The seed-to-seedling transition in Arabidopsis can occur following multiple spatiotemporal patterns of cell expansion, including hypocotyl-based growth promoted by the transcription factor ATHB5.
Abstract
Gibberellic acid (GA)-mediated cell expansion initiates the seed-to-seedling transition in plants and is repressed by DELLA proteins. Using digital single-cell analysis, we identified a cellular subdomain within the midhypocotyl, whose expansion drives the final step of this developmental transition under optimal conditions. Using network inference, the transcription factor ATHB5 was identified as a genetic factor whose localized expression promotes GA-mediated expansion specifically within these cells. Both this protein and its putative growth-promoting target EXPANSIN3 are repressed by DELLA, and coregulated at single-cell resolution during seed germination. The cellular domains of hormone sensitivity were explored within the Arabidopsis (Arabidopsis thaliana) embryo by putting seeds under GA-limiting conditions and quantifying cellular growth responses. The middle and upper hypocotyl have a greater requirement for GA to promote cell expansion than the lower embryo axis. Under these conditions, germination was still completed following enhanced growth within the radicle and lower axis. Under GA-limiting conditions, the athb5 mutant did not show a phenotype at the level of seed germination, but it did at a cellular level with reduced cell expansion in the hypocotyl relative to the wild type. These data reveal that the spatiotemporal cell expansion events driving this transition are not determinate, and the conditional use of GA-ATHB5-mediated hypocotyl growth under optimal conditions may be used to optionally support rapid seedling growth. This study demonstrates that multiple genetic and spatiotemporal cell expansion mechanisms underlie the seed to seedling transition in Arabidopsis.
Plant growth and development occurs in 3D space over time through a combination of cell expansion and division (Roeder et al., 2011; Petricka et al., 2012; Schiessl et al., 2012). Tightly regulated gene expression programs coordinate these processes through the output of the activity of gene regulatory networks (Bassel et al., 2012; Chae et al., 2012; Sparks et al., 2013). These regulatory network interactions enable signals from the environment to be integrated with and modulate downstream gene expression programs and control growth associated with developmental transitions.
The expansion of plant cells is a mechanically driven process facilitated by the loosening of the surrounding cell wall (Peaucelle et al., 2015) and their yielding to internal turgor pressure (Lockhart, 1965). Plant development is ultimately regulated at the level of changes in the mechanical properties of the cell wall and has been investigated in developmental transitions including the initiation of organs at the shoot meristem (Fleming et al., 1997; Hamant et al., 2008; Peaucelle et al., 2011; Kierzkowski et al., 2012; Nakayama et al., 2012) and lateral root initiation (Lee and Kim, 2013; Lucas et al., 2013; Vermeer et al., 2014; Vilches-Barro and Maizel, 2015).
The seed-to-seedling transition is initiated by the phytohormone gibberellic acid (GA) (Ogawa et al., 2003; Claeys et al., 2014) and driven exclusively by cell expansion (Bassel et al., 2014). Nuclear-localized DELLA proteins repress the action of GA and seed germination. Many classes of gene expression are repressed by DELLA proteins, including cell expansion-associated gene expression such as expansin (Lee et al., 2002; Cao et al., 2006; Harberd et al., 2009; Stamm et al., 2012). This reversible growth switch plays a central role in signal integration and the regulation of plant growth in response to the environment (Achard et al., 2006).
The signaling downstream of GA perception and DELLAs that functions to regulate gene expression controlling plant growth remains poorly defined (Claeys et al., 2014). The transcription factors (TFs) GNC and GNL (Richter et al., 2010), and SCARECROW-LIKE3 (Zhang et al., 2011) have been shown to act downstream of DELLA; however, mechanistic links between DELLA-mediated growth repression and the gene expression that alters biophysical properties of plant cells represents a key gap in our understanding of plant development. The dynamic regulation of these TFs downstream of DELLA within the context of 3D cell growth that mediates developmental transitions also remains poorly understood (Fernandez et al., 2010; Schiessl et al., 2012; Montenegro-Johnson et al., 2015; Shapiro et al., 2015).
To address these gaps, we investigated the seed to seedling transition in Arabidopsis (Arabidopsis thaliana) and the relationship between a growth-promoting TF and spatiotemporal cell expansion dynamics.
RESULTS
Quantification of Organ-Wide Single-Cell 3D Anisotropy Driving Arabidopsis Embryo Germination
The germination of Arabidopsis seeds is a two-step process. The first involves the rupture of the surrounding testa, the second being the elongation of the embryo axis and penetration of the endosperm by the radicle (Weitbrecht et al., 2011). Both steps are driven exclusively by cell shape changes in the absence of division (Bassel et al., 2014).
To gain a detailed understanding of cellular growth patterns driving the embryo to seedling transition, we analyzed organ-wide 3D anisotropy in the embryo at single-cell resolution. Using 3DCellAtlas (Montenegro-Johnson et al., 2015), it is possible to quantify both changes in 3D cell anisotropy and reporter abundance within individual cells of the germinating embryo over time. We focused on the contribution of cell anisotropy in epidermal and cortical cell layers toward the promotion of the two steps of Arabidopsis seed germination.
Cell growth was calculated relative to the initial cell size at 3 h after imbibition (HAI; unexpanded state) across four developmental stages during step 1 (testa rupture) and step 2 (protrusion of the radicle through the seed coat) of seed germination. We sampled seeds before testa rupture (BTR), during early (ETR), and late testa rupture (LTR), corresponding to approximate time points of 16 HAI, 25 HAI, and 28 HAI, and once they had just germinated (JG), at approximately 32 HAI (Fig. 1; Supplemental Fig. S1; Supplemental File S1).
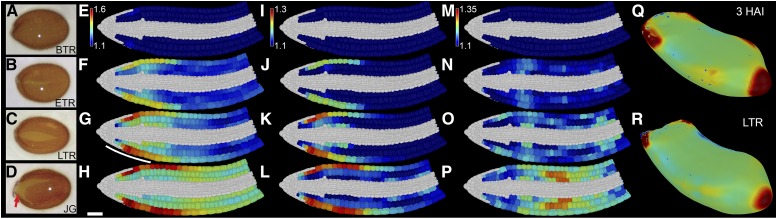
Figure 1.
Distinct steps of seed germination occur in discrete cellular locations. Quantification of 3D growth across embryo germination at single-cell resolution. Shown are Arabidopsis seeds at the sampled stages of germination (A–D), BTR (A, E, I, and M), during ETR (B, F, J, and N), during LTR (C, G, K, O, and R), and having JG (D, H, L, and P). E to H, Mean relative increase in cell surface area. I to L, Mean relative increase in radial cell length. M to P, Mean relative increase in longitudinal cell length. Data were derived from four embryos at each time point and calculated relative to unexpanded embryos at 3 HAI. Q and R, Mesh showing the surface curvature of the Arabidopsis embryo axis at 3 HAI (Q) and LTR false-colored for Gaussian curvature (R). Scale bar for all in H is 50 µm. The line in G indicates the position of visible testa rupture. See also Supplemental File S1, Supplemental Figures S1 and S2, and Supplemental Movie S1.
Increases in cell volume can be used as a measure for cell growth, yet the volumetric expansion of plant cells is promoted by the weakening and stretching of the cell wall in response to internal turgor pressure (Lockhart, 1965). To more accurately capture this aspect of plant cell growth, we focused our analyses on changes in cell surface area (Fig. 1, A–D) as a proxy to quantify how much cell wall stretch leading to cell growth occurs across different cell types during Arabidopsis seed germination (Bassel et al., 2014).
Following the discrete induction of growth in imbibed nondormant Arabidopsis seeds, cell surface area increases principally in the epidermis and moves progressively to inner layers over the course of step 1 of germination (Fig. 1, A–C; Supplemental Fig. S1, A–C; Supplemental Movie S1). The reduced growth of cortical cells in the radicle and the wave of growth along the length of the axis were both consistent with previously reported patterns of early embryo growth (Bassel et al., 2014). Neither 3D anisotropy nor growth of cell layers other than the outer cortex have been studied previously and were investigated further.
Step 1 of seed germination is manifest by the swelling of the embryo leading to the rupture of the surrounding seed coat (Bassel et al., 2004; Weitbrecht et al., 2011). Expansion of cells in the radial direction, principally in the epidermis, occurs in the cellular subdomain underlying the region of the seed where the testa ruptures (Fig. 1, C and E–H; white line in Fig. 1G). Principal directions of 3D cell anisotropy were quantified by calculating elongation factors for each direction (Montenegro-Johnson et al., 2015; Supplemental Fig. S2, A–L). Expansion in the radial direction within the lower embryo axis provides the largest contributor to overall cell size increase in this domain and promotes the testa-rupturing phase of step 1 of germination (Fig. 1, E–H; Supplemental Fig. S1, A–D).
Radial expansion also leads to a change in the overall shape of the embryo axis, which can be visualized by quantifying the Gaussian curvature of the surface of this organ (Fig. 1, M and N; Kierzkowski et al., 2012; Barbier de Reuille et al., 2015). The domain of positive curvature in the radicle tip decreases following radial expansion of the radicle and lower embryonic axis. The radial growth of the axis during step 1 of germination is primarily limited to the radicle and lower hypocotyl and was predicted previously using a 3D mechanical model of the germinating Arabidopsis embryo (Bassel et al., 2014), suggesting that active regulation of cell shape does not drive this anisotropy (Peaucelle et al., 2015) and may occur for simple mechanical reasons.
The transition from step 1 to step 2 of germination involves an increase in the domain of cell expansion along the length of the embryo axis into the hypocotyl region in a wave-like fashion (Fig. 1D; Bassel et al., 2014). Increases in cell surface area are driven primarily by an increase in longitudinal cell length, particularly in the middle region of the hypocotyl (Fig. 1L). This increase in longitudinal cell length appears to be smaller in the epidermis than that in the underlying cortical cell layers. This may be explained by the greater variability in epidermal cell length within the unexpanded embryo (Supplemental Fig. S2, M–O). This makes the quantification of subsequent differences in epidermal cell length, and their amplification following growth, a technical limitation. Given the coordination of growth between cell layers, it can be concluded that longitudinal growth in the midhypocotyl is responsible for driving step 2 of germination, the final phase of the seed-to-seedling transition (Fig. 1P; Supplemental Movie S1).
Identification of ATHB5 as a Hypocotyl-Specific Growth Regulatory Factor
The tip of the radicle is the seminal site where growth-promoting gene expression is induced in a germinating embryo (Bassel et al., 2014) and is consistent with the initiation of growth in this subdomain and induction of step 1 of seed germination (Fig. 1, E–H).
The observation that further expansion of the midhypocotyl drives step 2 of germination (Fig. 1L) raised the possibility that additional localized growth-promoting gene expression patterns are present within this cellular subdomain. Cell expansion-associated genes induced during Arabidopsis seed germination were previously identified using a cluster of coexpressed cell wall modifying genes within the gene correlation network SeedNet (Bassel et al., 2011, 2014). The spatial and temporal induction of additional members of this cell wall localized coexpressed gene cluster was examined by creating promoter::GUS reporters (Fig. 2A). From this analysis, EXPANSIN A3 (EXPA3), and the cell wall localized protease SUBTILISIN4.11 (SBT4.11) were identified as being late induced at BTR (Fig. 2S) and spatially within the middle of the hypocotyl (Fig. 2, D and I). From this initial induction site, the domain of promoter activity of these two genes spread up the embryo axis prior to the completion of seed germination and expansion of the upper hypocotyl (Fig. 2, E–G and J–L). This pattern of promoter activity for EXPA3 and SBT4.11 demonstrated the presence of spatially distinct, late-induced gene expression programs within the midhypocotyl of the germinating embryo prior to the expansion of this region.
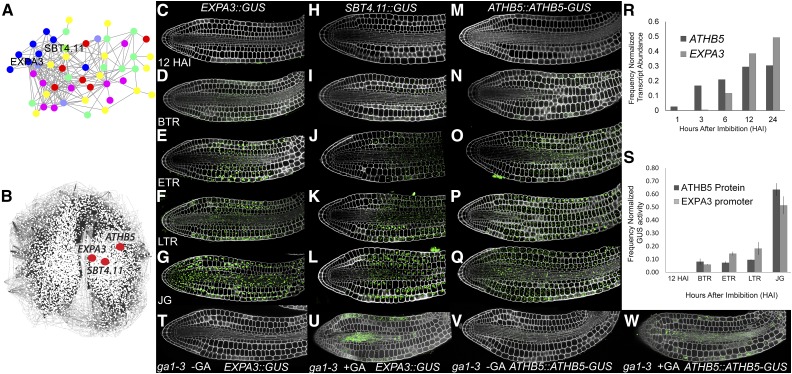
Figure 2.
Identification of late-induced midhypocotyl gene expression and protein accumulation patterns in germinating Arabidopsis embryos. A, Coexpressed genes whose protein products are cell wall targeted. Different classes of wall-modifying activity are indicated by different colors with expansin in blue and the subtilisin in green. B, Positions of EXPA3, SBT4.11, and ATHB5 within the gene coexpression network SeedNet. C to Q, EXPA3::GUS promoter activity (C–G), SBT4.11::GUS promoter activity (H–L), and ATHB5::ATHB5-GUS protein localization (M–Q) at 12 HAI (C, H, and M), BTR (D, I, and N), ETR (E, J, and O), LTR (F, K, and P), and JG (G, L, and Q). R, Transcript levels of EXPA3 and ATHB5 during imbibition based on previously published microarray data (Nakabayashi et al., 2005). S, Quantification of GUS activity reflecting EXPA3::GUS promoter activity, and ATHB5::ATHB5-GUS protein abundance within germinating embryos. Absolute activities for each reporter were normalized to the total activity of each detected throughout the time course for comparison. T to W, EXPA3::GUS and ATHB5::ATHB5-GUS reporters in the ga1-3 mutant background at 24 HAI in the presence or absence of GA.
cis-Element analysis of their promoter regions (O’Connor et al., 2005) of each EXPA3 and SBT4.11 revealed a significant overrepresentation of the ATHB5 HD-ZIP binding site (CAATNATTG; Johannesson et al., 2001) in their promoters (P < 0.005, respectively). The location of the gene encoding the ATHB5 TF was examined within SeedNet and found to be proximal to both EXPA3 and SBT4.11 (Fig. 2B). The transcript of ATHB5 has previously been demonstrated to be DELLA repressed (Cao et al., 2006), and the gene functionally linked to the ABA-mediated repression of germination and seedling root growth (Johannesson et al., 2003; Stamm et al., 2012). These observations collectively pointed to ATHB5 as a candidate upstream regulatory factor of EXPA3 and SBT4.11 expression.
We generated a C-terminal GUS translational fusion to ATHB5 under its native promoter to identify the spatial and temporal abundance of this protein during seed germination. Similar to EXPA3 and SBT4.11, the ATHB5 protein appeared at BTR (Fig. 2S) specifically within the hypocotyl (Fig. 2N). This de novo protein accumulation and promoter activity pattern suggests these factors promote cell expansion within the cellular subdomain associated with step 2 of germination.
The spatiotemporal pattern of the RGL2 protein distribution during Arabidopsis seed germination was examined by generating a C-terminal fusion to this gene using a genomic fragment with 2 kb of promoter sequence. Despite the generation of multiple independent transgenic lines, a positive GUS signal was not detected in nondormant germinating seeds over a germination time course (Supplemental Fig. S3). This is in contrast to previous reports of RGL2 protein abundance during seed germination using antibodies and western blotting, where protein was detected, while contrasting patterns of protein abundance were reported (Lee et al., 2010; Richter et al., 2010). It was therefore not possible to link the spatiotemporal dynamics of DELLA protein abundance to that of ATHB5 protein accumulation or EXPA3 promoter activity.
The expression pattern of SBT4.11 is spatiotemporally similar to that of EXPA3 and aided in the identification of ATHB5 as a putative upstream regulator of this previously undescribed hypocotyl-induced gene expression pattern during Arabidopsis seed germination. While this Ser subtilisin protease protein has a predicted extracellular localization, a functional role for this class of protein in plant cell expansion has not been demonstrated previously. As we sought to identify mechanisms of cell growth downstream of DELLA, we did not pursue further investigation of the regulation of this gene.
Gene expression data indicate increases in transcript levels of EXPA3 and ATHB5 during germination (Fig. 2R; Nakabayashi et al., 2005), and EXPA3 promoter activity and ATHB5 protein abundance follow a similar relationship over the course of germination (Fig. 2S).
The EXPA3::GUS and ATHB5::ATHB5-GUS reporters were introduced into the GA-deficient ga1-3 genetic background. This nongerminating mutant requires the exogenous application of GA to stimulate the germination program (Koornneef and van der Veen, 1980). The EXPA3 and ATHB5 transcripts had previously been reported to be dependent upon GA and DELLA signaling (Cao et al., 2006). By imbibing these seeds in the absence and presence of GA, we established that the activation of the EXPA3 promoter and accumulation of the ATHB5 protein are also GA dependent (Fig. 2, T–W). The conditional induction of the germination program and these components downstream of DELLA signaling are therefore all dependent upon the presence of GA.
ATHB5 Partially Contributes to the Induction of EXPA3 Expression
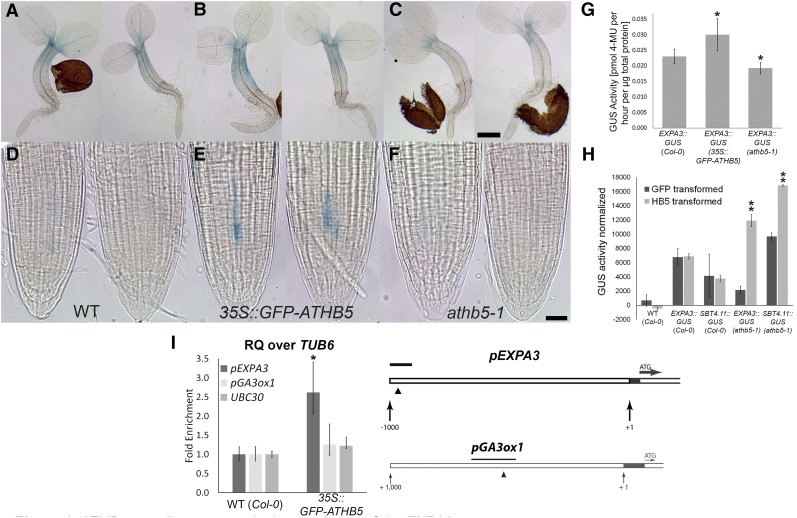
ATHB5 has been shown to be both an activator and repressor of promoter activity in plants (De Smet et al., 2013), and we examined whether it is a direct upstream regulator of the EXPA3 promoter. Some ectopic activity of the EXPA3::GUS reporter in a 35S::GFP-ATHB5 background was observed in the hypocotyl (Fig. 3A) and root (Fig. 3E), and a reduction in the domain of EXPA3::GUS activity in a null athb5-1 mutant was also observed (Fig. 3C). These microscopic observations are supported by a significant increase in EXPA3::GUS reporter in a 35S::GFP-ATHB5 background (Fig. 3G). Transactivation of the EXPA3 promoter by ATHB5 was observed using leaf protoplasts from the athb5-1 mutant carrying the EXPA3::GUS reporter. This induction was, however, not observed in the wild-type background, which contains native ATHB5 protein. Chromatin immunoprecipitation-quantitative real-time PCR (ChIP-qRT-PCR) was performed to examine the in vivo biding of the ATHB5 promoter to the EXPA3 promoter. A small but statistically significant enrichment of this interaction was detected (Fig. 3I).
Figure 3.
ATHB5 contributes toward the regulation of the EXPA3 promoter. A to F, Light microscopic images of GUS-stained 2-d-old EXPA3::GUS seedlings in wild type (Col; A and D), 35S::GFP-ATHB5 (B and E), and athb5-1 (C and F) background. Whole seedlings (A–C) and their root tips (D–F) are shown. Scale bars are 100 μm (C) and 20 μm (F). G, GUS activity in protein extracts from EXPA3::GUS seedlings in wild type (Col-0), 35S::GFP-ATHB5, and athb5-1 background at 48 HAI determined by fluorometric assay. Data are mean ± sd of biological quadruplicates. Asterisks indicate significant difference, according to a two-tailed Student’s t test (*P < 0.05). H, GUS activity in mesophyll protoplasts from EXPA3::GUS and SBT4.11::GUS reporter lines transfected with a 35S::ATHB5 construct. Asterisks indicate significant difference, according to a two-tailed Student’s t test (**P < 0.001). I, ChIP-qRT-PCR analysis of the ATHB5 bindings. The triangle shows the position of an ATHB5 binding site in the EXPA3 promoter (Johannesson et al., 2001), and the black bar indicates the position of fragment amplified by qRT-PCR. Graphs indicate relative enrichment of amplified fragments over TUBULIN6 (TUB6) in chromatin isolated from germinating 35S::GFP-ATHB5 seedlings, normalized to chromatin from wild-type (WT) embryos. Amplification of UBIQUITIN CONJUGATING ENZYME E230 (UBC30; Gibbs et al., 2014) and GA3-OXIDASE1 (GA3ox1) promoter sequences were included as controls. The GA3ox1 promoter contains an ATHB5 consensus binding site as indicated by the black arrowhead. RQ, Relative quantity. Data are mean ± sd of biological triplicates. Asterisks indicate significant difference, according to a two-tailed Student’s t test (P < 0.05).
These observations indicate that ATHB5 partially regulates EXPA3 expression. We proceeded to use the ATHB5-EXPA3 relationship as a proxy for hypocotyl-specific gene expression promoting cell expansion during step 2 of germination.
ATHB5 Does Not Interact Directly with DELLA
We examined whether the DELLA protein RGL2 and ATHB5 physically interact, directly linking GA signaling to the regulation of downstream growth-promoting gene expression. Yeast two-hybrid interaction screening failed to detect an interaction (Supplemental Fig. S4). This suggests these proteins do not interact, a result that is consistent with the observation that the ATHB5 transcript is repressed by DELLA (Cao et al., 2006), and additional TFs that physically interact with DELLA may mediate this transcriptional repression.
Quantification of the ATHB5 and EXPA3 Dynamics within the Context of Cell-Type-Specific 3D Anisotropy
The spatial and temporal relationship between the ATHB5 protein and EXPA3 promoter activity was quantified at single-cell resolution across the time course of seed germination using 3DCellAtlas (Barbier de Reuille et al., 2015; Montenegro-Johnson et al., 2015). A strong linear relationship between the detection of mean GUS reporter concentration and staining time is observed within individual cells of the embryo facilitating these analyses (Supplemental Fig. S5). Reporter concentration data were pooled from at least four samples per time point, and the activity of GUS reporters across all cells of each embryo axis was scaled using quantitative values derived from fluorometric enzyme assays (Fig. 2S). This enabled a semiquantitative comparison of reporter abundance to be performed in the absence of an internal standard for the microscopic quantification of reporter abundance (Federici et al., 2012).
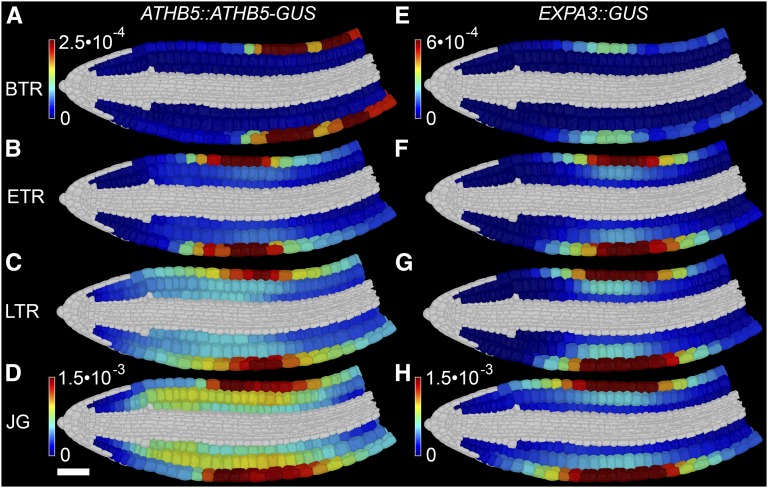
The accumulation of ATHB5 protein and EXPA3 promoter activity both occurred principally in the epidermis of the midhypocotyl of the embryo axis at ETR, preceding the growth of this region (Fig. 4, A–H; Supplemental File S2). Both ATHB5 protein abundance and EXPA3 promoter activity progressively increased and spread radially into the cortical cell layers, and longitudinally along the length of the embryo axis, with the EXPA3 promoter showing a stronger bias to the epidermal cell layer.
Figure 4.
ATHB5 protein abundance and EXPA3 promoter activity are spatiotemporally coregulated during Arabidopsis seed germination. Shown is digital single-cell quantification of ATHB5 protein abundance (A–D) and EXPA3 promoter activity (E–H), at BTR (A and E), ETR (B and F), LTR (C and G), and JG (D and H). Reporter quantification data were derived from four embryos each. The mean GUS quantity in each cell was calculated and normalized to GUS activity as determined by quantitative fluorometric assays for each sample. White scale bar for all in D is 50 µm. See also Supplemental Figure S4, Supplemental File S2, and Supplemental Movie S2.
The spatial and temporal coregulation between each the ATHB5 protein and EXPA3 promoter was determined by performing a linear regression revealing a significant positive relationship across all cell types of the hypocotyl at ETR, LTR, and JG (Supplemental Table S1) with an R2 > 0.8, with the exception of the epidermis at ETR, which was R2 = 0.61. This protein and its promoter target are therefore coregulated at single cell resolution in epidermal and cortical cells during the later stage of Arabidopsis seed germination.
The relationship between each the ATHB5 protein and EXPA3 promoter activity and cell growth was much weaker during germination (Supplemental Table S1), reflecting the induction of these growth-promoting components in advance of observed cell expansion.
ATHB5 Promotes EXPA3 Expression and GA-Mediated Cell Expansion Specifically in the Hypocotyl
GA promotes the induction of cell expansion in germinating seeds (Koornneef and van der Veen, 1980), though the cellular distribution of sensitivity to this hormone within the embryo remains unknown. To investigate this, we made use of the GA synthesis inhibitor paclobutrazol (PAC), which is capable of blocking seed germination at concentrations above 10 µm. At lower concentrations including 5 μm PAC, GA levels are reduced and germination is delayed (Piskurewicz et al., 2009; Nieuwland et al., 2016), but not blocked. We performed digital single cell analysis on embryos during the final step of seed germination (LTR to JG) under GA-limited conditions to identify which cells had the greatest sensitivity to GA-promoted cell expansion during this developmental transition. The pattern of EXPA3 promoter activity was also examined as a proxy to understand the regulation of hypocotyl-specific growth-promoting gene expression.
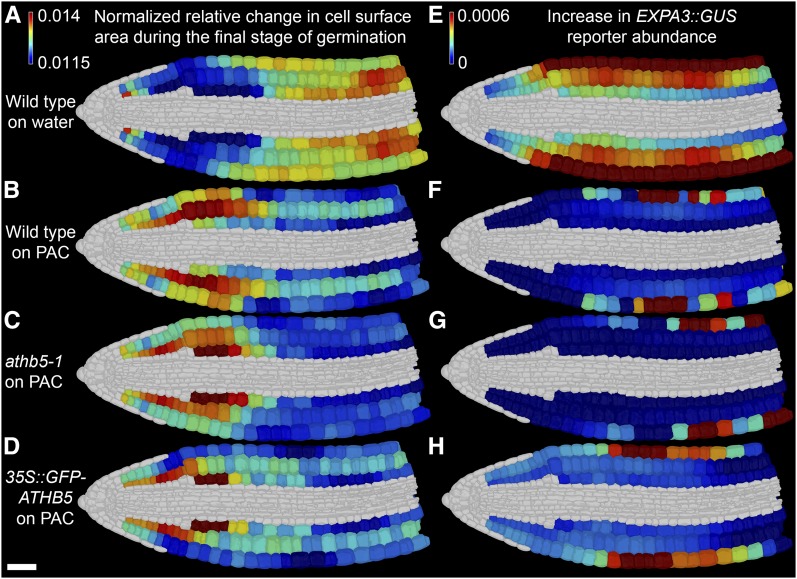
Displaying relative changes in cell surface area between LTR and JG during wild-type germination (Fig. 5A), the promotion of step 2 of germination by the middle and upper hypocotyl becomes apparent. Concurrently, changes in EXPA3 promoter activity across these final stages of germination are broadly distributed across the hypocotyl epidermis and to a lesser extent outer cortex (Fig. 5E).
Figure 5.
ATHB5 correlates with EXPA3 promoter activity and GA-mediated growth specifically in the embryonic hypocotyl. Shown is relative increase in cell surface area (A to D) and increase in EXPA3 promoter activity (E–H) in wild-type, athb5-1 knockout, and 35S::GFP-ATHB5 embryos between the final germination stages (LTR to JG) on water (A and E) and on 5 µm PAC (B–D and F–H). A to D, Changes in cell surface area were determined as the ratio between LTR and JG and frequency normalized to enable comparisons across genotypes. E to H, Change in EXPA3 promoter activity between LTR and JG. The mean GUS quantity in each cell was calculated, and the difference between these means at JG and LTR was determined. All growth and reporter quantification data were derived from four embryos at each stage. White scale bar in D is 50 µm.
Wild-type seeds germinating under GA-limiting conditions (5 μm PAC) completed step 2 of germination differently from water-imbibed controls, showing a growth pattern biased toward the lower axis (Fig. 5B; Supplemental Fig. S5, D and E). Changes in the pattern of EXPA3 promoter activity were observed in the hypocotyl and were much lower than in the water control (Fig. 5F, Supplemental Fig. S5F; Supplemental File S2), consistent with its reported upstream regulation by DELLA abundance and GA signaling (Cao et al., 2006).
This identified the cells of the lower embryo axis as having the greatest sensitivity to GA-promoted cell expansion, while the middle and upper hypocotyl have the greatest GA requirement for their cells to expand. These observed differences in cell growth patterns also demonstrate the ability for the embryo to germinate using different patterns of cell expansion, indicating that spatiotemporal cellular growth dynamics driving this developmental transition are not determinate.
Germination of the athb5-1 mutant embryo imbibed in the nonlimiting 5 μm PAC concentration showed a lower axis-biased growth pattern promoting step 2 of seed germination as in the wild type (Fig. 5C). Cell growth in the outer cortical cells of the hypocotyl was, however, reduced, as was the increase in EXPA3 promoter activity within these cells over this period (Fig. 5G; Supplemental Fig. S5G) relative to wild-type controls under the same treatment. The native ATHB5 gene therefore acts to promote both GA-mediated cell expansion and EXPA3 promoter activity specifically within the hypocotyl.
Seeds overexpressing ATHB5 also showed lower axis-enhanced germination on 5 μm PAC (Fig. 5D), and embryos grew more over the final phase of germination than either wild type or the athb5-1 mutant over this same time period (Supplemental Fig. S4C). This enhanced overall growth during step 2 of germination occurred both in the radicle and the hypocotyl (Supplemental Fig. S5, D and E). Accompanying this enhanced growth in the radicle and hypocotyl was ectopic activity of the EXPA3 promoter in the radicle (Fig. 5H).
These observations demonstrate that ATHB5 acts specifically within the midhypocotyl to promote both cell expansion and EXPA3 promoter activity downstream of GA. However, the hypocotyl has the greatest GA requirement to grow, and expansion of this region and ATHB5-mediated promotion of this growth are not necessary to undergo the seed to seedling transition.
DISCUSSION
The ability to link regulatory network dynamics at single-cell resolution to whole-organism development remains a key challenge toward a comprehensive understanding of plant growth and development (Roeder et al., 2011; Uyttewaal et al., 2012; Chew et al., 2014). Whole-organ digital single-cell analysis provides the ability to understand and capture both cellular growth in 3D and the dynamics of genetically encoded components within individual cells (Montenegro-Johnson et al., 2015). We used this approach to characterize a spatially distinct growth module in the midhypocotyl of the embryonic axis whose expansion is responsible for the completion of the seed to seedling transition under optimal conditions. Using network inference, the ATHB5 protein and putative growth-promoting targets were identified and were localized and coregulated specifically within this cellular subdomain. These findings provide the identification of a subdomain of cells driving a developmental transition in plants under optimal conditions, and the presence of genetically encoded regulatory factors specifically expressed therein.
This late-induced pattern of growth-promoting gene expression in the midhypocotyl immediately prior to step 2 of germination described here follows the radicle-derived pattern spatiotemporally correlated with step 1 of germination in the lower axis (Bassel et al., 2014). The ATHB5 protein accumulates prior to the induction of step 2, and contributes to the induction of the promoter of the growth-promoting gene EXPA3 downstream of the GA signal. This represents a spatially and temporally regulated interaction that promotes gene expression associated with localized biomechanical changes driving a developmental transition from seed to seedling within a specialized cellular subdomain.
While ATHB5 was sufficient to induce ectopic EXPA3 promoter activity, the absence of this TF did not abolish the activation of this expansin promoter (Fig. 3, C and G). The regulation of the multigene families of cell wall remodeling families is highly complex and the emergent property of the combinatorial activities of multiple TFs. Likewise, single knockouts of expansin genes have thus far failed to yield developmental phenotypes (Cosgrove, 2015). ATHB5 therefore represents one of many upstream regulatory factors of the EXPA3 gene.
ATHB5 Enhances GA-Mediated Growth in High-GA-Requiring Cells
The spatiotemporal cellular growth patterns of germinating embryos under low GA conditions demonstrated the middle and upper hypocotyl to undergo the greatest expansion during the seed-to-seedling transition and also have the highest requirement for GA to promote cell expansion (Fig. 5B). The targeted expression of ATHB5 within the midhypocotyl therefore acts to locally promote cell expansion gene expression, including EXPA3, downstream of the reduced GA-mediated promotion of cell expansion within this region.
The lower hypocotyl-based germination pattern under GA-limited conditions also demonstrates that the spatiotemporal cell growth sequence driving germination is not determinate (Lee et al., 2012), and phenotypical plasticity at a cellular level can facilitate the completion of this developmental transition. The expansion of the midhypocotyl is therefore not necessary for the completion of Arabidopsis seed germination. ATHB5 can therefore promote the expansion of the middle hypocotyl downstream of GA, but the activity of this protein in this cellular subdomain is also not required, as the growth of other regions of the embryo axis are sufficient to drive the completion of the seed-to-seedling transition. Consequently, the athb5-1 mutant does not show an organ-level phenotype in terms of reduced germination under GA-limited conditions (Stamm et al., 2012) but does show a cellular level phenotype in terms of the spatiotemporal sequence of cell expansion events during the seed-to-seedling transition (Fig. 5C). This cellular level plasticity in growth pattern highlights the presence of multiple mechanisms by which the Arabidopsis embryo is able to transition into a seedling under different conditions and versatility in the employment of the ATHB5 hypocotyl cell expansion module. This is in contrast to other characterized growing systems in plants, such as root growth, where a consistent gradient of cell expansion is observed (Ubeda-Tomás et al., 2012). Similar to this reported plasticity in cell expansion, a nondeterminate sequence of cellular patterning during lateral root formation has been reported (Lucas et al., 2013; von Wangenheim et al., 2016).
A Mechanically Advantageous Site to Enhance Embryo Growth
The conditional use of a regulated gene expression program raises questions as to why it is present. We previously demonstrated that larger cells have a greater capacity to grow in response to growth-promoting gene expression than those that are smaller (Bassel et al., 2014). The promotion of cell expansion and growth-promoting gene expression by ATHB5 within the middle and upper hypocotyl, where cells are larger than those in the radicle, can provide a more profound impact upon the mechanics of whole axis growth under optimal conditions. This makes targeting the hypocotyl with growth-promoting gene expression a mechanically advantageous region of the embryo for the promotion of step 2 of seed germination to support robust growth under ideal circumstance. In absolute terms, larger cells grow faster than smaller ones, enabling more rapid growth under ideal conditions. These principles may explain the conditional use of this gene expression pattern under favorable growth conditions.
Cell-Type-Specific Growth and Gene Expression Is Mediated by ATHB5
During all stages of germination, embryo epidermal cells show the highest relative increase in cell size, in terms of surface area and volume. This may be due to higher mechanical constraint on cells in inner cell layers, with the epidermis playing a key regulatory role in the control of plant organ growth (Dyson et al., 2014). In leaves and meristems, this cell layer has been shown to both drive and restrict overall organ growth, possibly through intercellular feedback (Savaldi-Goldstein et al., 2007; Uyttewaal et al., 2012).
The activity of the EXPA3 promoter was least affected in the embryo hypocotyl epidermis, where promoter activity was clearly detected under all conditions tested (Figs. 4 and 5). Promoter activity in the cortical cell layers of the hypocotyl was however reduced in athb5-1 knockouts treated with PAC (Fig. 5G), together with reduced cell growth in this cellular subdomain (Fig. 5C). This suggests the redundancy in the control of the EXPA3 promoter is greater in the epidermis and that the unique role of ATHB5 in regulating EXPA3 promoter activity is greatest in the cortex of the germinating embryo hypocotyl. The presence of an epidermis-specific homeodomain gene regulatory network mediated by AtML1 and PDF2 demonstrates the presence of cell-type-specific gene regulatory interactions that may be redundantly regulating the EXPA3 promoter within this cell type (Rombolá-Caldentey et al., 2014).
CONCLUSION
The identification of a specific cellular subdomain responsible for driving the completion of the seed-to-seedling transition in Arabidopsis, and a growth-promoting regulatory interaction specifically targeted to this region, represents an advance in our understanding of both mechanisms of plant development and its spatiotemporal regulation. The ability of the embryo axis to modulate spatiotemporal cell expansion patterns driving this developmental transition indicates that seeds have developed multiple mechanisms for the completion of germination, and this process is nondeterministic at a cellular level. The application of these findings in the context of embryo cell expansion can provide spatial and genetic targets for the manipulation and enhancement of crop establishment (Finch-Savage and Bassel, 2016).
MATERIALS AND METHODS
Plant Growth and Germination Conditions
Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia) were grown to maturity in environmentally controlled cabinets, using 16 h light (23°C) and 8 h dark (18°C). Seeds were harvested once plants had ceased flowering and siliques began to brown. Seeds were cleaned through a 500 μm mesh and stored in glassine bags at 22°C for 1 month to remove primary dormancy. All germination experiments were performed on nondormant seeds.
Reporter lines used in this study were pEXPA3::GUS (At2g37640), pSBT4.11::GUS (At5g59130), pRGL2::RGL2-GUS (At3g03450), and pATHB5::ATHB5-GUS (At5g65310; Col-0 background). For EXPA3 (At2g37640) and SBT4.11 (At5g59130) promoter fusions, a region 2 kb upstream of the transcriptional start sequence was cloned using GATEWAY into pKANGWF7 to drive GUS expression. For ATHB5 and RGL2, a region 2 kb upstream of the transcriptional start sequence plus genomic sequence of ATHB5 and RGL2 was cloned into pGWB433 using GATEWAY to drive the expression of an ATHB5-GUS and RGL2-GUS fusion protein.
Embryos from imbibed Arabidopsis seeds were dissected on moist filter paper with scalpel and forceps, using a binocular microscope. Seeds were dissected at 3 HAI (unexpanded state), at approximately 16 HAI when no testa rupture had occurred yet (BTR), at approximately 25 HAI when testa rupture was visible and had progressed for less than half the length of the seed (ETR), at approximately 28 HAI when the testa was ruptured along the whole length of the seed (LTR), and at approximately 32 HAI when endosperm rupture had just occurred (JG), marking the completion of germination (Bassel et al., 2014).
Hormone Treatments
Arabidopsis seeds were surface-sterilized and placed onto half-strength Murashige and Skoog medium with 0.8% agar (w/v), supplemented with 5 µm PAC (Fluka), and incubated at 22°C with a 16 h light/8 h dark-cycle. Embryos from PAC-treated seeds were dissected at LTR (∼40 HAI) and JG (∼48 HAI).
GUS Staining and Enzyme Assays
Samples were stained for GUS activity in staining buffer (sodium phosphate buffer, pH 7.0, 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA [Sigma], 1 mm potassium ferro- and ferricyanate), at 37°C until a blue product was visible. Embryos and seedlings from the same line were stained for the same amount of time at each stage.
To quantify GUS activity, 30 embryos were dissected and thoroughly homogenized in 30 μL protein extraction buffer (50 mm Na-phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% [v/v] Triton X-100, 10 mm β-mercaptoethanol). After centrifugation at 20,000g at 4°C for 10 min, the supernatant was used for analysis. 1:20 diluted protein extracts were mixed 1:1 with substrate buffer (2 mm 4-methylumbelliferone glucuronide [MUG] in protein extraction buffer), and incubated at 37°C. Twenty-microliter aliquots were taken each after 6, 18, and 24 h. Each aliquot was immediately mixed with 180 μL STOP buffer (200 mm sodium carbonate buffer, pH 9.5). To normalize for fluorescence quenching of the protein extracts, the same reactions were set up as blanks, without substrate, and treated in the same way as the actual samples. Fluorescence was measured in 96-well plates in a plate reader (Infinite 200 PRO [Tecan]) at 460 nm, with excitation at 360 nm, against a standard curve with 0, 0.25, 0.5, 1.5, 2.5, 4, 5, and 10 μm 4-methylumbelliferone. Fluorescence of samples was normalized to blanks. GUS activity was normalized and calculated as pmol substrate per hour per μg total protein. Total protein amount in each sample was determined by Bradford assay.
Sample Preparation and Imaging
For light microscopy, GUS-stained seedlings were fixed in ethanol:acetic acid (3:1) with 1% Tween 20 overnight. Samples were mounted on microscope slides in Hoyer’s medium and imaged using a Leica DM500 microscope.
Samples for confocal microscopy were prepared as described previously (Montenegro-Johnson et al., 2015). Confocal Z-stacks were imaged using a Zeiss LSM 710 and 488 nm laser with the Zen 2010 software. Typically, confocal stacks were taken with a pinhole of 0.6 AU, slice interval of 0.3 µm, and the highest possible signal-to-noise ratio (pixel saturation in cell walls, low to no signal inside the cells).
Image Analysis
Confocal stacks of embryos were analyzed using MorphoGraphX (Barbier de Reuille et al., 2015). Imaged Z-stacks were imported as tiff files, preprocessed by applying a Gaussian blur (0.5 µm diameter), and segmented using the 3D watershed algorithm. Segmented stacks were manually edited to correct oversegmentation, and a cellular mesh was generated with the 3D Marching Cubes 3D algorithm (Lorensen and Cline, 1987), with a cube size of 2 µm and no smoothing. A second mesh describing the surface of the whole axis was generated by blurring the original image at a diameter of 5 µm, and generating a surface mesh using the Marching Cubes Surface algorithm, at a cube size of 5 µm. A Bezier spline with five handle points was placed to describe the center of the embryo axis. Annotation of cell types was performed as described in Montenegro-Johnson et al. (2015) with CellAtlas3D, using Bezier spline, segmented mesh, and surface mesh for each sample. Misannotated cells were corrected manually. Cell data, containing cell type, position, and geometric data for each individual cell, were exported as csv files.
GUS crystals were imaged using “reflectance” settings on a second channel for each Z-stack. MorphoGraphX was used to quantify total GUS abundance within each cell, relative to its volume, and export these data as a csv file.
Normalization of Imaged Reporter Data Using Quantitative Enzyme Assays
For comparison, GUS signal detected within each cell was normalized for each stack. Total GUS abundance within each individual cell was expressed as a fraction of the total amount of GUS signal detected in all cells. This was then scaled to the GUS activity determined by the fluorometric assay for the corresponding sample.
ChIP
ChIP was performed according to Kaufmann et al. (2010), with changes described previously (Stamm et al., 2012) using 2-d-old 35S::GFP-ATHB5 seedlings derived from 100 mg (dry weight) of seeds. Pull-down of protein complexes was performed with anti-GFP-sepharose beads (0.1 mg/mL; Abcam). Protein complexes were eluted, and after reverse cross-linking, chromatin was purified using the QIAquick PCR Purification Kit (Qiagen) and eluted in 35 µL.
Yeast Two-Hybrid
The yeast two-hybrid Matchmaker Gold system was used as per the manufacturer’s instructions (Clontech). In brief, yeast strain AH109 was transformed with corresponding bait and prey combinations (Supplemental Fig. S4) by PEG-mediated transformation and grown on synthetic dropout medium lacking Leu and Trp (double dropout medium) as transformation control, medium lacking Leu, Trp, and His (triple dropout medium), and medium lacking Leu, Trp, His, and Ade (quadruple dropout medium) as test for protein-protein interactions. Assays were performed in triplicate.
Protoplast Assay
Arabidopsis mesophyll protoplast isolation and transformations were performed as described previously (Yoo et al., 2007). In brief, protoplasts from 3- to 4-week-old plants were transformed with 35S::ATHB5 plasmid or 35S::GFP as a control. After 16 h incubation at 20°C in the dark, protoplasts were lysed in protoplast lysis buffer (Luciferase kit; Promega), and GUS activity was determined by incubating the lysate in MUG buffer for 6 h at 37°C. Fluorescence was measured in a plate reader (360 nm excitation, 465 nm emission). Promoter activity of each sample was normalized to the MUG-only background.
Statistical Analysis of Growth and Promoter and Protein Expression
This was performed as previously reported (Montenegro-Johnson et al., 2015). Means for each metric were derived from pooled data of at least four embryos based on cell type and position. Corresponding data from six unexpanded embryos (3 HAI) were used as reference for analysis of each developmental stage and treatment. Total growth at each stage was calculated as a ratio to unexpanded embryo cells. GUS reporter abundance data were pooled in the same way. Each metric is given with its corresponding 95% confidence interval based on the t-distribution (see Supplemental Files S1 and S2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantification of changes in 3D cell shape of the Arabidopsis embryo during germination at single-cell resolution.
Supplemental Figure S2. 3D anisotropy of cell shape change driving Arabidopsis germination, and variability of cell lengths.
Supplemental Figure S3. Distribution of the RGL2 protein during nondormant seed germination.
Supplemental Figure S4. A yeast two-hybrid assay was performed to test for interaction of ATHB5 with RGL2.
Supplemental Figure S5. Relationship between GUS staining duration and GUS quantity measured in MorphoGraphX.
Supplemental Table S1. Linear regression examining relationships between ATHB5 protein abundance, EXPA3 promoter activity, and changes in cell surface area across different cell types of the Arabidopsis embryonic hypocotyl (cell positions 11–35).
Supplemental File S1. Cellular growth data.
Supplemental File S2. Cellular GUS reporter abundance data.
Supplemental Movie S1. Dynamics of cell growth across Arabidopsis seed germination.
Supplemental Movie S2. Dynamics of cell growth, EXPA3 promoter activity, and ATHB5 protein abundance across Arabidopsis seed germination.
Supplementary Material
Acknowledgments
We thank Andrew Wood, University of Nottingham, for helpful advice on statistical analyses and Sonya Hill, University of Birmingham, for technical assistance.
Glossary
- GA
gibberellic acid
- TF
transcription factor
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/L010232/1 (A.T.T. and G.W.B.), BBSRC Grant BB/J017604/1 (P.S. and G.W.B.), BBSRC Grant BB/N009754/1 (G.W.B.), BBSRC doctoral training partnership BB/M01116X/1 MIBTP (M.D.B.J.), and BBSRC Grant BB/G015066/1 (D.F.A.T. and J.L.B.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, et al. (2015) MorphoGraphX: a platform for quantifying morphogenesis in 4D. eLife 4: 05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Gaudinier A, Brady SM, Hennig L, Rhee SY, De Smet I (2012) Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Stamm P, Mosca G, Barbier de Reuille P, Gibbs DJ, Winter R, Janka A, Holdsworth MJ, Smith RS (2014) Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc Natl Acad Sci USA 111: 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Zielinska E, Mullen RT, Bewley JD (2004) Down-regulation of DELLA genes is not essential for germination of tomato, soybean, and Arabidopsis seeds. Plant Physiol 136: 2782–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae L, Lee I, Shin J, Rhee SY (2012) Towards understanding how molecular networks evolve in plants. Curr Opin Plant Biol 15: 177–184 [DOI] [PubMed] [Google Scholar]
- Chew YH, Wenden B, Flis A, Mengin V, Taylor J, Davey CL, Tindal C, Thomas H, Ougham HJ, de Reffye P, et al. (2014) Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proc Natl Acad Sci USA 111: E4127–E4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, De Bodt S, Inzé D (2014) Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci 19: 231–239 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25: 162–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Lau S, Ehrismann JS, Axiotis I, Kolb M, Kientz M, Weijers D, Jürgens G (2013) Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. J Exp Bot 64: 3009–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson RJ, Vizcay-Barrena G, Band LR, Fernandes AN, French AP, Fozard JA, Hodgman TC, Kenobi K, Pridmore TP, Stout M, et al. (2014) Mechanical modelling quantifies the functional importance of outer tissue layers during root elongation and bending. New Phytol 202: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici F, Dupuy L, Laplaze L, Heisler M, Haseloff J (2012) Integrated genetic and computation methods for in planta cytometry. Nat Methods 9: 483–485 [DOI] [PubMed] [Google Scholar]
- Fernandez R, Das P, Mirabet V, Moscardi E, Traas J, Verdeil JL, Malandain G, Godin C (2010) Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolution. Nat Methods 7: 547–553 [DOI] [PubMed] [Google Scholar]
- Finch-Savage W, Bassel G (2016) Seed vigour and crop establishment: extending performance beyond adaptation. J Exp Bot 67: 567–591 [DOI] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C (1997) Induction of leaf primordia by the cell wall protein expansion. Science 276: 1415–1418 [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP, et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Heisler MG, Jönsson H, Krupinski P, Uyttewaal M, Bokov P, Corson F, Sahlin P, Boudaoud A, Meyerowitz EM, et al. (2008) Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H, Wang Y, Engström P (2001) DNA-binding and dimerization preferences of Arabidopsis homeodomain-leucine zipper transcription factors in vitro. Plant Mol Biol 45: 63–73 [DOI] [PubMed] [Google Scholar]
- Johannesson H, Wang Y, Hanson J, Engström P (2003) The Arabidopsis thaliana homeobox gene ATHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Mol Biol 51: 719–729 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D, Nakayama N, Routier-Kierzkowska AL, Weber A, Bayer E, Schorderet M, Reinhardt D, Kuhlemeier C, Smith RS (2012) Elastic domains regulate growth and organogenesis in the plant shoot apical meristem. Science 335: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim J (2013) EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol 54: 1600–1611 [DOI] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turečková V, Carat S, Chappuis R, Strnad M, Fankhauser C, Lopez-Molina L (2012) Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev 26: 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turečková V, Strnad M, Lopez-Molina L (2010) A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc Natl Acad Sci USA 107: 19108–19113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart JA. (1965) An analysis of irreversible plant cell elongation. J Theor Biol 8: 264–275 [DOI] [PubMed] [Google Scholar]
- Lorensen WE, Cline HE (1987) Marching cubes: a high resolution 3D surface construction algorithm. In Stone MC, ed, Proceedings of the 14th Annual Conference on Computer Graphics and Interactive Techniques (SIGGRAPH ’87). ACM, New York, pp 163–169 [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Voβ U, Swarup K, De Smet I, Van Damme D, Lawrence T, Péret B, Moscardi E, et al. (2013) Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 110: 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro-Johnson TD, Stamm P, Strauss S, Topham AT, Tsagris M, Wood ATA, Smith RS, Bassel GW (2015) Digital single-cell analysis of plant organ development using 3DCellAtlas. Plant Cell 27: 1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakayama N, Smith RS, Mandel T, Robinson S, Kimura S, Boudaoud A, Kuhlemeier C (2012) Mechanical regulation of auxin-mediated growth. Curr Biol 22: 1468–1476 [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Stamm P, Wen B, Randall RS, Murray JAH, Bassel GW (2016) Re-induction of the cell cycle in the Arabidopsis post-embryonic root meristem is ABA-insensitive, GA-dependent and repressed by KRP6. Sci Rep 6: 23586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook SA, Le Guillou L, Bron E, Kuhlemeier C, Höfte H (2011) Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol 21: 1720–1726 [DOI] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Höfte H (2015) The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol 25: 1746–1752 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Turecková V, Lacombe E, Lopez-Molina L (2009) Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AHK, Tarr PT, Tobin C, Zhang X, Chickarmane V, Cunha A, Meyerowitz EM (2011) Computational morphodynamics of plants: integrating development over space and time. Nat Rev Mol Cell Biol 12: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombolá-Caldentey B, Rueda-Romero P, Iglesias-Fernández R, Carbonero P, Oñate-Sánchez L (2014) Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. Plant Cell 26: 2905–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schiessl K, Kausika S, Southam P, Bush M, Sablowski R (2012) JAGGED controls growth anisotropy and coordination between cell size and cell cycle during plant organogenesis. Curr Biol 22: 1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BE, Tobin C, Mjolsness E, Meyerowitz EM (2015) Analysis of cell division patterns in the Arabidopsis shoot apical meristem. Proc Natl Acad Sci USA 112: 4815–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E, Wachsman G, Benfey PN (2013) Spatiotemporal signalling in plant development. Nat Rev Genet 14: 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P, Ravindran P, Mohanty B, Tan EL, Yu H, Kumar PP (2012) Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol 12: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S, Beemster GT, Bennett MJ (2012) Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci 17: 326–331 [DOI] [PubMed] [Google Scholar]
- Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykręt D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, et al. (2012) Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149: 439–451 [DOI] [PubMed] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N (2014) A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343: 178–183 [DOI] [PubMed] [Google Scholar]
- Vilches-Barro A, Maizel A (2015) Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol 23: 31–38 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, Maizel A (2016) Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr Biol 26: 439–449 [DOI] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo JO, Lim J, Kamiya Y, Yamaguchi S, Sun TP (2011) Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.