Interlinking GWAS of Debranched Starch Structure with Gene Regulatory Networks Uncovers a Regulatory Region for Amylose Synthesis in Rice (Oryza sativa).
Abstract
A deeper understanding of the regulation of starch biosynthesis in rice (Oryza sativa) endosperm is crucial in tailoring digestibility without sacrificing grain quality. In this study, significant association peaks on chromosomes 6 and 7 were identified through a genomewide association study (GWAS) of debranched starch structure from grains of a 320 indica rice diversity panel using genotyping data from the high-density rice array. A systems genetics approach that interrelates starch structure data from GWAS to functional pathways from a gene regulatory network identified known genes with high correlation to the proportion of amylose and amylopectin. An SNP in the promoter region of Granule Bound Starch Synthase I was identified along with seven other SNPs to form haplotypes that discriminate samples into different phenotypic ranges of amylose. A GWAS peak on chromosome 7 between LOC_Os07g11020 and LOC_Os07g11520 indexed by a nonsynonymous SNP mutation on exon 5 of a bHLH transcription factor was found to elevate the proportion of amylose at the expense of reduced short-chain amylopectin. Linking starch structure with starch digestibility by determining the kinetics of cooked grain amylolysis of selected haplotypes revealed strong association of starch structure with estimated digestibility kinetics. Combining all results from grain quality genomics, systems genetics, and digestibility phenotyping, we propose target haplotypes for fine-tuning starch structure in rice through marker-assisted breeding that can be used to alter the digestibility of rice grain, thus offering rice consumers a new diet-based intervention to mitigate the impact of nutrition-related noncommunicable diseases.
Polished rice (Oryza sativa) is a staple food for billions of people worldwide, where the majority is from developing countries. It serves as a rich source of energy because it is composed of up to 90% (w/w) starch. Rice starch is a complex carbohydrate made up of amylose and amylopectin. Amylose is made up of a long helical homopolymer of d-Glc units linked together by α-1,4 glycosidic bonds with occasional branching at α-1,6 branch points (Hanashiro, 2015). Amylopectin is a highly branched polymer of short α-1,4-linked d-Glc units with reducing ends interlinked by α-1,6-glycosidic bonds that form a very large native biopolymer that is up to two orders-of-magnitude bigger than amylose (Bertoft, 2015). These two components are arranged in alternating crystalline and amorphous lamellae to form starch granules (Vandeputte and Delcour, 2004). The properties of these two starch polymers are associated with the cooking and eating quality of rice, which are routinely screened in rice breeding programs using apparent amylose content (AAC), gelatinization temperature, and gel consistency as proxy measures. Because of the difficulty in characterizing native starch structure, starch is commonly modeled by measuring the molecular weight distribution (MWD) of debranched starch by size exclusion chromatography (SEC; Castro et al., 2005a, 2005b). Previous studies have shown that mechanistic information deciphered from MWD of debranched starch associates well with activities of starch biosynthetic enzymes (Castro et al., 2005a, 2005b; Ward et al., 2006; Hernández et al., 2008).
When rice is cooked, the semicrystalline structure is disrupted, leaching out amylopectin and facilitating the entry of water that eventually leads to the gelatinization of the starch granule (Miller et al., 1992). The digestion of cooked milled rice is usually complete upon consumption (Strocchi and Levitt, 1991). Starch hydrolysis begins in the mouth upon mixing with salivary α-amylases during the mastication of rice grains. When the chewed rice reaches the small intestine, starch is further digested by pancreatic α-amylases and converted into simple sugars that are mostly maltose. These oligosaccharides are converted by maltase, isomaltase, and glucoamylase in the gut endothelial cells into Glc molecules that are then released into the bloodstream (Yokoyama, 2004). Nearly 85% to 90% of the calories are released from starch. In terms of nutrition, starch is classified as either rapidly or slowly digestible (Lehmann and Robin, 2007). If starch digestion is sufficiently slow, the starch survives until the end of the small intestine, with the residual material considered as resistant starch. Resistant starch enters the colon, where it is fermented in a manner analogous to dietary fiber with benefits for gut health (Topping, 2007).
Increased sedentary lifestyle and the consumption of a carbohydrate-rich diet including excess consumption of cooked milled rice is linked to nutrition-related diseases such as diabetes, which affects both developed and developing countries (Hu et al., 2012). Slowing down starch digestion and retarding the rate with which Glc becomes bioavailable are important in the management of diabetes and other nutrition and lifestyle diseases (Wolever et al., 1994). The relationship between amylose content as measured by the conventional iodine method (Juliano et al., 1981) and glycemic impact as assessed by an in vitro starch hydrolysis index (HI) of milled rice grains was found to be essentially inversely linear and is associated with alleles of Granule-Bound Starch Synthase I (GBSSI; Fitzgerald et al., 2011). However, this inverse linear relationship breaks down when lines with intermediate to higher amylose content are considered. In addition, RNA silencing or stop codon mutation of the Starch Branching Enzyme IIb (SBEIIb) gene was also identified to reduce raw starch granule amylolysis (Dhital et al., 2015a, 2015b) and cooked grain digestibility using an in vitro starch HI assay (Butardo et al., 2011). Mutagenesis and genetic engineering of the starch biosynthetic pathway were successful in producing rice grains with very high amylose and low digestibility. This was accomplished in rice by increasing the proportion of amylose and long-chain amylopectin at the expense of reduced short-chain amylopectin using mutants and transgenic plants of key starch biosynthesis genes (Yano et al., 1985; Nishi et al., 2001; Butardo et al., 2011; Man et al., 2013). However, these extreme phenotypes with elevated amylose levels lead to hard-textured rice upon cooking, thus reducing consumer acceptance and eating quality. In addition, these mutants and transgenic lines quite often exhibited shriveled seeds and have low grain yield. Therefore, there is a need to look at the natural gene pool to identify alleles that rice breeders can use to develop rice varieties that have slower digestibility without compromising acceptable grain quality.
With the availability of 3000 sequenced rice genomes (The 3000 Rice Genomes Project, 2014) and genotype data from the High-Density Rice Array project (McCouch et al., 2016), phenotyping has become the major bottleneck in screening for rice grains with altered digestibility properties. The in vivo glycemic index estimation is low throughput and requires adherence to complex clinical and ethical requirements. Therefore, a viable alternative is to invest in an expensive analytical instrument such as the in vitro HI method used in previous studies (Butardo et al., 2011; Fitzgerald et al., 2011). However, this method cannot be readily adapted as a routine screening method in the majority of rice breeding programs. We therefore used a method based on SEC (Castro et al., 2005a, 2005b) in parallel with amylose content estimation by iodine (Juliano et al., 1981) to unravel the genetic basis of starch structure through a GWAS approach using a diverse set of indica varieties. We then further linked the molecular genetic information obtained to the gene regulatory network influencing starch structure. The SEC-based screening method provided a viable approach to leverage the rich genetic diversity of indica rice in identifying, to our knowledge, novel genes and alleles from both major and minor quantitative trait loci (QTLs). We also demonstrate using a systems-genetics approach how transcriptionally active genes interact in a regulatory network that influences starch structure in rice grain.
RESULTS
The Indica Diversity Panel Shows Huge Variation in Starch Structure
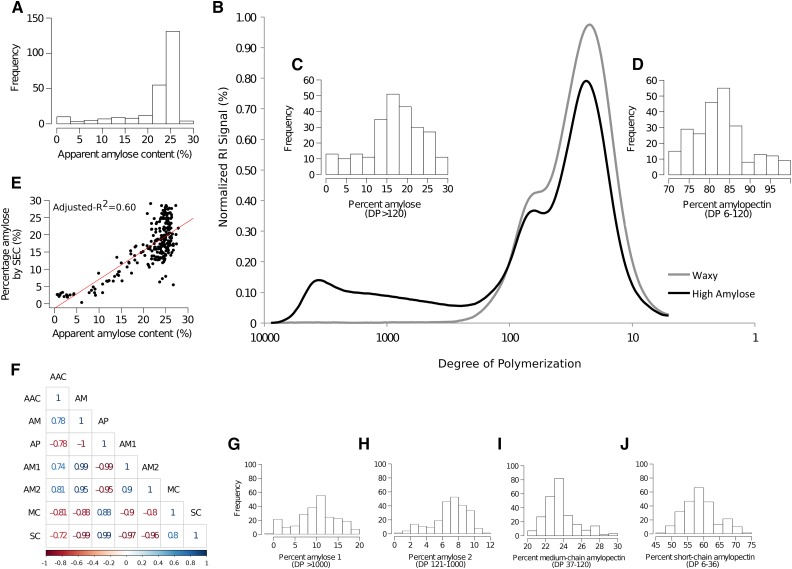
A total of 244 accessions remained after GWAS quality control filtering (Table I). The phenotypic diversity inherent in these accessions represented the full range of amylose classes known in rice cultivars and also demonstrated interesting variations in starch structure (Fig. 1). The AAC of these accessions using the conventional iodine method ranged from 0.6% to 27.8% (Fig. 1A), where the majority were classified as having intermediate (20% to 24%) to high amylose (>25%; Supplemental Table S1). In addition, the debranched starch structure data generated using the SEC method showed substantial diversity for both percentage amylose (PAM) and percentage amylopectin (PAP) within the indica diversity panel (Fig. 1). PAM in this study is defined by debranched amylose chains with degree of polymerization (DP) higher than 120 (Fig. 1C), which is the difference between the chromatogram of glutinous and nonglutinous rice (Fig. 1B) as defined by Fitzgerald et al. (2009) and validated in high-amylose mutants (Butardo et al., 2011). On the other hand, PAP is defined by debranched amylopectin chains with DP ranging from 6 to 120 (Fig. 1D). PAM ranges from 0.4% to 27.7% (Fig. 1C), while PAP ranges from 71.9% to 99.3% (Fig. 1D) within the indica diversity panel studied. Traditionally, breeders use AAC in screening for amylose content because it correlates well with the quality of cooked rice and it is relatively high in throughput. In comparison, the SEC method provides a more accurate measure of amylose content (Butardo et al., 2011) but is low throughput and requires an expensive enzyme for debranching. The linear regression line fitted to the AAC-versus-PAM scatter plot has an adjusted-R2 = 0.60 (Fig. 1E), with deviation from the linear relationship occurring when AAC reaches 22% and beyond. This is due to the complex formed by long-chain amylopectin and iodine that falsely classifies as an amylose-iodine complex using the colorimetric method. These results indicated that AAC cannot resolve structural variations in the high-amylose class (Fig. 1F), and is concordant with a previous study that demonstrated that the iodine assay cannot distinguish true amylose chains from long-chain amylopectin (Butardo et al., 2011).
Table I. Summary of indica diversity panel and genotypic polymorphisms used.
| Parameter | Indica Diversity Panel |
|---|---|
| Phenotypic accessions | 320 |
| GWAS accessions | 244 |
| Total biallelic SNPs | 673,483 |
| Polymorphic SNPs | 662,526 |
| SNPs with MAF > 0.05 | 122,785 |
| Total genotyping rate | 95.85% |
Figure 1.
Characterization of starch structure in a diversity panel with variable AAC by iodine (A) is verified by size-exclusion chromatography (B) using debranched amylose (C) and amylopectin (D). The frequency distribution for (C) PAM and (D) PAP is shown as insets in the representative sections of the SEC chromatogram for waxy (glutinous) and high-amylose-content rice lines in the population under study to graphically show the delineation between debranched amylose (DP > 120) and amylopectin chains (DP 6-120). E, Correlations established between AAC and PAM method are depicted. F, All-pairs Pearson’s correlation r values with the iodine method and values obtained using the SEC method are shown. The debranched SEC plot was further subdivided into (G) percent AM1 (DP > 1000), (H) percent AM2 (DP 1000-121), (I) percent MCAP (DP 120-37), and (J) percent SCAP (DP 6-36). The DP delineations are based on the method described in Butardo et al. (2011).
The MWD of debranched starch based on SEC results was further subdivided into four fractions to account for percent amylose 1 (AM1 or true amylose chains; DP > 1000, Fig. 1G), percent amylose 2 (AM2 or long chain amylopectin; DP 1000-121, Fig. 1H), percent medium-chain amylopectin (MCAP; DP 120-37, Fig. 1I), and percent short-chain amylopectin (SCAP; DP 36-6, Fig. 1J). These fractions were identified based on shifts in MWD distribution of very high amylose kernel mutants or transgenic lines compared with their parental rice lines (Butardo et al., 2011, 2012). The quantified four fractions correspond to AM1, AM2, MCAP, and SCAP were subjected to GWAS analysis.
Genomewide Association Study Highlights Genomic Loci That Associate with Starch Structure
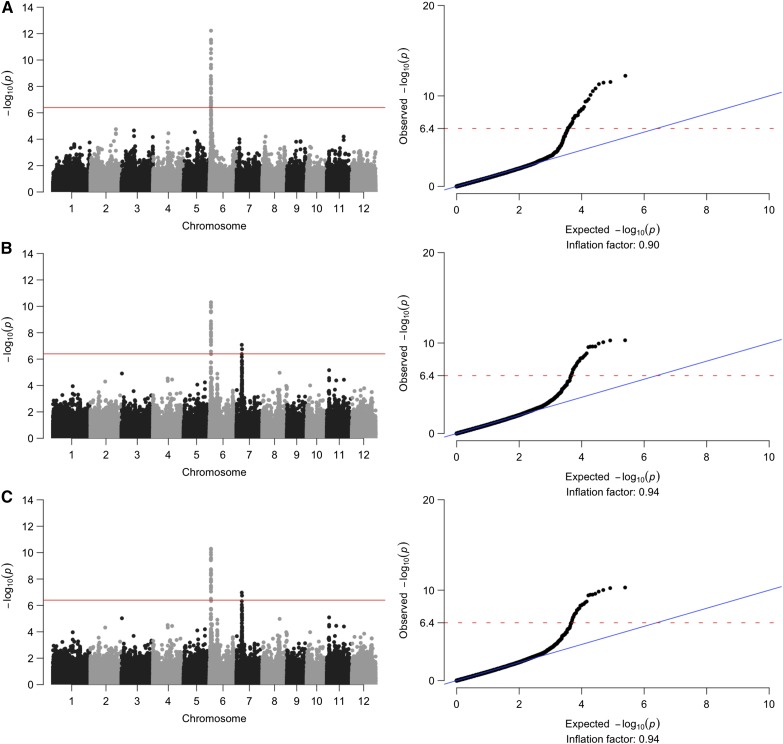
A total of 122,785 quality-assured biallelic SNP markers matching 244 germplasm accessions with a total genotyping rate of 95.85% were used to pinpoint which regions in the genome have significant association with AAC using the iodine method and subsequently to starch structure phenotypes using the SEC method. Most of the accessions were composed of indica (n = 229), while the rest were admixed-indica (n = 7), tropical japonica (n = 6), and admixture (n = 2; McCouch et al., 2016). GWAS on AAC revealed a prominent peak on chromosome 6 (Fig. 2A). Partitioning the SEC chromatograms into four fractions revealed that all four were associated with the same region on chromosome 6 (Supplemental Fig. S1) but interestingly, only AM1 and SCAP were associated with the GWAS peak on chromosome 7 (Supplemental Fig. S1). Subsequent structural genomic analyses for debranched starch structure focused on the GWAS peaks for PAM and PAP (Fig. 2, B and C). It revealed that distinct haplotypes were found to elevate PAM and PAP (Fig. 3). GWAS on AAC (Fig. 2A) has already been reported for rice (Huang et al., 2010) and barley (Hordeum vulgare; Shu and Rasmussen, 2014), although at much lower SNP marker densities.
Figure 2.
Manhattan plots of the genomewide association studies on AAC determined using the iodine colorimetric method (A) and PAM (B) and PAP (C) that were determined using SEC. To the right are their respective quantile-quantile plots (Q-Q plots).
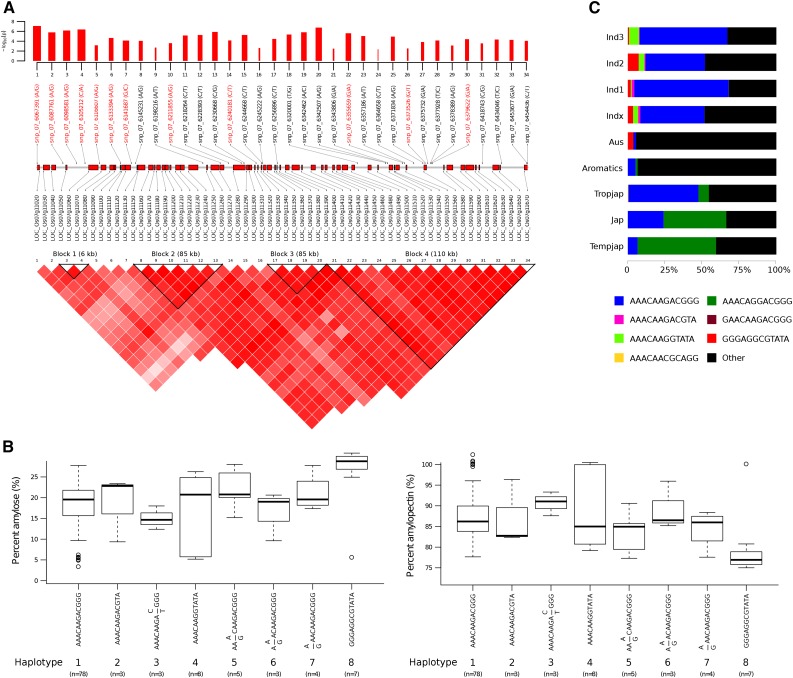
Figure 3.
Mapping of the GWAS peak on chromosome 6 associated with debranched starch structure showing (A) the linkage disequilibrium plot of the 38 SNPs most significantly associated with PAM and PAP measured using the SEC method. A scaled section of the chromosome underlying this region is shown where the positions of the genes are labeled with red boxes sized according to their annotation in the Nipponbare reference genome (release 7). The positions of the 38 SNPs are also indicated where the names of the 11 tag SNPs are highlighted in red. The log10-scaled association P values of these 38 SNPs are shown in the bar plot where bars reflect their relative effect sizes. Also shown are (B) the eight haplotypes formed from the 11 tag SNPs where specific phenotypic ranges for both PAM and PAP are explained by specific haplotypes. The distribution of these eight haplotypes (C) is then checked with data from The 3000 Rice Genomes Project (2014) in which it was shown that haplotypes representing higher PAM values appear prominently among the indica and aus subspecies, while the haplotypes representing lower PAM values appear prominently among the japonica, predominantly in temperate japonica, and haplotypes belonging to intermediate PAM are represented in aromatics subspecies.
Haplotypes on Chromosome 6 Explain Phenotypic Variations in the Proportion of Amylose and Amylopectin
A total of 38 SNPs were found to be in linkage disequilibrium (LD) with index SNPs that mark the region on chromosome 6 associated with PAM (Fig. 2B), which is also the same region that associates with AAC (Fig. 2A). These SNPs were positioned in the intergenic, exonic, intronic, and 5′- or 3′-untranslated regions (UTRs) of structural genes within this region (Supplemental Table S2). Among these intergenic SNPs were four that have the most significant associations with both PAM and PAP (Supplemental Table S2). Eleven tag SNPs (highlighted in red in Fig. 3A) were identified to form haplotypes on chromosome 6 that explained specific PAM and PAP ranges (Fig. 3B and Supplemental Fig. S2). Haplotypes 1 and 5 (T/CGGCCCTTTCT) represented those accessions whose median PAM values were approximately 20%, although 25% of these values overlap with those explained by haplotypes 2, 3, and 6. Haplotypes 2 (TGGCCCTTTCA), 3 (TGGCCCATATA), and 6 (CGGCCGATATA) were found to cover from approximately 10% to 18% PAM. Haplotypes 4 (TGGGTGACATA) and 7 and 8 (CCTGTGACATT/A) represented lines that had very low PAM values.
The topmost significant SNP in the GWAS peak on chromosome 6 (snp_06_1704413) formed an LD block from 1,535,452 to 1,831,347 basepairs (bp). Among those SNPs within this 295.9-kb LD block is snp_06_1765448 (Table II) that lies in the promoter region of GBSSI, which is known to be a major gene involved in amylose synthesis in the grain (Shimada et al., 1993). In addition to this SNP that implicated GBSSI, five other significantly associated SNPs were found and annotated to cause nonsynonymous amino acid substitution for their respective genes. These genes are expressed in the endosperm and code for an uncharacterized transcription factor (TF; LOC_Os06g04010), transmembrane glycosyl hydrolase (LOC_Os06g04169), and ubiquitination protein (LOC_Os06g03840; Table II). The SNP on exon 4 of the transmembrane glycosyl hydrolase causes a C→T mutation that leads to an Arg→Gln amino acid substitution. The contributory role of these target genes on the starch phenotype and its resulting effect on digestibility were not directly explored in this study and requires further evidence by functional characterization.
Table II. List of upstream and exonic nonsynonymous linked SNP mutations detected in QTL locus of chromosomes 6 and 7 associated with amylose content.
| SNP location | Allele | Locus | Gene Ontology | Functional Annotation | Exon Location: Amino Acid Change | %Amylose | |||
|---|---|---|---|---|---|---|---|---|---|
| Ref | Alt | Beta | P Value | −log10(P) | |||||
| A. Chromosome 6 | |||||||||
| 1545370 | C | T | LOC_Os06g03840a | Protein ubiquitination | H-BTB4, Bric-a-Brac, Tramtrack, Broad Complex BTB domain with H family conserved sequence, expressed (protein-binding protein) | Exon2: G64E | 2.5434 | 0.0000 | 4.9249 |
| 1587997 | C | G | LOC_Os06g03940 | Hydrolase activity | Spastin, AAA-type ATPase protein, ATP binding, nucleoside-triphosphatase, nucleotide binding, hydrolase activity | NA | 3.4151 | 0.0000 | 5.3047 |
| 1637153 | A | C | LOC_Os06g04010a | DNA binding | GAGA-binding transcriptional activator BBR-d/RNA regulation of transcription BBR/BPC | Exon3: D235E | 4.0685 | 0.0000 | 9.5475 |
| 1691751 | G | C | LOC_Os06g04080 | Hydrolase activity, plasma membrane | Glycosyl hydrolases family 17, putative, expressed, putative beta-1,3-glucanase | NA | 3.5921 | 0.0000 | 8.8629 |
| 1736459 | C | T | LOC_Os06g04169 | Hydrolase activity, transmembrane | Glycosyl hydrolase family 17, alpha/beta fold family domain-containing protein, expressed | Exon4: R387Q | 4.0010 | 0.0000 | 7.9898 |
| 1765448 | C | T | LOC_Os06g04200a | Glycosyl transferase, plastidial | Granule-bound starch synthase I, glycosyl transferase family 5 | NA | 2.6206 | 0.0000 | 5.1138 |
| 1783773 | T | C | LOC_Os06g04230 | Q-rich domain protein | NA | 3.4509 | 0.0000 | 8.3407 | |
| 1858522 | A | T | LOC_Os06g04360a | Unknown | Expressed protein | Exon4: L227H | 2.1262 | 0.0004 | 3.3532 |
| 1945281 | T | G | LOC_Os06g04520a | Endosomal transport/ organization | Expressed protein | Exon19: F738L Exon19: F593L | 1.3493 | 0.0155 | 1.8104 |
| B. Chromosome 7 | |||||||||
| 6067391 | A | G | LOC_Os07g11020a | Various | bHLH transcription factor regulating proanthocyanidin production in seeds, expressed | Exon5: I299M | −2.8028 | 0.0000 | 7.0796 |
| 6133394 | G | A | LOC_Os07g11100 | Unclassified | NAD-dependent epimerase/dehydratase family protein, putative/NAD-dependent epimerase/dehydratase family | NA | −2.1421 | 0.0000 | 4.6531 |
| 6141687 | G | C | LOC_Os07g11120 | Hydrolase activity | Hydrolase, NUDIX family (hydrolysis P-containing substrates; MutT-like protein) | NA | −2.3994 | 0.0000 | 4.1238 |
| 6211855 | A | G | LOC_Os07g11270a | Unclassified | Retrotransposon protein, putative, Ty3-gypsy subclass, expressed (aminotransferase-like) | Exon7: M835T | −1.8315 | 0.0003 | 3.5924 |
| 6240181 | C | T | LOC_Os07g11310 | Unclassified | LTPL166, protease inhibitor/seed storage/LTP family protein precursor | NA | −1.7520 | 0.0000 | 4.1538 |
| 6244668 | C | T | LOC_Os07g11330 | Unclassified | Alpha-amylase inhibitor/RAL2, seed allergenic protein RA5/RA14/RA17 precursor | Exon1: R29H | −2.2093 | 0.0000 | 5.2574 |
| 6256896 | C | T | LOC_Os07g11370 | Unclassified | Expressed uncharacterized protein | Exon1: P116L | −1.9208 | 0.0000 | 4.4529 |
| 6434046 | T | C | LOC_Os07g11650 | Unclassified | LTPL164, protease inhibitor/seed storage/LTP family protein precursor, expressed | NA | −1.8559 | 0.0000 | 4.3326 |
| 6453677 | G | A | LOC_Os07g11670 | Unclassified | Retrotransposon protein, putative | Exon3: V390I | −1.8530 | 0.0001 | 4.2608 |
| 6454436 | C | T | LOC_Os07g11670 | Unclassified | Retrotransposon protein, putative | Exon4: P596L | −1.8011 | 0.0001 | 4.0678 |
DEG interacting in the same gene regulatory network hub based on rice grain transcriptome data.
Mining the 3000 rice genomes for these eight haplotypes revealed that haplotype 1 was very common among the indica and aus subspecies (Fig. 3C). In contrast, haplotype 8 was present in higher frequency in the subgroups of japonica, particularly among the temperate varieties. Haplotype 6, whose median PAM is approximately 12%, was underrepresented in indicas but was very prominent in tropical japonicas and among the aromatics. Haplotype 2, whose median PAM is approximately 15%, was observed among a few indica varieties as well as with some aromatics. Interestingly, haplotype 3, which also has a PAM median of approximately 15%, appeared only among the few indica varieties.
GBSSI Allele Mining Revealed Additional SNPs Associated with Amylose Content
Targeted haplotyping of starch biosynthesis genes has previously provided insights into the functional properties of rice grains (Kharabian-Masouleh et al., 2011, 2012). A major starch biosynthesis gene found in this study to be in LD with the topmost significant SNP is GBSSI. There were eight SNPs identified and located on exons, introns, and the 5′-UTR of GBSSI (Supplemental Fig. S3A). Three of these SNPs are located in the exonic regions responsible for causing nonsynonymous amino acid substitution on exon 5, 8, and 10 (Supplemental Table S3). A newly identified linked SNP (snp_06_1765448, Table 2) located in the promoter region of GBSSI (−769 bp) could potentially play a role in the transcriptional regulation of the gene considering its proximity to a previously identified and experimentally validated 31-bp transcription factor binding site located upstream (−840 to −810 bp) of the start codon (Zhu et al., 2003). The SNP at 5′-UTR (snp_06_1766647) and the two in the exon 5 and 8 regions (snp_06_1768006 and snp_06_1768724) strongly influenced amylose content (Supplemental Table S3). Haplotypes based on these SNPs were associated with an increased proportion of amylose in the rice endosperm at the cost of decreased amylopectin (Supplemental Fig. S3B). Haplotypes 1, 3, and 4 were found to be diagnostic for intermediate to high amylose classes (Supplemental Fig. S3B). These haplotypes were very frequent in indica and aus subpopulations (Supplemental Fig. S3C). Among all these, haplotype 1 (AACCAA) increases PAM because of elevations in AM1 and AM2 fractions, which were probably preferentially selected during domestication in indica, along with haplotype 4. In contrast, the haplotype AATCAA is preferentially represented in japonica subtypes (Supplemental Fig. S4C), which is not found in the GWAS analysis in this study as the panel mostly represents indica subtypes. The SNP causes a C→T mutation on exon 8 of GBSSI, which introduces, to our knowledge, a novel allele influencing variation between amylose versus amylopectin among the subtypes. These diagnostic haplotypes defined using constituent alleles of GBSSI can be used in targeted breeding to distinguish specific amylose classes within the indica subspecies. Other previously identified allelic variations in the structural gene of GBSSI (Supplemental Table S3) were not represented in this population.
Phenotypic Variations in AM1 and SCAP Structures Are Explained by Haplotypes on Chromosome 7
A total of 34 SNPs were found to be in LD (Fig. 4A) with the GWAS peak on chromosome 7 (Fig. 2C). These SNPs are located at 6,067,391 to 6,379,622 bp of chromosome 7 and are positioned in intergenic, exonic, intronic, and 5′-or 3′-UTR of putative structural genes (Fig. 4A). Twelve of these SNPs were tag SNPs (Fig. 4 and Supplemental Table S2) defined as distinct haplotypes on chromosome 7 (Fig. 4B), which explains phenotypic variation in %AM1 and SCAP content. The most significant SNP (snp_07_6067391) is found on exon 5 of a gene (LOC_Os07g11020) that codes for basic helix-loop-helix (bHLH) transcription factor (Table 2). It causes an A→G nonsynonymous amino acid substitution from Ile→Met. Genotypes formed by alleles of this SNP could classify specific phenotypic ranges of AM1 and SCAP (Supplemental Fig. S4). Another most significant SNP in the region is snp_07_6342507 that associates with all traits measured by SEC except AM2 and MCAP (Supplemental Table S2). It is an intergenic SNP located in between two genes: one codes for a putative α-amylase inhibitor (seed allergenic protein) and the other is a putative transposon. In addition, five SNPs on chromosome 7 have been identified to cause nonsynonymous amino acid changes (Table II). Several genes have unknown function except for one that codes for a seed allergenic protein (LOC_Os07g11330), which also putatively functions as an α-amylase inhibitor (Table II). Interestingly, this region on chromosome 7 (6,067,391 to 6,379,622 bp) is enriched with α-amylase inhibitors, most probably due to tandem duplications. Whether these amylase inhibitors have a functional role in starch digestibility and palatability remains to be seen and requires further functional evidence.
Figure 4.
Mapping of the GWAS peak on chromosome 7 associated with debranched starch structure that shows (A) the linkage disequilibrium blocks formed by the 34 significantly associated SNPs at the genetic interval flanked by LOC_Os07g11020 and LOC_Os07g11670 structural genes (shown as red horizontal bars). These SNPs are based on the Nipponbare reference genome (rel. 7). Also shown together with the SNP IDs are the alternative alleles. SNP IDs highlighted in red are the 12 tag SNPs from which the haplotypes were formed (B). The bar chart shows the −log10 of the association P values of the SNPs; the relative thickness of the bars reflects the effect size of each SNP. The bars are colored red to mean that these SNPs have negative allelic effects. The haplotype distribution in the 3000 rice genomes is shown (C).
Haplotypes defined from the designated 12 tag SNPs of the fine mapped genetic region of chromosome 7 show distinct associations with specific PAM classes (Fig. 4B). Haplotype 1 (AAACAAGACGGG) forms the biggest group of accessions with a median PAM value of approximately 17%, and where 75% of these accessions have PAM values of less than approximately 19%. Other haplotypes appear to represent specific phenotypic ranges with PAM of 10% to 20% (Fig. 4B). It is apparent that haplotype 8 (GGGAGGCGTATA) is an important haplotype whose sequence of alleles is the exact complement to haplotype 1, which explains association with PAM value of 25% and above. Although it forms a smaller group of accessions, it is quite evident that it represents accessions with high PAM values. Most of these rare haplotypes were found in rice accessions from countries in South Asia such as Bangladesh and Sri Lanka, where high amylose is preferred and is probably favorably selected during the course of domestication.
Allele mining studies conducted in The 3000 Rice Genomes Project (2014) suggested that haplotype 1 appeared prominently in indica subgroups, but was very underrepresented in aus and aromatics (Fig. 4C). Interestingly, this haplotype can also be seen at an abundant frequency in tropical japonicas but not in temperate japonicas (Fig. 4C). Additionally, haplotype 8 is very rare in indica subgroups and aus but is not observed at all in the other subpopulations of rice, further proving the rarity of this allele.
bHLH Allele Mining Reveals Distinct Regulation for Starch Biosynthesis in Rice Grain
A major regulatory gene found to be in LD with the topmost significant SNP is bHLH (LOC_Os07g11020), a gene that codes for a TF. Targeted haplotyping of the bHLH gene showed significant association with the functional properties of rice starch structure. Ten SNPs were located on exons, introns, and the 3′-UTR of bHLH (Supplemental Fig. S5A). Haplotype 4 (GTGTCGGGAC) and haplotype 9 (TTGTCTAGAC) showed an increased proportion of AM1 in the rice endosperm at the cost of SCAP (Supplemental Fig. S5B). These are rare haplotypes found in aus and indica and they are completely missing in all other subtypes (Supplemental Fig. S5C). The most significant SNP (snp_07_6067391) found on exon 5 of bHLH causes an A→G nonsynonymous amino acid substitution, which was found to strongly influence amylose content (Supplemental Fig. S5C) and red seed color (see below). This diagnostic SNP defined using alternative allele A→G of bHLH can potentially be used in targeted breeding to develop high-amylose rice within the indica subspecies. It must be pointed out, however, the effect of this allele on seed color can adversely impact consumer acceptance unless the health benefits of red pigments in rice are simultaneously promoted alongside reduced digestibility.
Integration of Genetic and Transcriptional Variations Identified Key Pathways Influencing Starch Structure and Isoflavonoid Metabolism through a Network-Based System Genetic Validation Method
The genetic variants identified by the GWAS approach highlight the important genetic region on chromosome 7 influencing the AM1 target region of starch structure. However, a mechanistic understanding of how this genetic region can potentially modulate gene expression and alter the target phenotype is lacking. We therefore implemented methods that integrate genetic and transcriptional variations from an indica diversity collection of 185 lines to unravel complex traits such as starch structure through a gene regulatory network-based system genetic validation method using whole-genome transcriptome data.
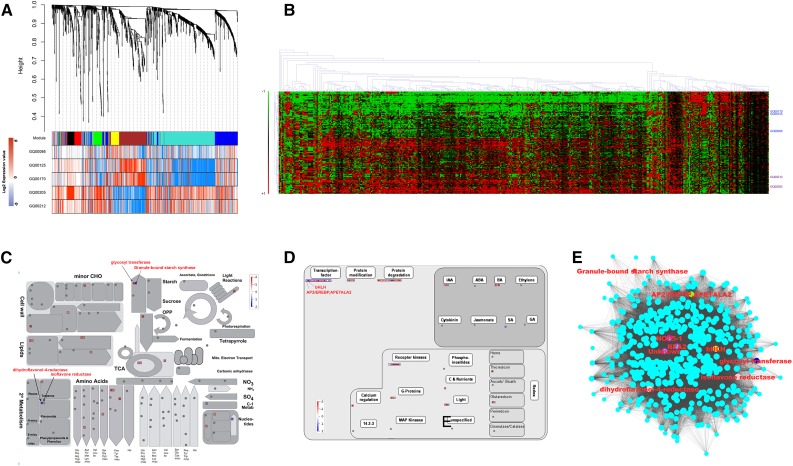
While haplotype 1 of chromosome 7 explains the genetic variation for intermediate amylose content with a median PAM value of approximately 17%, lines containing haplotype 8, whose allelic sequence is the exact complement to haplotype 1, were identified and found to elevate PAM (Fig. 4B). In addition, another subset of lines with alleles belonging to haplotype 4 was identified that contains low to waxy (glutinous or no amylose) indica accessions (Fig. 4B). The 60 K normalized gene expression data were obtained from 16 d after fertilization (16 DAF) of developing seed considered from three lines representing high PAM from haplotype 8 of chromosome 7 (Group 1) and two lines representing glutinous accessions from haplotype 4 of chromosome 7 (Group 2). They were used to identify 1242 significantly differentially expressed genes (DEGs) by performing differential group analysis (Supplemental Table S7). To test the 1242 DEGs whose expression is significantly associated with genetic variation measured by the chromosome 7 region through GWAS, the expression matrix was imputed on 184 lines whose expression and genotyping data matched to derive mutual information based on gene regulatory networks. This resulted in the identification of six major modules (Fig. 5A). The expression heat map of six modules belonging to selected contrasting lines from waxy haplotype 4 and high-amylose haplotype 8 revealed a prominent difference in gene expression profile (Fig. 5A).
Figure 5.
Gene regulatory network and pathway analyses of differentially expressed genes from developing seeds at 16 DAF carrying contrasting haplotypes on chromosome 7. A, Gene expression modules derived from the dendrogram of DEGs between selected lines belonging to haplotypes 4 and 8 show prominent contrast in the turquoise module. B, Heatmap validation of DEGs within the developing grains of the indica population (n = 184) reveal the same trend. C and D, Pathway depiction of DEGs (ratio of log normalized expression values) involved in primary and secondary metabolism (C) and regulatory pathway (D) with major up-regulated genes in haplotype 8 lines represented in blue and down-regulated in haplotype 8 lines labeled in red. E, Gene regulatory network derived based on DEGs in the turquoise module showing three central hub genes connected with key genes related to starch metabolism and isoflavonoid enzymes and key regulators.
Among the six modules, the turquoise module represents 441 genes with the maximum interaction node identified for LOC_Os07g11020 bHLH transcription factor (Fig. 5E), where the most significant SNP_07_6067391 found on exon 5 of the BHLH gene was located on chromosome 7 (Fig. 4A and Supplemental Table S2). Analyses of the gene regulatory network of the turquoise module showed that key genes contributing to significant associations with PAM detected on chromosomes 6 and 7 were found in the network hub (Fig. 5E). In particular, GBSSI involved in starch biosynthesis, dihydroflavonol-4-reductase (DFR), and isoflavone reductase (IFR) from isoflavanoid metabolism and the key regulator bHLH were found to belong within an adjacent network neighborhood defined by three central hub genes putatively involved in RNA transcription: NOP5-1 (LOC_Os03g22730), RPA2 (LOC_Os10g35290), and an uncharacterized gene (LOC_Os03g01016; Fig. 5E). DFR is the enzyme responsible for the conversion of dihydroflavonols to leucoanthocyanidins, which then leads to downstream biochemical pathway producing proanthocyanidins, the red pigment in rice grain (Furukawa et al., 2007). Interestingly, within the 1000-bp promoter region of GBSSI (highlighted in Supplemental Table S5) and DFR (highlighted in Supplemental Table S6) genes, we found a cis-acting element for bHLH that was not found in the promoter region of the other glycosyl hydrolase or glycosyl transferase genes (Supplemental Table S6). In addition, genes involved in sugar sensing, signaling, and transport; carbohydrate synthesis and degradation; and cell wall synthesis and modification were very prominently enriched in the turquoise gene regulatory network (Supplemental Table S8). Known homologous genes coding for auxin and brassinosteroid hormone metabolism, protein degradation and protein modification genes, signaling receptor kinases and G-proteins, and chromatin remodeling were also abundantly represented (Supplemental Table S8).
Heatmap comparison of DEGs of the turquoise module from 184 indica lines that signified association through GWAS on chromosome 7 (Fig. 2B) exhibited differential regulation of 441 genes with two distinct clusters (Fig. 5B). One cluster representing lines with haplotype 8 exhibited prominently up-regulated genes at 16 DAF, while the other group of lines represented in haplotype 4 demonstrated a reverse trend in which most of the genes were down-regulated (Fig. 5B). Moreover, analyses of differentially expressed transcription factor genes revealed that bHLH is the only up-regulated transcription factor found in haplotype 8-related lines (Fig. 5D). MapMan visualization of metabolic pathways showed that GBSSI in the starch biosynthetic pathway was prominently up-regulated in haplotype 8-containing lines in comparison with haplotype 4-related lines, while a gene coding for a member of the glycosyl transferase family 8 (LOC_Os02g41520) was down-regulated in haplotype 8 lines (Fig. 5C). This uncharacterized glycosyl transferase codes for a major gene involved in carbohydrate metabolism in rice homologous to Arabidopsis (Arabidopsis thaliana) plant glycogenin-like starch initiation proteins (PGSIP6; AT5G18480). Additionally, two genes involved in secondary metabolism of flavonoid biosynthesis in rice grains were differentially expressed. In particular, DFR was found up-regulated in haplotype 8-containing lines, while the IFR gene was down-regulated (Fig. 5C). Summing up, the genomic and transcriptomic results generated in this study showcased the power of integrating genomic and gene expression data to explain the influence of genetic variants in expression-trait associations related to starch structure.
Associating Starch Structure with Digestibility
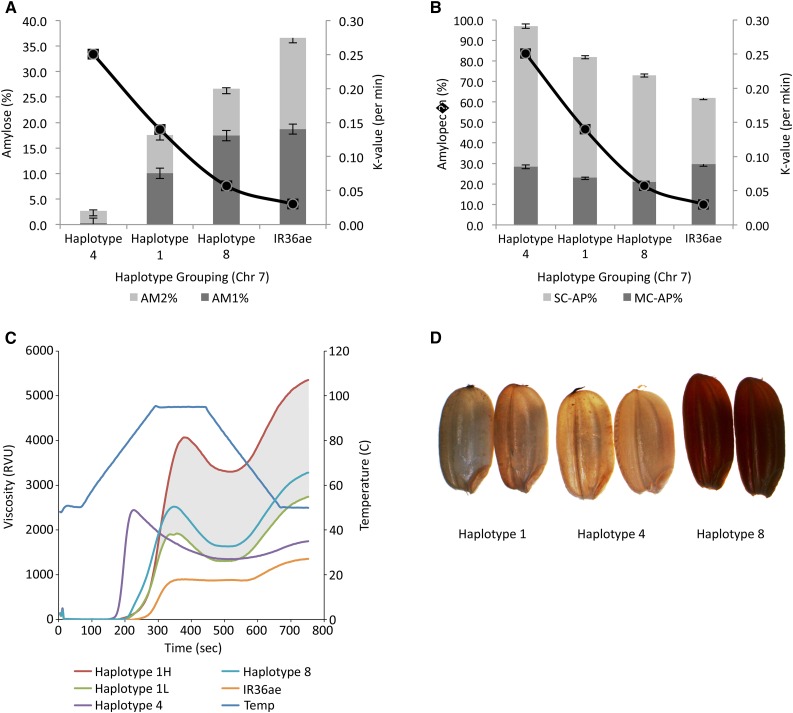
To determine the correlation between starch structure and digestibility, a few lines covering the minimum-maximum value of PAM from haplotypes 8, 4, and 1 were identified representing the entire range of diversity in debranched starch structure from waxy to high amylose (Fig. 1). In addition, IR36ae was used as a high-amylose kernel mutant (Juliano et al., 1990) to represent the least digestible control with extreme starch structural modification. This rice grain mutant has a stop codon mutation in SBEIIb leading to elevated amylose content of up to 35% (Butardo et al., 2012). Characterizing digestibility kinetics based on the extent of starch hydrolysis (%) of cooked starch granules revealed that the first hour is most crucial in differentiating digestibility. Starch hydrolysis is complete on the second hour, as indicated by the plateau or saturation point in the enzyme digestibility kinetics. The rate of starch hydrolysis based on the k value routinely used to estimate glycemic index of starchy foods (Goñi et al., 1997) revealed that digestibility decreases with increasing amylose content, with the very high amylose mutant IR36ae exhibiting the highest resistance to starch hydrolysis (Fig. 6A). The k value from cooked grain amylolysis revealed that the low amylose accessions of haplotype 4 were the most highly digestible, followed by the abundantly represented intermediate amylose class of haplotype 1, with the rare high-amylose class of haplotype 8 exhibiting slower digestibility (Fig. 6B). Results further confirmed that an increase in AM1 chains directly leads to reduced digestibility kinetics for lines belonging to haplotype 8 on chromosome 7 (Fig. 6A). In addition, haplotype 8-containing lines exhibit red color phenotype (Fig. 6D) because of the differential expression of flavonoid metabolism and bHLH transcripts (Fig. 5, C and D). In contrast to all haplotypes, the reduction in digestibility for IR36ae is due to an elevation in AM1 (i.e. true amylose chain) as well as the elevation of AM2 (i.e. long-chain amylopectin), which is corroborated by a previous study (Butardo et al., 2011). The reduction in digestibility based on the k value can also be attributed to a decrease in the proportion of SCAP in haplotype 8 as well as in IR36ae (Fig. 6B).
Figure 6.
Characterization of digestibility and cooking property of selected rice lines belonging to different haplotypes represented from chromosome 7. A and B, The k values from cooked milled rice flour of representative lines from haplotypes 1, 4, and 8 compared with IR36ae as a line graph. The k values were overlaid with (A) amylose and (B) amylopectin fractions. C, The starch paste viscosity profiles of the three groups of haplotypes were also shown in comparison with IR36ae. Note that haplotype 1 represented lines showing a range of relative viscosity units as exemplified by the shaded gray region with the viscosity profile of haplotype 8 matching within the gray zone and the lowest observations depicted for glutinous lines represented from haplotype 4 (green). D, Representative seeds from haplotypes 1, 4, and 8.
Promotion of more slowly digestible rice varieties can be successful only when cooked rice quality matches with what consumers prefer. Although previous studies were successful in producing high-amylose rice with reduced digestibility because of the increased proportion of resistant starches, these rice grains will tend to have low consumer acceptance because of cooked rice hardness (Butardo and Sreenivasulu, 2015). The starch paste viscosity profiles of selected haplotypes were compared with IR36ae to assess cooking quality through visco-elastic property characterization. Results revealed that rice lines belonging to haplotype 4 cooked faster and demonstrated a typical waxy (glutinous) starch paste viscosity for which the peak viscosity is higher than the final viscosity (Fig. 6C). In contrast, those belonging to the common haplotype 1 showed a wide range of viscosity profiles characterized by lower peak viscosity than the final viscosity (Fig. 6C shaded). This is typical for rice lines with intermediate to high amylose. Although haplotype 8 demonstrated a lower rate of starch hydrolysis, the final viscosity is similar to that of haplotype 1, but is clustering toward the lower range (Fig. 6C). In contrast, IR36ae demonstrated an atypical starch viscosity profile characterized by the slowest pasting onset and inability to swell, with a lack of gelatinized paste properties (Fig. 6C). Cooked rice grains from this viscosity profile are expected to produce extremely hard texture, which was experimentally validated by our rice grain sensory profiling study (data not shown).
DISCUSSION
Starch Structure Implication to Tweak Cooking Quality and Starch Digestibility
The proportion of amylose and amylopectin in rice endosperm starches is influenced by the activity of many biosynthetic enzymes that synergistically act during grain filling to influence several levels of starch structural organization (Lourdin et al., 2015; Nakamura, 2015). The details of the primary structure of amylose and amylopectin are well characterized through functional characterization of the target genes by either overexpressing genes for amylose biosynthesis (GBSSI) or by suppressing the expression of key genes for amylopectin biosynthesis (SSI, SSIIa, SSIIIa, BEI, and BEIIb). This usually leads to increased amylose or altered amylopectin branching, which in turn alter grain functional properties (Nishi et al., 2001; Umemoto et al., 2002; Nakamura et al., 2005; Hanashiro et al., 2008, 2011; Butardo et al., 2011; Zhu et al., 2012; Man et al., 2013). The structures of starches unraveled from these rice mutants and/or transgenic plants suggested that GBSSI is crucial in amylose synthesis but additional carbohydrate-active enzymes might play a role in influencing the overall structure and proportion of starch fractions. In this study, a prominent association peak on chromosome 6 (Fig. 2) linked with distinct alleles identified within GBSSI (Fig. 3 and Supplemental Fig. S3) further strengthened the importance of this gene in influencing amylose content and overall starch structure. The GBSSI gene codes for an enzyme exclusively responsible for synthesizing the amylose fraction of starch (Ball et al., 1998; Smith et al., 2001). It is also implicated in the synthesis of long-chain amylopectin of DP greater than 100 (Hanashiro et al., 2008). Our results corroborated these findings based on the indica rice diversity panel. Amylose content defined from the SEC data suggests the dual role of GBSSI in the synthesis of true amylose chains (AM1) and long-chain amylopectin (AM2). In addition, the SEC method (Fig. 1C) provided more accurate quantification of amylose content (Fitzgerald et al., 2009) than the iodine method (Fig. 1A) because it is capable of differentiating true amylose from longer amylopectin chains (Butardo et al., 2011). This is evident from the correlation plot comparing iodine versus SEC methods, which shows that there is high correlation between the two methods for the ranges of low to intermediate amylose classes, but the correlation significantly drops in the high-amylose class (Fig. 1E and Supplemental Table S1). The iodine method suffers interference from the complex formed by long-chain amylopectin and iodine, leading to overestimation of the amylose content when measured at 620-nm absorbance (Butardo et al., 2011).
The large-effect SNPs in the QTL region of the GBSSI locus were previously reported to influence amylose content in rice (McNally et al., 2009). Candidate genes from QTLs and eQTLs under positive selection during the course of domestication were also reported in rice (House et al., 2014). This included a selective sweep in the rice GBSSI QTL region (Olsen et al., 2006). Haplotype 1 of chromosome 6 that explains intermediate to high amylose range is abundantly represented in the rice diversity panels (Fig. 3, B and C). These results support the possibility that haplotype 1 of chromosome 6 is favorably selected during the course of domestication on the basis of good grain quality traits. GBSSI was also linked as one of the major genetic determinants of glycemic impact upon rice consumption (Fitzgerald et al., 2011). In addition, we have identified haplotypes that mostly explain intermediate digestibility from regions that were found to associate with PAM and PAP (Fig. 3). Other genes such as SBEIIb (Butardo et al., 2011) and Starch Synthase IIa (Morell et al., 2003; Hogg et al., 2013) were also found to influence slower digestibility in rice and other cereals by increasing the amount of amylose and resistant starch fractions. However, we did not find any direct association between these genes and starch structural variation within the indica diversity panel in this study. It is possible that the alleles coding for extreme starch phenotypes were not favorably selected during the course of domestication because they significantly lower the cooking and eating qualities of rice grains.
Introducing subtle modifications in starch phenotypes arising from naturally occurring allelic variations accumulated during the course of domestication is better for the purpose of breeding rice with slower digestibility. This approach is expected to produce rice grains with acceptable grain quality compared with extreme phenotypes artificially induced by mutagenesis or transgenesis. In this study, a genetic region on chromosome 7 was fine-mapped and the identified haplotype 8 was found to be associated with increased AM1 (Figs. 4B and 6A) and with reductions in SCAP (Fig. 6B). This increase in the ratio of true amylose chains at the expense of shorter amylopectin chains resulted in slower digestibility based on a reduction in cooked grain amylolysis as measured by the k value (overlaid line plot in Fig. 6, A and B). Moreover, rice lines containing haplotype 8 of chromosome 7 have better textural attributes than the SBEIIb mutant IR36ae based on RVA profile (Fig. 6C). In addition, lines belonging to this haplotype are tightly linked to red seed color phenotype (Fig. 6D). This genetic region being linked to slower digestibility and red pigmentation in rice grains displays rare alleles (Fig. 6). Along with seed shattering and seed dormancy, pericarp pigmentation is not favorably selected during the course of rice domestication (Sweeney and McCouch, 2007; Gross et al., 2010). It is possible that ancient rice farmers used the red pericarp color as a visual cue to discard red rice seeds due to perceived association with inferior grain quality. In the process, white endosperm was artificially selected. We have demonstrated in this study that these rare alleles can still be recaptured by GWAS given a good rice panel representing diverse, a highly heritable grain quality trait and a very dense set of SNP markers. This rare haplotype can directly be used in marker-assisted breeding for rice with reduced digestibility without sacrificing grain quality acceptance. Because the reduced digestibility phenotype identified in this study is also linked with the formation of red pigments, a specialty market can be tapped to simultaneously promote the health benefits of flavonoids in colored rice grains with reduced glycemic response. To promote slower digestibility traits in white milled rice genotypes, we need to produce genome-edited rice mutants with stop codon mutation in DFR and combine them with slower digestible haplotype 8 from the fine mapped chromosome 7 region.
Automated in vitro systems that mimic digestion along the human gastrointestinal tract or fermentation in the human colon are available (MacFarlane et al., 1998; Woolnough et al., 2008; Hur et al., 2011; Tanner et al., 2014; Ou et al., 2015) but are still low in throughput and require very expensive capital investment. Hence, aside from debranched SEC, the cooked-grain amylolysis method developed in this study can be used, as a method for screening rice with low digestibility using k values (Fig. 6, A and B). The diagnostic haplotypes defined for various starch fractions obtained through the SEC method can supplement as an initial screening pipeline (Fig. 1) because they are associated with the k value (Fig. 6, A and B; Supplemental Fig. S6B). As shown using landrace varieties, the distinct alleles of haplotype 8 on chromosome 7 (Fig. 4, A and B) reported to increase true amylose chains (Fig. 1G) at the expense of reduced SCAP (Fig. 1J) lead to a decrease in the k value. The relative shifts in the proportion of these starch fractions were also previously reported to decrease starch hydrolysis index (Butardo et al., 2011; Fitzgerald et al., 2011), a proxy measure for estimated glycemic scores. We have therefore demonstrated in this study a highly efficient and streamlined grain quality phenotyping and genomics pipeline that can aid breeders in developing new rice cultivars with improved nutritional quality without compromising consumer acceptance by maintaining region-specific grain quality traits.
A Systems Genetics Approach Implemented to Unravel Starch Structure Helps in Identifying Distinct Alleles in the Diverse Pool and in Simultaneously Providing Comprehensive Links to Functional Regulatory Pathways
Systems genetics approaches were useful in dissecting obesity-related diseases in mouse (Gusev et al., 2016) and in unraveling complex metabolism in maize (Zea mays) kernels and brassica rape (Brassica rapa) seeds (Basnet et al., 2016; Wen et al., 2016). Such approaches have not yet been exploited in rice. As exemplified in this study, the rapid developments in next-generation sequencing technologies enabled the construction of high-density SNP-based linkage maps that can be associated with starch structural traits through GWAS. Furthermore, linking this genetic information (significant SNPs associated with starch structure identified in target genes) to transcript profiling of 16 DAF developing seeds of the same diversity panel helped in identifying functional pathway evidences through gene network-based analysis (Fig. 5). Using this strategy, we confirmed that GBSSI is the primary large-effect causal gene on chromosome 6 (Fig. 2A). In addition and more importantly, a highly significant association peak on chromosome 7 (Fig. 2B) linked with a significant SNP on exon 5 of a gene that codes for a basic helix-loop-helix (bHLH) TF (Table II and Supplemental Fig. S5) was identified as a potential regulator of starch structure. Interestingly, the expression of this gene was identified to belong in the same gene regulatory network hub as GBSSI (Fig. 5). We presume that this TF confers a transregulatory influence toward a cis-acting element located upstream of the 5′-regulatory region of the structural gene for GBSSI. These gene regulatory network data (Supplemental Tables S7 and S8) are supported by the identification of a putative binding site in the 5′-promoter region of the structural gene of GBSSI identifying bHLH as one of the cis-regulatory elements (Supplemental Table S5). In comparison, no transcription binding site has been identified for bHLH in the other starch-related genes identified in this study (Supplemental Table S6). This hypothesis is also supported by a previous study experimentally demonstrating that a paralogous bHLH transcription factor (OsBP-5) can transcriptionally enhance GBSSI expression in the promoter region (Zhu et al., 2003). It is possible that transcriptional regulation of GBSSI by bHLH can affect the proportion of amylose and amylopectin as demonstrated by our GWAS (Fig. 2 and Supplemental Fig. S2) and gene regulatory network data (Fig. 5 and Supplemental Tables S7 and S8). This influence on starch structure can have profound effects on the functional property of rice grains considering that the dry weight of milled rice is composed of starch (Juliano, 1979). This was demonstrated in this study by investigating the kinetics of digestibility of cooked starch granules (Fig. 6 and Supplemental Fig. S6A) using various haplotypes on chromosome 7 (Fig. 4 and Supplemental Figs. S4, S5, and S6B) containing structural variations in the AM1 and SCAP starch fractions (Fig. 6).
GBSSI belongs to glycosyl transferase (EC 2.4.1.21) family 5 (GT5) based on carbohydrate-active enzyme database classification (Lombard et al., 2014). GBSSI enzyme facilitates the transfer of glucosyl residue of ADP-Glc into the nonreducing end of malto-oligosaccharides (MOS) such as maltotriose and maltohexose, which probably act as the primer for amylose synthesis in plants (Denyer et al., 1996, 1999; Zeeman et al., 2002). How MOS are generated in heterotrophic plant cells such as rice endosperm and which enzyme catalyzes their formation is still unknown. Although GBSSI is the sole glycosyltransferase (GT) identified by previous studies to be directly involved in amylose synthesis in cereals, to our knowledge, a novel plastidial PROTEIN TARGETING TO STARCH, which contains an N-terminal coiled-coil domain and a C-terminal carbohydrate binding module, is identified as a regulatory scaffold protein (Lohmeier-Vogel et al., 2008) crucial in amylose synthesis in Arabidopsis leaves by directing GBSSI into the starch granules (Seung et al., 2015). We identified the GBSSI gene possessing glycosyltransferase 5 domain in the network neighborhood of the turquoise module (Fig. 5E). In addition, a glycosyltransferase belonging to family 8 (GT8)-containing genes was identified by GWAS (Supplemental Table S2) and gene regulatory network analyses (Fig. 5E), respectively. The GT8-domain-containing gene (LOC_Os02g41520) belongs to the glycogenin glucosyltransferase family (EC 2.4.1.186; Cao et al., 2008; Lombard et al., 2014). It is homologous to the PGSIP based on MSU rice genome annotation (Ouyang et al., 2007). It is believed that PGSIP is possibly involved in the initiation of the synthesis of amylopectin in a manner analogous to glycogenin enzymes (Yin et al., 2010). Glycogenins are self-glycosylating proteins in fungal and mammalian systems crucial in initiating glycogen synthesis by forming short MOS primers (Smythe and Cohen, 1991; Roach et al., 2012). Self-glycosylating proteins in plants have been previously identified to be involved in plant cell wall matrix polysaccharide synthesis (Langeveld et al., 2002; Sandhu et al., 2009) but no homologous enzyme has been implicated in the initiation step of starch biosynthesis. The GT8 domain-containing gene identified in this study by a transcriptomic approach is therefore interesting because the down-regulation of this gene in haplotype 8 accessions (Fig. 5C) is accompanied by an up-regulated expression of the GBSSI gene. Whether this gene is important as a self-glycosylating enzyme that generates MOS remains to be established.
The intricacies of starch biosynthesis and its regulation are just starting to be unraveled. What is known so far is that the amount of amylose in rice grain is primarily due to allelic variations in the structural gene of GBSSI, its transcriptional regulation by cis-regulatory elements, the level of transcripts after alternative mRNA splicing, and the enzymatic activity of its protein product GBSSI (Sano, 1984; Shimada et al., 1993; Bligh et al., 1995; Ayres et al., 1997; Hirano et al., 1998; Larkin and Park, 2003; Mikami et al., 2008; Zhu et al., 2011). Any mutations influencing the synthesis and regulation of GBSSI lead to significant alterations in the proportion of amylose and long-chain amylopectin with a concomitant impact on the ratio of medium- and short-chain amylopectin. This in turn affects grain functional properties. The null alleles of GBSSI produce amylose-free grain phenotype in tropical glutinous cultivars usually because of the introduction of a 23-bp in-frame duplication leading to a premature stop codon (Wanchana et al., 2003). Additionally, a G/T SNP mutation in the intron 1-exon 1 boundary leads to a reduction in the premRNA splicing efficiency of GBSSI (Wang et al., 1995; Frances et al., 1998; Cai et al., 1998; Isshiki et al., 1998; Larkin and Park, 1999). This aberrant splicing results in decreased concentration of functional GBSSI enzymes, which explains the glutinous to low-amylose phenotypes commonly observed in japonica lines. Lastly, two exonic SNPs can differentiate between intermediate- and high-amylose rice grains: one is present on exon 6 (A/C) and the other on exon 10 (C/T; Larkin and Park, 2003; Dobo et al., 2010; Kharabian-Masouleh et al., 2011). Although not functionally validated, these two nonsynonymous SNP mutations are believed to change the polarity and therefore enzymatic activity of GBSSI. All these previously observed SNPs and insertion mutations, including other previously published GBSSI alleles summarized in Supplemental Table S3, were not represented in the high density rice array (McCouch et al., 2016) and therefore were not detected in this study. Instead, a highly significant SNP was identified in the promoter region of GBSSI located at the −769 bp of the start codon (Table II and Supplemental Fig. S3). This SNP at the 5′-untranslated region (UTR) appears to be functionally relevant because it is proximal to a 31-bp bHLH cis-acting element in the promoter region of GBSSI. This promoter region has been functionally demonstrated to interact with a bHLH paralogue (OsBP-5) in tandem with OsEBP-89, another TF belonging to the APETALA2 and ethylene-responsive element-binding proteins (AP2/EREBPs) to synergistically regulate the expression of GBSSI (Zhu et al., 2003). In this study, the up-regulation of bHLH is accompanied by a down-regulation of the expression of a gene coding for an AP2/EREBP TF (Fig. 5D). The implication of this regulatory interaction needs further functional validation. On the other hand, the highly associated gene LOC_Os07g11020 detected by GWAS in this study (Fig. 2, B and C) codes for a bHLH TF that has been previously implicated in modulating the formation of red pigments in rice grains via the proanthocyanidin biosynthesis pathway (Sweeney et al., 2006; Furukawa et al., 2007). Interestingly, all the rice accessions that belong to the rare haplotype 8 have red pigments (Fig. 6D). Based on our transcriptome data, the up-regulation of DFR in the secondary metabolic pathway (Fig. 5C) is accompanied by the up-regulation of bHLH (Fig. 5D). Additionally, a binding motif for bHLH was also detected in DFR (Supplemental Table S6), indicating the role of this TF in activating the expression of DFR. In this study, we propose another putative role for this bHLH TF: binding in the promoter region of GBSSI and modulating the expression of its transcript. This TF can potentially influence the starch biosynthetic pathway given its homology with another well-characterized bHLH shown to regulate the promoter region of GBSSI (Zhu et al., 2003). In other plant systems, the phenylpropanoid metabolism responsible for the production of red and purple potatoes (solanum tuberosum) is tightly linked with bHLH TF regulation and Suc metabolism (Payyavula et al., 2013). Suc can enhance the anthocyanin biosynthesis pathway in Arabidopsis (Solfanelli et al., 2006). The accumulation of carotenoids in citrus is also linked with starch and anthocyanin metabolism (Cao et al., 2015). Whether a homologous interconnected pathway also exists in rice endosperm and how secondary metabolism related to the anthocyanin pathway can influence the primary metabolic pathway related to starch biosynthesis will be the subject of future metabolomic and functional genomic validation studies.
CONCLUSION
This study unraveled the genetic basis of rice starch structure revealing a fine mapped genetic region in chromosomes 6 and 7. The GWAS peak on chromosome 6 is strongly associated with, to our knowledge, novel alleles in the 5′-untranslated and coding regions of GBSSI, which is potentially responsible for defining starch structure by influencing the proportion of amylose versus amylopectin fractions. In addition, the peak in chromosome 7 is associated with the proportion of long amylose fractions (DP > 1000) at the cost of a reduced proportion of SCAP (DP 6-36). In particular, a haplotype defined from the region covering the bHLH transcription factor seems to influence elevated levels of amylose 1 fraction at the expense of a reduced proportion of SCAP aside from influencing the red pericarp phenotype. Selected lines carrying targeted haplotypes from chromosomes 6 and 7 confirmed its influence on starch structure and grain digestibility. Grain quality genomics and systems genetics approaches were employed in this study to identify new haplotypes and regulatory networks responsible for starch structural variations in rice grain, which can be tapped to modulate grain digestibility in rice breeding pipelines using cutting-edge postgenomic marker-assisted technologies (Anacleto et al., 2015). This approach led to the discovery of rare allelic variations in the natural germplasm pool that code for more subtle alterations in grain digestibility, which can potentially be tapped to develop less digestible rice grains with superior grain quality to ensure consumer acceptance. Introgressing, unique QTLs conferring low digestibility into high-yielding rice mega-varieties can provide value-added traits to increase the market value of premium rice as well as aid in the faster deployment of healthier rice grains to consumers without sacrificing grain quality (Butardo and Sreenivasulu, 2015).
MATERIALS AND METHODS
Plant Materials
A total of 320 accessions (Table I) were selected from the 498 indica landraces identified by McCouch et al. (2016) for Rice Diversity Panel 2. Paddy rice samples of five additional varieties used in a previous study to represent diversity in amylose classes (Fitzgerald et al., 2011) were also obtained from the IRRI Genetic Resources Center. They were all grown in randomized replicated plots at the IRRI experimental field station during the wet and dry seasons of 2013 to 2014 in standard irrigated rice conditions. Seeds were harvested at maturity and allowed to air-dry at room temperature to 12% to 14% moisture content.
Sample Processing
Rough rice was dehulled using a THU35A Test Husker (Satake). Brown rice was ground into brown flour by passing twice through a Udy Cyclone Sample Mill (Seedburo Equipment Company) with an installed mesh 25 sieve. Milled rice was obtained by polishing brown rice using a Grainman 60-230-60-2AT (Grain Machinery Manufacturing Corporation). White flour was obtained by pulverizing in Udy Cyclone Sample Mill model 3010-20, as already described.
Amylose Content Estimation by Iodine and SEC Methods
AAC determination by iodine colorimetry was done on white flour using the standard method (Juliano et al., 1981) adapted to high-throughput screening using a Scalar Continuous Flow Analyzer (San++ System, Netherlands). For size-exclusion chromatography, brown rice flour (55 mg) was gelatinized and debranched with isoamylase as previously described in Ward et al. (2006) using glass scintillation vials containing 400 μL of 95% ethanol and 1 mL of 0.25 m NaOH. The debranched sample was desalted in a 2-mL microtube containing approximately 320 mg ion-exchange resin [Bio-Rad AG 501-X8 (D)] and incubated for 30 min at 50°C with mixing every 10 min. The solution was then pipetted into a 150-µL mandrel insert and placed in a sample vial (Waters) with a septum with a slit. An aliquot of the debranched solution (40 µL) was analyzed using SEC (Alliance 2695; Waters), fitted with an Ultrahydrogel 250 column (Waters) and a model no. 2414 Refractive Index detector (Waters), using 0.05 m ammonium acetate, pH 4.75, with 0.02% sodium azide as mobile phase. The column was calibrated for Mr using individually injected pullulan standards (P800, P400, P200, P100, P50, P20, P10, and P5; P-82 Shodex; Showa Denko). SEC plots were obtained using the Mark-Houwink-Sakaruda equation and universal calibration (Castro, 2005; Ward et al., 2006). Four zones of debranched SEC plots were defined based on a previous publication (Butardo et al., 2011) to correspond to AM1, AM2, MCAP, and SCAP. Results of amylose estimation by iodine and SEC analyses were used for GWAS, as described below.
Genomewide Association Study and Structural Genomics Analyses
The genomewide association study used the SNP genotype data from the High-Density Rice Array project (McCouch et al., 2016). Nonbiallelic SNP markers were removed and the remaining markers were filtered in a stepwise manner. First, variants with missing call rates that exceeded 10% were removed. Then, samples whose missing call rates exceeded 10% were also removed. Finally, the SNP genotyped data were filtered to retain only those SNP markers with minimum allele frequency of at least 5%. The result is SNP genotype data with 122,785 biallelic markers and 244 samples with a total genotyping rate of 95.85% (Table I). These filtering steps were done using PLINK (Purcell et al., 2007; Chang et al., 2015). Linear mixed model GWAS was then carried out using the EMMAX suite (Kang et al., 2010). EMMAX-kin was used to calculate the Balding-Nichols kinship matrix to account for cryptic relatedness among the samples, after which EMMAX associations with the phenotype were calculated. The threshold value for significance is P value < 4.072e-7, indicated by a red horizontal line in the Manhattan plot at –log10(P value) = 6.39.
The peaks were then processed using the “–clump” function in PLINK. Index SNPs were identified to be among those with P values smaller than the threshold value for significance. Mutually exclusive clumps of SNPs were then formed around an index SNP. Any SNP within a clump has a P value < 0.01 and an LD value of r2 > 0.5 with the index SNP and within 200 kb of the index SNP. Tag SNPs were then identified from each clump using De Bakker’s algorithm implemented in Haploview (Barrett et al., 2005). The linkage disequilibrium blocks of these tag SNPs were then formed using Gabriel’s algorithm (Gabriel et al., 2002) and visualized using Haploview (Barrett et al., 2005). These SNPs were linked to the annotated genes using annovar (Wang et al., 2010) with data from the Rice Genome Annotation Project (MSU v7; Ouyang et al., 2007). The genes were annotated from public databases such as Interproscan (https://code.google.com/p/interproscan/wiki/HowToDownload), GOMapMan (http://mapman.gabipd.org/web/guest), SWISSPROT (http://www.ebi.ac.uk/uniprot), and NR (https://www.ebi.ac.uk/patentdata/nr).The phytozome database contains genome annotations of Sorghum, Arabidopsis (Arabidopsis thaliana), Brachypodium, and Maize (Zea mays; http://phytozome.jgi.doe.gov/pz/portal.html), BRENDA (http://www.brenda-enzymes.org/), KINASES (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi), Transcription Factor (http://bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi), Transporter (http://plantst.genomics.purdue.edu/families.shtml), Phosphatase (http://plantsp.genomics.purdue.edu/), and ARAMEMNON (http://aramemnon.uni-koeln.de/) databases. All gene annotations were accessed in January 2014.
Transcriptome and Gene Regulatory Network Analyses
Total RNA was extracted from developing (16 d post antithesis, dpa) grains for transcriptome analyses using a genomewide microarray platform (Agilent Technologies). Gene expression profiling was conducted by hybridizing onto a newly developed genomewide microarray slide for rice based on the manufacturer’s protocols (Agilent Single Color; Agilent Technologies). A total of 200 indica lines were selected from the original 320 (Table I) for transcriptome analyses. They were grown in triplicate during the 2013 dry season at IRRI using a glasshouse maintained with ambient light, 29°C/21°C d/n temperature, and 75% to 85% relative humidity. Soil preparation, fertilizer regime, pesticide application, and glasshouse maintenance were as previously described (Jagadish et al., 2011).
Individual rice accessions were grown until panicle emergence. Individual panicles were labeled to track DPA (dpa). Individual panicles were harvested at 16 dpa, temporarily placed in dry ice while other panicles were being harvested, and immediately stored in an ultra-low freezer. The panicles were quickly threshed in dry ice and the seeds were ground under cryogenic conditions maintained using liquid nitrogen. Grinding was individually done in a model no. MM400 (Retsch) with a cryo-kit using a 50-mL stainless steel grinding jar with a 25-mm grinding ball. Total RNA was extracted in a 2.0-mL RNAse-free tube from 200 mg pulverized seeds using 750 μL RNA extraction buffer (100 mm Tris-HCl pH 8.0, 150 mm LiCl, 50 mm EDTA and 1.5% SDS plus 1.5% β-mercaptoethanol freshly added before use) and 500 μL phenol:chloroform:isoamyl (25:24:1). RNA was precipitated from the supernatant at −20°C overnight using 700 μL isopropanol and 400 μL 1.2 m NaCl in a fresh 2-mL microfuge tube. The RNA pellet was resuspended in 450 µL RLT buffer (Qiagen) with 10% β-mercaptoethanol and further purified using Qiagen RNeasy Plant Mini Kit based on the manufacturer’s recommendation (Qiagen). Purified total RNA extract was treated with RNAse-free DNAse (Qiagen) to ensure that no residual genomic DNA was present in the sample. The quality and quantity of the total RNA extract were evaluated using Nanodrop (Thermo Scientific) and BioAnalyzer (Agilent Technologies). Only samples with an RNA integrity number of at least 7.0 and a concentration of 100 ng/µL were used. All RNA extracts were immediately stored in an ultra-low freezer until the microarray experiment.
A microarray experiment was conducted strictly following the One-Color Microarray-Based Gene Expression Analysis protocol using a custom 8 × 60 K microarray slide for rice (Agilent, Germany). Briefly, cDNA synthesis and cRNA labeling were done using a single-color Low Input Quick Amp Labeling Kit. Microarray hybridization was conducted in a SureHyb chamber assembly using the large-volume Hi-RPM Gene Expression Hybridization Kit. Hybridization was done in a hybridization oven set at 60°C rotating at 10 rpm for exactly 17 h. Up to four microarray slides were processed and washed per d using the Gene Expression Wash buffer Kit with 0.005% Triton X-102. Microarray slides with an ozone barrier slide cover were read by a SureScan Microarray Scanner controlled by Scan Control software (Agilent Technologies) using a scan resolution of 3 µm double pass. Raw data were generated from the TIFF file using the Feature Extraction software (Agilent Technologies). The quality of the data were judged per batch using the PDF QC report and as a group using QC validation software. The data were normalized using GeneSpring GX (Agilent Technologies) following the quantile normalization algorithm.
After QC validation, a total of 184 normalized transcriptome data with 59,732 probes were selected for further analysis. DEGs between haplotype 8 (high amylose) and haplotype 4 (glutinous) with the most contrasting samples were analyzed. The Limma R package was used following the empirical Bayes method to shrink the probewise sample variances toward a common value and to augment the degrees of freedom for the individual variances (Ritchie et al., 2015). The P values adjustment method used was either Bonferroni or Benjamini and Hochberg (1995). The top-ranked genes were selected having an adjusted P value below 0.05 and fold change above ±1 and a few genes known to be involved in starch biosynthesis were included with relaxed parameter to check their interaction in the network. A set containing 1242 DEGs were obtained (Supplemental Table S7). The weighted gene correlation network analysis method (Langfelder and Horvath, 2008) was used to identify clusters (modules) of densely connected correlated genes and to derive the coexpression networks describing the pairwise relationships (Pearson) among gene transcripts. The rationale behind the correlation network methodology is to use network language to explore the system-level functionality of genes. The 1242 DEGs with their expression along all 184 samples were selected and subjected to unsigned weighted gene coexpression analysis with PCC ≥ 0.75 and unsigned soft power of 4. A total of eight modules were obtained: turquoise, yellow, red, gray, green, brown, blue, and black. The most distinct expression pattern was observed in the turquoise module consisting of 441 nodes (Supplemental Table S8). This module also codes for most of the genes of interest identified from the GWAS results. Visualization of the coexpression network was done by using Cytoscape (Shannon et al., 2003). The nodes of interest were marked with a different color and central hub nodes with a bordered pattern. The log2-fold changes between the genes of haplotype 8 versus haplotype 4 were visualized using MapMan software (Thimm et al., 2004; Usadel et al., 2005). The genes were mapped with the metabolic and regulation pathways and their transcriptional responses were highlighted.
Cooked Grain Amylolysis
Cooked grain amylolysis by starch hydrolysis (Supplemental Fig. S6A) as a proxy measure for estimated glycemic score was determined using a modification of previous methods (Goñi et al., 1997; Dhital et al., 2015a, 2015b). Rate of digestibility using first-order kinetics was quantified using the k value employing the log-of-slope method (Butterworth et al., 2012). Selected haplotypes belonging to haplotypes 1, 4, and 8 underwent cooked grain amylolysis as a proxy measure for digestibility. Total starch was determined using the Megazyme Total Starch Assay Kit (Megazyme K-TSTA 07/11) adapted for microfuge screening. Absorbance was determined at 510 nm against a blank solution consisting of Milli-Q water (Millipore) and GOPOD. For each sample, 500 mg of rice granules (400 μm to 600 μm particle size) were cooked for 30 min in a boiling water bath using a 50-mL glass tube (Pyrex) based on the absorption cooking method normalized depending on amylose content (Butardo, 2011). After cooking, tubes were placed in a 37°C water bath over a stirring plate. Exactly 6 mL of water were added to dislodge the cooked sample. Samples were digested in 5 mL pepsin (cat. no. P-6887 from gastric porcine mucosa; Sigma-Aldrich) solution (1 mg/mL in 0.01 m HCl) for 30 min while stirring (350 rpm) with a 1/2 × 5/16” stir bar. The solution was neutralized with 5 mL of 0.02 m NaOH and 20 mL of 20 mm sodium acetate buffer (pH 6.0), followed by hydrolysis using pancreatin/amyloglucosidase mixture [2 mg/mL cat. no. P1750 pancreatin and 28 U/mL cat. no. 10115 amyloglucosidase (both Sigma-Aldrich) in 20 mm sodium acetate buffer, pH 6, with 4 mm CaCl2 and 0.49 mm MgCl2] at 37°C with stirring. At the desired time point (0, 5, 10, 20, 30, 45, 60, 90, 120, and 180 min), an aliquot was obtained and immediately immersed in an ice bath. This was centrifuged at 13,000 rpm, 4°C, to remove undigested starch. A 50-μL aliquot of the supernatant was reacted with 50 µL of 300 U/mL amyloglucosidase (Megazyme E-AMGDF 101006a) for 30 min and the Glc was quantified according to the Megazyme procedure (K-GLUC). Correlation of the k value was tested against percent amylose (PAM using the SEC method).
Rapid Visco Analysis
Selected rice grains with contrasting haplotypes also underwent rapid visco analysis to characterize the visco-elastic properties of flour using RVA-4 (Newport Scientific). Exactly 3.0 g of flour were placed in an RVA canister and mixed with 25.0 g of distilled deionized water weighed using an analytical balance. An RVA paddle was placed inside the canister to disperse the mixture. The prepared canister with paddle was inserted into the RVA machine and run immediately using a profile designed to mimic the old Brabender Amylograph. The protocol used in this study is approved as a standard method of the AACC (No. 61-02) and RACI (No. 06-05). Thermocline software (Newport Scientific) was used to operate the RVA machine and to generate and then analyze the results. The maximum viscosity during heating (peak viscosity), minimum viscosity after the peak (trough), final viscosity (FV), and their interrelationships were taken into account to provide insight into the pasting properties of each flour sample under study. Of particular importance in this study is the setback (FV ‒ peak viscosity) as it is commonly used as a proxy measure of the firmness of cooked rice, with higher values indicating firmer texture.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Amylose classification of indica diversity panel using iodine and SEC estimation methods.
Supplemental Table S2. List of most significant and linked (tag) SNPs detected on chromosomes 6 and 7 and their significance and allelic effects based on six fractions defined for debranched SEC. MS, most significant.
Supplemental Table S3. Novel SNPs (to our knowledge) identified at the structural gene of GBSSI using a subset of the 700 K SNP data.
Supplemental Table S4. SNPs and other structural variations in GBSSI previously reported in the literature. The major allelic variations are shaded in gray.
Supplemental Table S5. Identification of putative cis-regulatory elements binding to the 5′-promoter region of the structural gene of GBSSI using an on-line JASPAR 2016 Plant Database search tool (Mathelier et al., 2016).
Supplemental Table S6. Motif analysis for starch-related genes and bHLH done using locally installed Meme package (Bailey et al., 2009) with the JASPAR Plant Database (Mathelier et al., 2016).
Supplemental Table S7. List of differentially expressed genes identified from contrasting lines of haplotype 8 (high amylose) and haplotype 4 (waxy).
Supplemental Table S8. The turquoise module showing the most distinct expression pattern consisting of 441 nodes that code for most of the genes of interest.
Supplemental Figure S1. Genomewide association study for: A, AM1 (DP > 1000); B, AM2 (DP 1000-121); C, MCAP (DP 120-37); and D, SCAP (DP 6-36) as depicted by Manhattan (left) and quantile-quantile (right) plots.
Supplemental Figure S2. Mapping of the GWAS peak on chromosome 6 associated with debranched starch structure showing the linkage disequilibrium plot of the most significant SNPs associated with four subfractions of starch corresponding to (A) amylose and (B) amylopectin measured using the SEC method.
Supplemental Figure S3. Targeted haplotyping of waxy gene (LOC_Os06g04200.3) coding for GBSSI showing SNPs detected from the 700 K data (McCouch et al., 2016).
Supplemental Figure S4. Mapping of the GWAS peak on chromosome 7 associated with AM1 and SCAP fractions.
Supplemental Figure S5. Targeted haplotyping for gene coding for bHLH TF (LOC_Os07g11020) showing tagged SNPs detected from the 700 K SNP data (McCouch et al., 2016).
Supplementary Material
Acknowledgments
Markus Kuhlmann, Christianne Seiler, and Mandy Pueffeld (IPK Gatersleben, Germany) are acknowledged for hosting and assisting VMB in the conduct of the grain transcriptome experiment by microarray; Sushil Dhital (University of Queensland, Australia) and Peter Butterworth (Kings College London, UK) are acknowledged for their helpful suggestions and insights during the conduct of the cooked grain amylolysis experiment; and Bienvenido Juliano (PhilRice, Philippines) is acknowledged for helpful discussion on debranched starch structure. We also acknowledge the following IRRI staff for their helpful technical contributions during the conduct of experiments related to this research publication: Adoracion Resurreccion for supervising the generation of SEC data; Lila Molina for supervising the collection of AAC and RVA data; Lenie Quiatchon-Baeza, Artemio Madrid, Roldan Ilagan, and Ferdie Salisi for their assistance in maintaining and planting the diversity population in the field and in the greenhouse; and Dmytro Chebotarov and Millicent Sanciangco for their insights and discussion on GWAS.
Footnotes
Articles can be viewed without a subscription.
References
- Anacleto R, Cuevas RP, Jimenez R, Llorente C, Nissila E, Henry R, Sreenivasulu N (2015) Prospects of breeding high-quality rice using post-genomic tools. Theor Appl Genet 128: 1449–1466 [DOI] [PubMed] [Google Scholar]
- Ayres NM, McClung AM, Larkin PD, Bligh HFJ, Jones CA, Park WD (1997) Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germplasm. Theor Appl Genet 94: 773–781 [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SG, van de Wal M, Visser RGF (1998) Progress in understanding the biosynthesis of amylose. Trends Plant Sci 3: 462–467 [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265 [DOI] [PubMed] [Google Scholar]
- Basnet RK, Del Carpio DP, Xiao D, Bucher J, Jin M, Boyle K, Fobert P, Visser RGF, Maliepaard C, Bonnema G (2016) A systems genetics approach identifies gene regulatory networks associated with fatty acid composition in Brassica rapa seed. Plant Physiol 170: 568–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bertoft E. (2015) Fine structure of amylopectin. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer Japan, Tokyo, pp 3-40 [Google Scholar]
- Bligh HFJ, Till RI, Jones CA (1995) A microsatellite sequence closely linked to the Waxy gene of Oryza sativa. Euphytica 86: 83–85 [Google Scholar]
- Butardo VM (2011) Exploring rice diversity and biotechnology to develop grains with novel starch properties and altered digestibility. PhD thesis. University of Queensland, Brisbane, Queensland, Australia [Google Scholar]
- Butardo VM Jr, Daygon VD, Colgrave ML, Campbell PM, Resurreccion A, Cuevas RP, Jobling SA, Tetlow I, Rahman S, Morell M, Fitzgerald M (2012) Biomolecular analyses of starch and starch granule proteins in the high-amylose rice mutant Goami 2. J Agric Food Chem 60: 11576–11585 [DOI] [PubMed] [Google Scholar]
- Butardo VM Jr, Fitzgerald MA, Bird AR, Gidley MJ, Flanagan BM, Larroque O, Resurreccion AP, Laidlaw HK, Jobling SA, Morell MK, Rahman S (2011) Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J Exp Bot 62: 4927–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo VM, Sreenivasulu N (2015) Tailoring grain storage reserves for a healthier rice diet and its comparative status with other cereals. In International Review of Cell and Molecular Biology. Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- Butterworth PJ, Warren FJ, Grassby T, Patel H, Ellis PR (2012) Analysis of starch amylolysis using plots for first-order kinetics. Carbohydr Polym 87: 2189–2197 [Google Scholar]
- Cai XL, Wang ZY, Xing YY, Zhang J-L, Hong M-M (1998) Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J 14: 459–465 [DOI] [PubMed] [Google Scholar]
- Cao H, Wang J, Dong X, Han Y, Ma Q, Ding Y, Zhao F, Zhang J, Chen H, Xu Q, Xu J, Deng X (2015) Carotenoid accumulation affects redox status, starch metabolism, and flavonoid/anthocyanin accumulation in citrus. BMC Plant Biol 15: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao PJ, Bartley LE, Jung KH, Ronald PC (2008) Construction of a rice glycosyltransferase phylogenomic database and identification of rice-diverged glycosyltransferases. Mol Plant 1: 858–877 [DOI] [PubMed] [Google Scholar]
- Castro JV (2005) The molecular weight distribution of starch. PhD thesis. University of Sydney, Sydney, Australia [Google Scholar]
- Castro JV, Dumas C, Chiou H, Fitzgerald MA, Gilbert RG (2005a) Mechanistic information from analysis of molecular weight distributions of starch. Biomacromolecules 6: 2248–2259 [DOI] [PubMed] [Google Scholar]
- Castro JV, Ward RM, Gilbert RG, Fitzgerald MA (2005b) Measurement of the molecular weight distribution of debranched starch. Biomacromolecules 6: 2260–2270 [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K, Clarke B, Hylton C, Tatge H, Smith AM (1996) The elongation of amylose and amylopectin chains in isolated starch granules. Plant J 10: 1135–1143 [Google Scholar]
- Denyer K, Waite D, Motawia S, Møller BL, Smith AM (1999) Granule-bound starch synthase I in isolated starch granules elongates malto-oligosaccharides processively. Biochem J 340: 183–191 [PMC free article] [PubMed] [Google Scholar]
- Dhital S, Butardo VM Jr, Jobling SA, Gidley MJ (2015a) Rice starch granule amylolysis--differentiating effects of particle size, morphology, thermal properties and crystalline polymorph. Carbohydr Polym 115: 305–316 [DOI] [PubMed] [Google Scholar]
- Dhital S, Dabit L, Zhang B, Flanagan B, Shrestha AK (2015b) In vitro digestibility and physicochemical properties of milled rice. Food Chem 172: 757–765 [DOI] [PubMed] [Google Scholar]
- Dobo M, Ayres N, Walker G, Park WD (2010) Polymorphism in the GBSS gene affects amylose content in US and European rice germplasm. J Cereal Sci 52: 450–456 [Google Scholar]
- Fitzgerald MA, Bergman CJ, Resurreccion AP, Moller J, Jimenez R, Reinke RF, Martin M, Blanco P, Molina F, Chen M-H, Kuri V, Romero MV, et al. (2009) Addressing the dilemmas of measuring amylose in rice. Cereal Chem 86: 492–498 [Google Scholar]
- Fitzgerald MA, Rahman S, Resurreccion AP, Concepcion JC, Daygon VD, Dipti SS, Kabir KA, Klingner B, Morell MK, Bird AR (2011) Identification of a major genetic determinant of glycaemic index in rice. Rice (N Y) 4: 66–74 [Google Scholar]
- Frances H, Bligh J, Larkin PD, Roach PS, Jones CA, Fu H, Park WD (1998) Use of alternate splice sites in granule-bound starch synthase mRNA from low-amylose rice varieties. Plant Mol Biol 38: 407–415 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Maekawa M, Oki T, Suda I, Iida S, Shimada H, Takamure I, Kadowaki K (2007) The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J 49: 91–102 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229 [DOI] [PubMed] [Google Scholar]
- Goñi I, Garcia-Alonso A, Saura-Calixto F (1997) A starch hydrolysis procedure to estimate glycemic index. Nutr Res 17: 427–437 [Google Scholar]
- Gross BL, Reagon M, Hsu S-C, Caicedo AL, Jia Y, Olsen KM (2010) Seeing red: the origin of grain pigmentation in US weedy rice. Mol Ecol 19: 3380–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, de Geus EJC, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, et al. (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48: 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashiro I. (2015) Fine structure of amylose. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer, Tokyo, Japan, pp 41-60 [Google Scholar]
- Hanashiro I, Higuchi T, Aihara S, Nakamura Y, Fujita N (2011) Structures of starches from rice mutants deficient in the starch synthase isozyme SSI or SSIIIa. Biomacromolecules 12: 1621–1628 [DOI] [PubMed] [Google Scholar]
- Hanashiro I, Itoh K, Kuratomi Y, Yamazaki M, Igarashi T, Matsugasako J, Takeda Y (2008) Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiol 49: 925–933 [DOI] [PubMed] [Google Scholar]
- Hernández JM, Gaborieau M, Castignolles P, Gidley MJ, Myers AM, Gilbert RG (2008) Mechanistic investigation of a starch-branching enzyme using hydrodynamic volume SEC analysis. Biomacromolecules 9: 954–965 [DOI] [PubMed] [Google Scholar]
- Hirano HY, Eiguchi M, Sano Y (1998) A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Mol Biol Evol 15: 978–987 [DOI] [PubMed] [Google Scholar]
- Hogg AC, Gause K, Hofer P, Martin JM, Graybosch RA, Hansen LE, Giroux MJ (2013) Creation of a high-amylose durum wheat through mutagenesis of starch synthase II (SSIIa). J Cereal Sci 57: 377–383 [Google Scholar]
- House MA, Griswold CK, Lukens LN (2014) Evidence for selection on gene expression in cultivated rice (Oryza sativa). Mol Biol Evol 31: 1514–1525 [DOI] [PubMed] [Google Scholar]
- Hu EA, Pan A, Malik V, Sun Q (2012) White rice consumption and risk of type 2 diabetes: meta-analysis and systematic review. BMJ 344: e1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, et al. (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42: 961–967 [DOI] [PubMed] [Google Scholar]
- Hur SJ, Lim BO, Decker EA, McClements DJ (2011) In vitro human digestion models for food applications. Food Chem 125: 1–12 [Google Scholar]
- Isshiki M, Morino K, Nakajima M, Okagaki RJ, Wessler SR, Izawa T, Shimamoto K (1998) A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J 15: 133–138 [DOI] [PubMed] [Google Scholar]
- Jagadish SVK, Muthurajan R, Rang ZW, Malo R, Heuer S, Bennett J, Craufurd PQ (2011) Spikelet proteomic response to combined water deficit and heat stress in rice (Oryza sativa cv. N22). Rice (N Y) 4: 1–11 [Google Scholar]
- Juliano BO. (1979) The chemical basis of rice grain quality. In Workshop on Chemical Aspects of Rice Grain Quality. International Rice Research Institute, Los Banos, Laguna [Google Scholar]
- Juliano BO, Perez CM, Blakeney AB, Castillo T, Kongseree N, Laignelet B, Lapis ET, Murty VVS, Paule CM, Webb BD (1981) International cooperative testing on the amylose content of milled rice. Starch 33: 157–162 [Google Scholar]
- Juliano BO, Perez CM, Kaushik R, Khush GS (1990) Some grain properties of IR36-based starch mutants. Starke 42: 256–260 [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42: 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharabian-Masouleh A, Waters DLE, Reinke RF, Henry RJ (2011) Discovery of polymorphisms in starch-related genes in rice germplasm by amplification of pooled DNA and deeply parallel sequencing. Plant Biotechnol J 9: 1074–1085 [DOI] [PubMed] [Google Scholar]
- Kharabian-Masouleh A, Waters DLE, Reinke RF, Ward R, Henry RJ (2012) SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Sci Rep 2: 557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld SMJ, Vennik M, Kottenhagen M, van Wijk R, Buijk A, Kijne JW, de Pater S (2002) Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Physiol 129: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin PD, Park WD (1999) Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol Biol 40: 719–727 [DOI] [PubMed] [Google Scholar]
- Larkin PD, Park WD (2003) Association of waxy gene single nucleotide polymorphisms with starch characteristics in rice (Oryza sativa L.). Mol Breed 12: 335–339 [Google Scholar]
- Lehmann U, Robin F (2007) Slowly digestible starch—its structure and health implications: a review. Trends Food Sci Technol 18: 346–355 [Google Scholar]
- Lohmeier-Vogel EM, Kerk D, Nimick M, Wrobel S, Vickerman L, Muench DG, Moorhead GB (2008) Arabidopsis At5g39790 encodes a chloroplast-localized, carbohydrate-binding, coiled-coil domain-containing putative scaffold protein. BMC Plant Biol 8: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42: D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourdin D, Putaux J-L, Potocki-Véronèse G, Chevigny C, Rolland-Sabaté A, Buléon A (2015) Crystalline structure in starch. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer, Tokyo, Japan, pp 61–90 [Google Scholar]
- MacFarlane GT, Macfarlane S, Gibson GR (1998) Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35: 180–187 [DOI] [PubMed] [Google Scholar]
- Man J, Yang Y, Huang J, Zhang C, Chen Y, Wang Y, Gu M, Liu Q, Wei C (2013) Effect of simultaneous inhibition of starch branching enzymes I and IIb on the crystalline structure of rice starches with different amylose contents. J Agric Food Chem 61: 9930–9937 [DOI] [PubMed] [Google Scholar]
- Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, et al. (2016) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44(D1): D110–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch SR, Wright MH, Tung C-W, Maron LG, McNally KL, Fitzgerald M, Singh N, DeClerck G, Agosto-Perez F, Korniliev P, Greenberg AJ, Naredo MEB, et al. (2016) Open access resources for genome-wide association mapping in rice. Nat Commun 7, doi:10.1038/ncoms10532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, Stokowski R, Ballinger DG, et al. (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. USA 106: 12273–12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami I, Uwatoko N, Ikeda Y, Yamaguchi J, Hirano H-Y, Suzuki Y, Sano Y (2008) Allelic diversification at the wx locus in landraces of Asian rice. Theor Appl Genet 116: 979–989 [DOI] [PubMed] [Google Scholar]
- Morell MK, Kosar-Hashemi B, Cmiel M, Samuel MS, Chandler P, Rahman S, Buleon A, Batey IL, Li Z (2003) Barley sex6 mutants lack starch synthase IIa activity and contain a starch with novel properties. Plant J 34: 173–185 [DOI] [PubMed] [Google Scholar]
- Nakamura Y. (2015) Biosynthesis of reserve starch. In Nakamura Y, ed, Starch: Metabolism and Structure. Springer Japan, Tokyo, pp 161–209 [Google Scholar]
- Nakamura Y, Francisco PB Jr, Hosaka Y, Sato A, Sawada T, Kubo A, Fujita N (2005) Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol Biol 58: 213–227 [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H (2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD (2006) Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics 173: 975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou JZ, Yao CK, Rotbart A, Muir JG, Gibson PR, Kalantar-zadeh K (2015) Human intestinal gas measurement systems: in vitro fermentation and gas capsules. Trends Biotechnol 33: 208–213 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, Orvis J, Haas B, et al. (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payyavula RS, Singh RK, Navarre DA (2013) Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J Exp Bot 64: 5115–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS (2012) Glycogen and its metabolism: some new developments and old themes. Biochem J 441: 763–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu APS, Randhawa GS, Dhugga KS (2009) Plant cell wall matrix polysaccharide biosynthesis. Mol Plant 2: 840–850 [DOI] [PubMed] [Google Scholar]
- Sano Y. (1984) Differential regulation of waxy gene expression in rice endosperm. Theor Appl Genet 68: 467–473 [DOI] [PubMed] [Google Scholar]
- Seung D, Soyk S, Coiro M, Maier BA, Eicke S, Zeeman SC (2015) PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Tada Y, Kawasaki T, Fujimura T (1993) Antisense regulation of the rice waxy gene expression using a PCR-amplified fragment of the rice genome reduces the amylose content in grain starch. Theor Appl Genet 86: 665–672 [DOI] [PubMed] [Google Scholar]
- Shu X, Rasmussen SK (2014) Quantification of amylose, amylopectin, and β-glucan in search for genes controlling the three major quality traits in barley by genome-wide association studies. Front Plant Sci 5: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Denyer K (2001) The synthesis of amylose. In Barsby TL, Donald AM, Frazier PJ, eds, Starch: Advances in Structure and Function. The Royal Society of Chemistry, London, UK, pp 150–163 [Google Scholar]
- Smythe C, Cohen P (1991) The discovery of glycogenin and the priming mechanism for glycogen biogenesis. Eur J Biochem 200: 625–631 [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strocchi A, Levitt MD (1991) Measurement of starch absorption in humans. Can J Physiol Pharmacol 69: 108–110 [DOI] [PubMed] [Google Scholar]
- Sweeney M, McCouch S (2007) The complex history of the domestication of rice. Ann Bot (Lond) 100: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner SA, Zihler Berner A, Rigozzi E, Grattepanche F, Chassard C, Lacroix C (2014) In vitro continuous fermentation model (PolyFermS) of the swine proximal colon for simultaneous testing on the same gut microbiota. PLoS One 9: e94123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 3000 Rice Genomes Project (2014) The 3000 Rice Genomes Project. Gigascience 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Topping D. (2007) Cereal complex carbohydrates and their contribution to human health. J Cereal Sci 46: 220–229 [Google Scholar]
- Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet 104: 1–8 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte GE, Delcour JA (2004) From sucrose to starch granule to starch physical behaviour: a focus on rice starch. Carbohydr Polym 58: 245–266 [Google Scholar]
- Wanchana S, Toojinda T, Tragoonrung S, Vanavichit A (2003) Duplicated coding sequence in the waxy allele of tropical glutinous rice (Oryza sativa L.). Plant Sci 165: 1193–1199 [Google Scholar]
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Zheng F-Q, Shen G-Z, Gao J-P, Snustad DP, Li M-G, Zhang J-L, Hong M-M (1995) The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J 7: 613–622 [DOI] [PubMed] [Google Scholar]
- Ward RM, Gao Q, de Bruyn H, Gilbert RG, Fitzgerald MA (2006) Improved methods for the structural analysis of the amylose-rich fraction from rice flour. Biomacromolecules 7: 866–876 [DOI] [PubMed] [Google Scholar]
- Wen W, Liu H, Zhou Y, Jin M, Yang N, Li D, Luo J, Xiao Y, Pan Q, Tohge T, Fernie AR, Yan J (2016) Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiol 170: 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolever TMS, Katzmanrelle L, Jenkins AL, Vuksan V, Josse RG, Jenkins DJA (1994) Glycemic index of 102 complex carbohydrate foods in patients with diabetes. Nutr Res 14: 651–669 [Google Scholar]
- Woolnough JW, Monro JA, Brennan CS, Bird AR (2008) Simulating human carbohydrate digestion in vitro: a review of methods and the need for standardisation. Int J Food Sci Technol 43: 2245–2256 [Google Scholar]
- Yano M, Okuno K, Kawakami J, Satoh H, Omura T (1985) High amylose mutants of rice, Oryza sativa L. Theor Appl Genet 69: 253–257 [DOI] [PubMed] [Google Scholar]
- Yin Y, Mohnen D, Gelineo-Albersheim I, Xu Y, Hahn MG (2010) Glycosyltransferases of the GT8 Family. In Annual Plant Reviews. Wiley-Blackwell, Hoboken, NJ, pp 167–211 [Google Scholar]
- Yokoyama WH. (2004) Nutritional properties of rice and rice bran. In Champagne ET, ed, Rice Chemistry and Technology. American Association of Cereal Chemists, Minneapolis, MN, pp 595–606 [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2002) The priming of amylose synthesis in Arabidopsis leaves. Plant Physiol 128: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Ye N, Yang J, Peng X, Zhang J (2011) Regulation of expression of starch synthesis genes by ethylene and ABA in relation to the development of rice inferior and superior spikelets. J Exp Bot 62: 3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Gu M, Meng X, Cheung SCK, Yu H, Huang J, Sun Y, Shi Y, Liu Q (2012) High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol J 10: 353–362 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cai XL, Wang ZY, Hong MM (2003) An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem 278: 47803–47811 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.