1 fragment (Fig. 6A). An electrophoretic mobility shift assay (EMSA) was conducted to confirm the interaction of RhHB6 and the RhPR10.1 promoter, and RhHB6 was found to bind to the biotin-labeled proRhPR10.1 probe; moreover, binding was gradually attenuated by increasing concentrations of unlabeled probe (Fig. 6B).
Figure 6.
RhHB6 binds to a cis-element in the promoter of RhPR10.1. A, Transactivation activity of RhHB6 to the RhPR10.1 promoter in yeast. Wild-type and mutant probes were derived from the RhPR10.1 promoter. The wild-type cis-element and its nucleotide substitutions in the mutant versions are underlined. B, EMSA was used to analyze the interaction of GST-RhHB6 and a biotin-labeled probe. Purified protein (3 µg) samples were incubated with 25 pm of the biotin-labeled wild-type probe. Nonlabeled probes at 10- and 100-fold concentrations were added for the competition test. Images are representative of three independent experiments.
Silencing of RhHB6 Accelerates Rose Flower Senescence
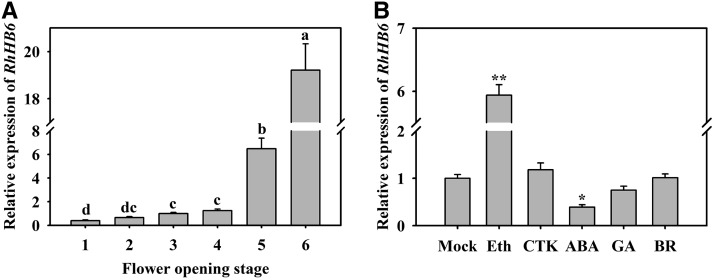
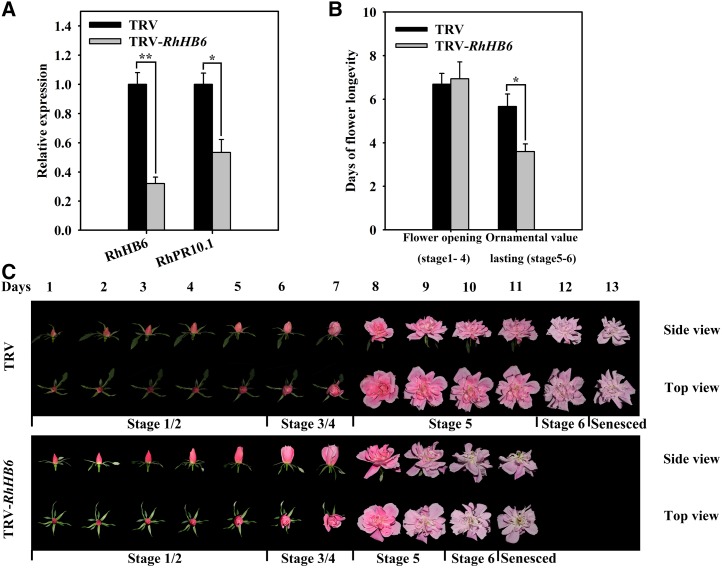
To investigate the potential function of RhHB6 in rose petal senescence, we evaluated its expression in different flowering stages and following various hormone treatments. As shown in Figure 7, the expression of RhHB6 increased during flower opening (Fig. 7A), and was induced by ethylene but inhibited by an ABA treatment (Fig. 7B). When RhHB6 was silenced in rose plantlets by VIGS, the expression levels of RhHB6 and RhPR10.1 were lower in petals of the silenced flowers than in those of the TRV control (Fig. 8A). The duration of the flower opening phase of the RhHB6-silenced flowers was similar to that of the TRV control (Fig. 8, B and C); however, RhHB6 silencing caused an accelerated senescence phenotype during the ornamental value phases, with stages 5 and 6 only lasting 3.6 ± 0.3 d compared with 5.7 ± 0.6 d for the TRV control (Fig. 8, B and C).
Figure 7.
Expression of RhHB6 in rose petals at different flower opening stages (A) and in response to exogenous hormones (B). Eth, 10 µL L−1 ethylene; CTK, 100 µm 6-BA; ABA, 100 µm ABA; GA, 80 µm GA3; BR, 5 µm brassinosteroid. The petals at stage 2 were used as material in B. The results are the means of three biological replicates with standard deviations. Letters indicate significant differences according to Duncan’s multiple range test (P < 0.05), and asterisks indicate significant differences according to Student’s t test (*P < 0.05, **P < 0.01).
Figure 8.
Silencing of RhHB6 promotes flower senescence. A, Expression of the RhHB6 and RhPR10.1 genes in RhHB6-silenced and TRV control petals was analyzed by qRT-PCR. B, The time of flower opening and senescence phases was recorded. C, The phenotypes of flowers were recorded and photographed every day. The results are the means of three biological replicates with standard deviations. Asterisks indicate statistically significant differences (Student’s t test, *P < 0.05, **P < 0.01).
DISCUSSION
RhPR10.1 Is Involved in Ethylene-Induced Rose Petal Senescence
Flower senescence is accompanied by changes in the levels of endogenous hormones, which regulate networks of signaling events that control the senescence program (van Doorn and Woltering, 2008; Zhang and Zhou, 2013). Ethylene is known to be a crucial accelerant of flower senescence. Based on the results generated in this study, we propose that RhPR10.1 is a member of the network of genes involved in the ethylene-induced senescence process in rose flowers. This hypothesis is supported by the expression pattern of RhPR10.1, which was found to be significantly induced by ethylene treatments (Fig. 1B), and by the accelerated flower senescence phenotype of the RhPR10.1 VIGS plants (Figs. 2 and 3).
The expression of PR-10 family members is known to increase during senescence, although their function in senescence is not clear. In bean (Phaseolus vulgaris), the transcript levels of the PR-10 protein Ypr10 were reported to increase at the onset of leaf senescence (Walter et al., 1996), and in yellow lupine, the expression of the PR-10 gene LlPR10.1A was induced in senescent leaves (Sikorski et al., 1999). Here, we showed that in rose, the transcript levels of several PR10 members also accumulated during flower senescence (Supplemental Fig. S2). Although we targeted RhPR10.1 with the highest accumulation among RhPR-10 members for analyzing the function in senescence, it is feasible that other RhPR-10 genes may also play roles in petal senescence, especially those showing trends of increasing expression during flower senescence. Therefore, additional studies are required to determine whether other RhPR-10 members have functions in flower senescence.
Intriguingly, functional analysis of the RhPR10.1 gene indicated that the effect of RhPR10.1 on senescence is antagonistic, with reduced RhPR10.1 expression promoting flower senescence (Figs. 2 and 3). In Arabidopsis, during the last step of petal senescence, abscission, the expression of BREVIPEDICELLUS (BP)/KNAT1 is induced, and the accelerated petal abscission phenotype of its knockout mutant, bp, indicates that BP/KNAT1 plays an antagonistic role in the petal abscission process (Shi et al., 2011). It has also been shown that in sunflower, increased expression of HaHB-4 suppresses flower senescence (Manavella et al., 2006). These represent antagonistic mechanisms that may be important in preventing premature organ senescence.
RhPR10.1 Inhibits Ethylene-Induced Rose Flower Senescence through Regulation of CTK Signaling and Content
Altered levels of CTKs and the expression of three CTK signaling pathway genes were observed in rose petals with reduced RhPR10.1 expression (Fig. 4), suggesting that RhPR10.1 may play a role in modulating CTK levels. Sequence alignment showed that RhPR10.1 has a conserved P-loop domain, which functions as a nucleotide binding site correlated to RNase activity (Supplemental Fig. S3A; Liu and Ekramoddoullah, 2006). Three conserved residues related to ribonucleolytic activity of PR-10 proteins also existed in RhPR10.1 protein, including Glu (E) at amino acid position 96, 148, and Tyr (Y) or His (H) at 150 (Supplemental Fig. S3A; Liu et al., 2006; Lebel et al., 2010). The two conserved features above suggest that RhPR10.1 may have RNase activity. In addition, RhPR10.1 belongs to the major pollen and food allergens subclass of the PR10 protein family (Supplemental Fig. S3B). It is known that PR-10 proteins from the major pollen and food allergens subclass acting as RNases can degradate certain types of tRNAs (Fernandes et al., 2013), which have been suggested to be a major source of CTK since isoprenoid CTKs have been identified in tRNA hydrolysates (Sakakibara, 2006). Moreover, although PR-10 protein has been reported to have capable of binding to trans-zeatin (Fernandes et al., 2008, 2009), here we found that the levels of trans-zeatin were undetectable in both TRV and RhPR10.1-silenced petals (Fig. 4C), suggesting that RhPR10.1 might not interact with trans-zeatin during petal senescence.
It has been suggested that PR-10-mediated tRNA degradation may result in the accumulation of different types of CTKs, depending on plant species and environmental conditions (Srivastava et al., 2006, 2007). Ectopic expression of the pea PR-10 family gene ABR17 in both B. napus and Arabidopsis enhanced endogenous CTK levels (Srivastava et al., 2006, 2007), and in ABR17-overexpressing B. napus seedlings, a significant increase in trans-zeatin riboside, but not iP or cis-zeatin, was observed (Srivastava et al., 2006). However, in ABR17-overexpressed Arabidopsis seedlings, increases in the concentrations of cis-zeatin and iP were observed in both Murashige and Skoog medium- and soil-grown plants, while trans-zeatin content was only increased in Murashige and Skoog-grown plants (Srivastava et al., 2007). In this study, we detected a significant reduction in the levels of iPA, but not trans-zeatin or iP, in RhPR10.1-silenced flowers (Fig. 4C), suggesting that different pathways may function in CTK accumulation in rose petals as a consequence of tRNA degradation. In carnation flower, among iso-pentenyladenine derivatives, iPA was most abundant during flower senescence, and application of iPA delayed the onset of flower senescence (van Staden et al., 1990). It suggests that iPA may play an important role in flower senescence.
RhHB6 Acts Upstream of RhPR10.1 to Regulate Flower Senescence
HD-Zip transcription factors have been grouped into four different classes (HD-Zip I–IV) based on sequence similarity criteria, and these groupings are further supported by their intron/exon patterns (Henriksson et al., 2005). Previous studies have reported that HD-Zip I transcription factors are involved in abiotic stress responses (Ariel et al., 2010), fruit ripening (Lin et al., 2008), and leaf and flower senescence (Manavella et al., 2006; Lü et al., 2014). Here we showed that RhHB6 acts as a regulator of RhPR10.1, and that its expression was also up-regulated in senescent rose petals and involved in ethylene-induced flower senescence (Figs. 7 and 8). A regulatory action of RhHB6/RhPR10.1 is suggested based on the observation that RhHB6 binds to a 9-bp CAATTATTA region of the RhPR10.1 promoter, as revealed by EMSA and yeast one-hybrid analysis (Fig. 6, A and B). In Arabidopsis, ATHB6 recognized the same 9-bp cis-element present in the promoters of its target genes (Himmelbach et al., 2002). In addition, ATHB6 specific targeting of this cis-element was revealed by the elimination of the interaction when a single nucleotide was mutated in this cis-element (Himmelbach et al., 2002). The effects of RhHB6 silencing in rose (Fig. 8) are consistent with those of RhPR10.1 silencing, in that both accelerated flower senescence (Fig. 2). We conclude that the ethylene-induced RhHB6-RhPR10.1 regulatory module inhibits ethylene-induced flower senescence through increasing the levels of CTKs (Supplemental Fig. S7).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
For expression pattern analyses of RhPR10.1 and RhHB6, cut rose (Rosa hybrida ‘Samantha’) flowers were harvested from a local commercial greenhouse and transported to the laboratory within 1h. Stems were then recut under water to 25 cm length and placed in vases with distilled water. The cut flower opening stages were defined as described previously (Ma et al., 2005). Floral organ samples were collected at different opening stages, and petal samples were collected from the same middle whorl of the flowers.
For hormone treatments, cut rose flowers at opening stage 2 were placed in vases containing 100 µm 6-BA, 100 µm ABA, 80 µm GA3, or 5 µm brassinosteroid for 24 h. Mock samples were placed in 0.1% dimethyl sulfoxide. For ethylene treatment, the cut rose flowers at opening stage 2 were exposed to 10 µL L−1 ethylene in an airtight chamber for 24 h. NaOH solution (1 m) was also applied in the chamber to prevent the CO2 accumulation (Lü et al., 2014).
For the VIGS assay, rose plantlets were propagated by in vitro culturing. Rose shoots of at least 2 cm, and including one node, were cultured on propagation medium comprising Murashige and Skoog medium supplemented with 1.0 mg L−1 6-BA, 0.05 mg L−1 α-naphthalene acetic acid, and 3 mg L−1 GA3 for 30 d, before being transferred to rooting medium, comprising half-strength Murashige and Skoog supplemented with 0.1 mg L−1 α-naphthalene acetic acid for 30 d. Flower opening stages were defined as follows: stage 1, bud with completely opened sepal; stage 2, bud with outmost petal layer loosened; stage 3, flower with outmost petal layer opened; stage 4, flower with inner petal layer loosened; stage 5, fully opened flower; and stage 6, flower with faded color and, consequently, reduced ornamental value.
Cloning, Plasmid Construction, and Plant Transformation
The full-length and promoter sequences of RhHB6 and RhPR10.1 were amplified using rapid amplification of cDNA ends (Clontech) and thermal asymmetric interlaced PCR (Takara) according to the manufacturer’s instructions, respectively. All PCR products were subcloned into the pGEM-T easy vector (Promega) and then transformed into Escherichia coli DH5α cells prior to sequencing.
The RhHB6 VIGS construct was created by inserting a 365-bp fragment of the RhHB6 3′-untranslated region into the pTRV2 vector, and the RhPR10.1 VIGS construct by inserting a 210-bp fragment (Supplemental Fig. S1) from the 3′-untranslated region into the pTRV2 vector.
To express the RhHB6 recombinant protein in E. coli for the EMSA (see below for details), the open reading frame of RhHB6 was inserted into the pGEX-4T-2 vector to produce the GST-RhHB6 fusion protein.
For the yeast one-hybrid assay (see below), the open reading frame of RhHB6 was cloned into the EcoRI and XhoI sites of the pJG4-5 vector (Clontech), resulting in the GAD-RhHB6 construct. To generate a construct expressing the LacZ reporter gene driven by the wild-type or mutant motif of the RhPR10.1 promoter, a 35-bp wild-type or mutant oligonucleotide was synthesized and ligated into the pLacZi2μ vector (Lin et al., 2007). All primers used above are listed in Supplemental Table S1.
RNA Extraction and qRT-PCR
Total RNA was extracted from rose floral organs as described previously (Xue et al., 2008). One microgram of DNase-treated RNA was used to synthesize cDNA in a 25-μL reaction volume by Superscript Reverse Transcriptase (Invitrogen). qRT-PCR reactions were performed using 1 μL cDNA as template and the Step One Plus real-time PCR system (Applied Biosystems) with the KAPA SYBR FAST quantitative PCR kit (Kapa Biosystems). RhACT5 was used as an internal control (Pei et al., 2013).
Sequence Analysis
Deduced amino acid sequences were aligned using ClustalX and DNAMAN, and phylogenetic analysis was performed using MEGA. The phylogenetic tree was computed using the Neighbor-Joining algorithm with 1,000 bootstrap replicates.
VIGS
Silencing of RhPR10.1 and RhHB6 in petal discs and plantlets by VIGS was conducted as described previously (Ma et al., 2008; Dai et al., 2012; Tian et al., 2014), but with some modifications. The pTRV1, pTRV2, pTRV-RhPR10.1, and pTRV-RhHB6 vectors were transformed into Agrobacterium tumefaciens strain GV3101. The transformed A. tumefaciens lines were cultured in Luria-Bertani medium supplemented with 10 mm MES, 20 mm acetosyringone, 50 μg mL−1 kanamycin, and 50 μg mL−1 gentamycin sulfate. The cultures were harvested by centrifugation at 4,000 rpm for 10 min, and resuspended in infiltration buffer (10 mm MgCl2, 200 mm acetosyringine, 10 mm MES, pH 5.6) to a final OD600 of approximately 1.5. Mixtures of cultures containing an equal ratio (v/v) of pTRV1 and pTRV2, pTRV1 and pTRV-RhPR10.1, or pTRV1 and pTRV-RhHB6, were used as TRV control, TRV-RhHB6, and TRV-RhPR10.1 experiments, respectively. The culture mixtures were placed at room temperature in the dark for 3 to 6 h before vacuum infiltration into the rose petal discs or plantlets.
For VIGS in rose petal discs, the petals were collected from the same middle whorl at stage 2 of flower opening, and 1-cm diameter discs were excised from the center of the petals using a hole punch. For VIGS in rose plantlets, plantlets were propagated by in vitro culturing as described above. The plantlets were washed in deionized water, and the roots were placed in water for 2 d to equilibrate. Vacuum infiltration was performed by immersing the discs or plantlets in the bacterial suspension and infiltrating under a vacuum at 0.7 MPa. After release of the vacuum, the discs or plantlets were washed in deionized water. Discs were placed in petri dishes, and plantlets were transplanted into a mixture of vermiculite and nutritive soil (1:1). The discs and plantlets were then placed in the dark at 8°C for 3 d. For RNA extraction, discs were kept in deionized water at 23°C until sampling. The phenotypes of the discs were observed daily until the onset of necrosis. Three independent experiments were performed with 40 petal discs in each experiment. The flowers of the rose plantlets were observed from stages 1 to 6. Three independent experiments were performed with 30 plantlets in each experiment.
Extraction and Quantification of Endogenous CTKs
The extraction and quantification of endogenous CTKs was performed as previously described (Pan et al., 2010). Rose petals (100 mg) were frozen in liquid nitrogen, ground to a powder in a mortar, and transferred into 2-mL tubes. Extraction solvent (2-propanol/H2O/concentrated HCl [2:1:0.002, v/v/v]) was added to each tube, keeping the ratio of sample:solvent at 1:10 (mg µL−1). Samples were shaken at 100 rpm for 30 min at 4°C. One milliliter dichloromethane was added to each sample, and then samples were shaken at 100 rpm for 30 min at 4°C. After centrifugation at 13,000g for 5 min (4°C), the solvent (approximately 1.5 mL) from the lower phase was transferred into a new 2-mL tube and concentrated (not completely to dryness) using a concentrator (Eppendorf), and redissolved in 0.1 mL methanol. Quantitative analysis of endogenous CTKs in crude rose flower extracts was performed by high-performance liquid chromatography electrospray ionization tandem mass spectrometry.
Measurement of Electrolyte Leakage Rates
Electrolyte leakage rates were measured as described previously (Lee et al., 2015), with minor modifications. Briefly, membrane leakage was determined by measurement of electrolytes leaking from rose petal discs. Sixteen rose petal discs from each treatment were immersed in 15 mL of 0.4 m mannitol at room temperature with gentle shaking for 3h, and initial conductivity of the solution was measured with a conductivity meter (DDBJ-350; LeiCi). Total conductivity was determined after sample incubation at 85°C for 20 min. The electrolyte leakage rates were calculated as the percentage of initial conductivity divided by total conductivity.
Yeast One-Hybrid Assay
The yeast one-hybrid assay was performed by transferring LacZ reporter gene constructs into yeast strain EGY48, as described in the Yeast Protocols Handbook (Clontech). Transformants were grown on synthetic dextrose plates lacking uracil and Trp, but containing X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to observe the color development of yeast colonies.
Purification of GST-RhHB6 Recombinant Protein and EMSA
The EMSA was performed as described previously (Dai et al., 2012). Expression of the GST-RhHB6 fusion protein was induced in 100-mL cultures of transformed E. coli BL21 cells (see above) by addition of isopropylthio-β-galactoside to a final concentration of 0.2 mm, before the cultures were incubated at 20°C for 6 h. The recombinant proteins were extracted from the cells and purified using glutathione Sepharose 4B beads according to the manufacturer’s instructions (GE Healthcare). EMSA was performed using a Light Shift chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instructions. Biotin-labeled DNA fragments (wild-type fragment in Fig. 6A) were synthesized and used as probes, while unlabeled DNA of the same sequence was used as a competitor.
Accession Numbers
The GenBank (http://www.ncbi.nlm.nih.gov) accession numbers for rose genes used in this study are as follows: RhPR10.1 (KX462848) and RhHB6 (KX462849).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of seven rose PR-10 family cDNA sequences.
Supplemental Figure S2. qRT-PCR analysis of the expression of seven PR-10 family genes in rose petals at different flower opening stages.
Supplemental Figure S3. Alignment of deduced amino acid sequences (A) and a phylogenetic tree (B) of the RhPR10.1 protein and representative PR-10 members from other species.
Supplemental Figure S4. Flower phenotypes of wild-type and TRV control plantlets.
Supplemental Figure S5. A pictorial representation of the promoter region of RhPR10.1 with potential cis-element binding sites.
Supplemental Figure S6. Transactivation activity of four HD-Zip transcription factors to the RhPR10.1 promoter in yeast.
Supplemental Figure S7. Proposed model for how the RhHB6-RhPR10.1 module regulates ethylene-induced flower senescence.
Supplemental Table S1. Primer list.
Supplementary Material
Acknowledgments
We thank PlantScribe (www.plantscribe.com) for careful editing of this article.
Glossary
- CTK
cytokinin
- VIGS
virus-induced gene silencing
- iP
N6-(∆2-isopentenyl)-adenine
- iPA
isopentenyl adenosine
- 6-BA
6-benzyl aminopurine
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31520103913, 31471903, and 31130048).
Articles can be viewed without a subscription.
References
- Agarwal P, Agarwal PK (2014) Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol Biol Rep 41: 599–611 [DOI] [PubMed] [Google Scholar]
- Ariel F, Diet A, Verdenaud M, Gruber V, Frugier F, Chan R, Crespi M (2010) Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22: 2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiadka J, Bujacz G, Sikorski MM, Jaskolski M (2002) Crystal structures of two homologous pathogenesis-related proteins from yellow lupine. J Mol Biol 319: 1223–1234 [DOI] [PubMed] [Google Scholar]
- Chang H, Jones ML, Banowetz GM, Clark DG (2003) Overproduction of cytokinins in petunia flowers transformed with P(SAG12)-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol 132: 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Zhang C, Jiang X, Kang M, Yin X, Lü P, Zhang X, Zheng Y, Gao J (2012) RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol 160: 2064–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger W. (1977) Role of cytokinins in carnation flower senescence. Plant Physiol 59: 707–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes H, Bujacz A, Bujacz G, Jelen F, Jasinski M, Kachlicki P, Otlewski J, Sikorski MM, Jaskolski M (2009) Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein. FEBS J 276: 1596–1609 [DOI] [PubMed] [Google Scholar]
- Fernandes H, Michalska K, Sikorski M, Jaskolski M (2013) Structural and functional aspects of PR-10 proteins. FEBS J 280: 1169–1199 [DOI] [PubMed] [Google Scholar]
- Fernandes H, Pasternak O, Bujacz G, Bujacz A, Sikorski MM, Jaskolski M (2008) Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin. J Mol Biol 378: 1040–1051 [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1997) Making sense of senescence: molecular genetic regulation and manipulation of leaf senescence. Plant Physiol 113: 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Nogué F, Belles-Boix E, Jublot D, Grandjean O, Traas J, Pautot V (2002) The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol 130: 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T (2004) A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol 45: 550–559 [DOI] [PubMed] [Google Scholar]
- Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E (2005) Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol 139: 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Höhener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts FA, van Doorn WG, Vorst O, Hall RD, van Wordragen MF (2007) Sucrose prevents up-regulation of senescence-associated genes in carnation petals. J Exp Bot 58: 2873–2885 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Yamada T, Yoshioka S, Pun U, Tanase K, Shimizu-Yumoto H, Ottosen C, Grout B, Mueller R (2009) Ethylene regulates programmed cell death (PCD) associated with petal senescence in carnation flowers. Acta Hortic 847: 185–190 [Google Scholar]
- Jones ML, Chaffin GS, Eason JR, Clark DG (2005) Ethylene-sensitivity regulates proteolytic activity and cysteine protease gene expression in petunia corollas. J Exp Bot 56: 2733–2744 [DOI] [PubMed] [Google Scholar]
- Lebel S, Schellenbaum P, Walter B, Maillot P (2010) Characterisation of the Vitis vinifera PR10 multigene family. BMC Plant Biol 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sakuraba Y, Lee T, Kim KW, An G, Lee HY, Paek NC (2015) Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J Integr Plant Biol 57: 562–576 [DOI] [PubMed] [Google Scholar]
- Lerslerwong L, Ketsa S, van Doorn WG (2009) Protein degradation and peptidase activity during petal senescence in Dendrobium cv. Khao sanan. Postharvest Biol Technol 52: 84–90 [Google Scholar]
- Li Z, Peng J, Wen X, Guo H (2013) Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ekramoddoullah AKM (2006) The family 10 of plant pathogenesis-related proteins: their structure regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68: 3–13 [Google Scholar]
- Liu X, Huang B, Lin J, Fei J, Chen Z, Pang Y, Sun X, Tang K (2006) A novel pathogenesis-related protein (SsPR10) from Solanum surattense with ribonucleolytic and antimicrobial activity is stress- and pathogen-inducible. J Plant Physiol 163: 546–556 [DOI] [PubMed] [Google Scholar]
- Lü P, Zhang C, Liu J, Liu X, Jiang G, Jiang X, Khan MA, Wang L, Hong B, Gao J (2014) RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. Plant J 78: 578–590 [DOI] [PubMed] [Google Scholar]
- Lukaszewska AJ, Bianco J, Barthe P, Le Page-Degivry MT (1994) Endogenous cytokinins in rose petals and the effect of exogenously applied cytokinins on flower senescence. Plant Growth Regul 14: 119–126 [Google Scholar]
- Ma N, Cai L, Lu W, Tan H, Gao J (2005) Exogenous ethylene influences flower opening of cut roses (Rosa hybrida) by regulating the genes encoding ethylene biosynthesis enzymes. Sci China C Life Sci 48: 434–444 [DOI] [PubMed] [Google Scholar]
- Ma N, Xue J, Li Y, Liu X, Dai F, Jia W, Luo Y, Gao J (2008) Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiol 148: 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella PA, Arce AL, Dezar CA, Bitton F, Renou JP, Crespi M, Chan RL (2006) Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J 48: 125–137 [DOI] [PubMed] [Google Scholar]
- Mayak S, Halevy AH (1970) Cytokinin activity in rose petals and its relation to senescence. Plant Physiol 46: 497–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor Y, Spiegelstein H, Halevy AH (1983) Inhibition of ethylene biosynthesis in carnation petals by cytokinin. Plant Physiol 71: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc 5: 986–992 [DOI] [PubMed] [Google Scholar]
- Pasternak O, Bujacz GD, Fujimoto Y, Hashimoto Y, Jelen F, Otlewski J, Sikorski MM, Jaskolski M (2006) Crystal structure of Vigna radiata cytokinin-specific binding protein in complex with zeatin. Plant Cell 18: 2622–263416998071 [Google Scholar]
- Pei H, Ma N, Tian J, Luo J, Chen J, Li J, Zheng Y, Chen X, Fei Z, Gao J (2013) An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol 163: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HJ. (2013) From models to ornamentals: How is flower senescence regulated? Plant Mol Biol 82: 563–574 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57: 431–449 [DOI] [PubMed] [Google Scholar]
- Shahri W, Tahir I (2011) Flower senescence-strategies and some associated events. Bot Rev 77: 152–184 [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA (2011) Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski MM, Biesiadka J, Kasperska AE, Kopcińska J, Łotocka B, Golinowski W, Legocki AB (1999) Expression of genes encoding PR10 class pathogenesis-related proteins is inhibited in yellow lupine root nodules. Plant Sci 149: 125–137 [Google Scholar]
- Somssich IE, Schmelzer E, Kawalleck P, Hahlbrock K (1988) Gene structure and in situ transcript localization of pathogenesis-related protein 1 in parsley. Mol Gen Genet 213: 93–98 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Emery RN, Kurepin LV, Reid DM, Fristensky B, Kav NN (2006) Pea PR 10.1 is a ribonuclease and its transgenic expression elevates cytokinin levels. Plant Growth Regul 49: 17–25 [Google Scholar]
- Srivastava S, Emery RN, Rahman MH, Kav NN (2007) A crucial role for cytokinins in pea ABR17-mediated enhanced germination and early seedling growth of Arabidopsis thaliana under saline and low-temperature stresses. J Plant Growth Regul 26: 26–37 [Google Scholar]
- Taverner E, Letham DS, Wang J, Cornish E, Willcocks DA (1999) Influence of ethylene on cytokinin metabolism in relation to Petunia corolla. Phytochemistry 51: 341–347 [Google Scholar]
- Tian J, Pei H, Zhang S, Chen J, Chen W, Yang R, Meng Y, You J, Gao J, Ma N (2014) TRV-GFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J Exp Bot 65: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SK, Tuteja N (2007) Integrated signaling in flower senescence: an overview. Plant Signal Behav 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG. (2001) Categories of petal senescence and abscission: a re-evaluation. Ann Bot (Lond) 87: 447–456 [Google Scholar]
- van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59: 453–480 [DOI] [PubMed] [Google Scholar]
- van Staden J, Upfold SJ, Bayley AD, Drewes FE (1990) Cytokinin in cut carnation flower. IX. Transport and metabolism of iso-pentenyladenine and the effect of its derivatives on flower longevity. Plant Growth Regul 9: 255–261 [Google Scholar]
- Wagstaff C, Leverentz MK, Griffiths G, Thomas B, Chanasut U, Stead AD, Rogers HJ (2002) Cysteine protease gene expression and proteolytic activity during senescence of Alstroemeria petals. J Exp Bot 53: 233–240 [DOI] [PubMed] [Google Scholar]
- Walter MH, Liu JW, Wünn J, Hess D (1996) Bean ribonuclease-like pathogenesis-related protein genes (Ypr10) display complex patterns of developmental, dark-induced and exogenous-stimulus-dependent expression. Eur J Biochem 239: 281–293 [DOI] [PubMed] [Google Scholar]
- Xu TF, Zhao XC, Jiao YT, Wei JY, Wang L, Xu Y (2014) A pathogenesis related protein, VpPR-10.1, from Vitis pseudoreticulata: an insight of its mode of antifungal activity. PLoS One 9: e95102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ishida H, Reisen D, Hanson MR (2006) Upregulation of a tonoplast-localized cytochrome P450 during petal senescence in Petunia inflata. BMC Plant Biol 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Li Y, Tan H, Yang F, Ma N, Gao J (2008) Expression of ethylene biosynthetic and receptor genes in rose floral tissues during ethylene-enhanced flower opening. J Exp Bot 59: 2161–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ichimura K, Kanekatsu M, van Doorn WG (2009) Homologs of genes associated with programmed cell death in animal cells are differentially expressed during senescence of Ipomoea nil petals. Plant Cell Physiol 50: 610–625 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhou C (2013) Signal transduction in leaf senescence. Plant Mol Biol 82: 539–545 [DOI] [PubMed] [Google Scholar]
- Zubini P, Zambelli B, Musiani F, Ciurli S, Bertolini P, Baraldi E (2009) The RNA hydrolysis and the cytokinin binding activities of PR-10 proteins are differently performed by two isoforms of the Pru p 1 peach major allergen and are possibly functionally related. Plant Physiol 150: 1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.