CmOr golden SNP dramatically affects carotenoid content and plastid fate in melon by inhibiting metabolism downstream of β-carotene.
Abstract
β-Carotene adds nutritious value and determines the color of many fruits, including melon (Cucumis melo). In melon mesocarp, β-carotene accumulation is governed by the Orange gene (CmOr) golden single-nucleotide polymorphism (SNP) through a yet to be discovered mechanism. In Arabidopsis (Arabidopsis thaliana), OR increases carotenoid levels by posttranscriptionally regulating phytoene synthase (PSY). Here, we identified a CmOr nonsense mutation (Cmor-lowβ) that lowered fruit β-carotene levels with impaired chromoplast biogenesis. Cmor-lowβ exerted a minimal effect on PSY transcripts but dramatically decreased PSY protein levels and enzymatic activity, leading to reduced carotenoid metabolic flux and accumulation. However, the golden SNP was discovered to not affect PSY protein levels and carotenoid metabolic flux in melon fruit, as shown by carotenoid and immunoblot analyses of selected melon genotypes and by using chemical pathway inhibitors. The high β-carotene accumulation in golden SNP melons was found to be due to a reduced further metabolism of β-carotene. This was revealed by genetic studies with double mutants including carotenoid isomerase (yofi), a carotenoid-isomerase nonsense mutant, which arrests the turnover of prolycopene. The yofi F2 segregants accumulated prolycopene independently of the golden SNP. Moreover, Cmor-lowβ was found to inhibit chromoplast formation and chloroplast disintegration in fruits from 30 d after anthesis until ripening, suggesting that CmOr regulates the chloroplast-to-chromoplast transition. Taken together, our results demonstrate that CmOr is required to achieve PSY protein levels to maintain carotenoid biosynthesis metabolic flux but that the mechanism of the CmOr golden SNP involves an inhibited metabolism downstream of β-carotene to dramatically affect both carotenoid content and plastid fate.
β-Carotene, a C-40 isoprenoid molecule, is the major source of vitamin A in the human diet (Maiani et al., 2009). Lack of dietary vitamin A is a common cause of premature death and child blindness in developing countries. β-Carotene is abundant in diverse edible plant tissues such as pumpkin (Cucurbita pepo) fruits, sweet potato (Ipomoea batatas) tubers, carrot (Daucus carota) roots, kale (Brassica oleracea) leaves, mango (Mangifera indica), and orange-fleshed melon (Cucumis melo) fruits (Howitt and Pogson, 2006; Yuan et al., 2015b). In green tissues, two β-carotene molecules are located in each PSII reaction center to facilitate electron transfer and to provide photoprotection by quenching singlet oxygen products (Telfer, 2002). In many fruits and flowers, β-carotene serves as a yellow-orange colorant and as a precursor for aromatic molecules to attract pollinators and seed dispersers (Walter and Strack, 2011).
β-Carotene is a metabolite in the carotenoid biosynthesis pathway that receives its precursors from the plastid-localized methyl-erythritol phosphate pathway. Carotenoid biosynthesis starts with the condensation of two geranylgeranyl diphosphate molecules, yielding the colorless carotene phytoene in a reaction catalyzed by phytoene synthase (PSY; Nisar et al., 2015). Two desaturases, phytoene desaturase (PDS) and ζ-carotene desaturase, introduce four double bonds into phytoene to convert phytoene into lycopene. The cis-configured carotene double bonds are isomerized to the trans-configuration by two enzymes, ζ-carotene isomerase (Z-ISO) and carotenoid isomerase (CRTISO). Z-ISO isomerizes the 15-cis-double bond in 9,15,9′-tri-cis-ζ-carotene, a reaction that can also be catalyzed by light in green tissues (Chen et al., 2010). CRTISO isomerizes all four cis-double bonds in prolycopene (7,9,7′,9′-tetra-cis-lycopene). These isomerizations yield all-trans-lycopene, the substrate for lycopene β- and ε-ring cyclases (Isaacson et al., 2002). CRTISO deficiency results in an arrest of pathway flux in tissues without a functional photosynthetic apparatus; for instance, CRTISO mutant tomato (Solanum lycopersicum) or melon fruits accumulate mainly prolycopene (Isaacson et al., 2002; Galpaz et al., 2013).
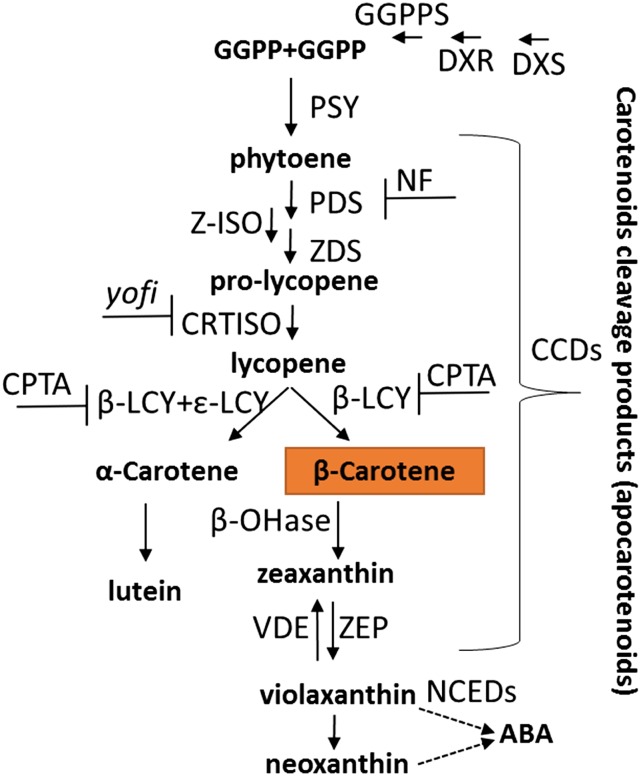
All-trans-lycopene can be cyclized twice by lycopene β-cyclase (β-LCY) into β-carotene or, alternatively, by β-LCY and lycopene ε-cyclase (ε-LCY) into α-carotene. β-Carotene is further hydroxylized and epoxidized to form xanthophylls. β-Carotene and other carotenoids also serve as substrates for various carotenoid cleavage dioxygenases to produce apocarotenoids, including the phytohormone abscisic acid and strigolactones, and volatiles and scent molecules (Nisar et al., 2015; Yuan et al., 2015b; Fig. 1)
Figure 1.
Schematic biosynthetic pathway of carotenoids and apocarotenoids. Uppercase abbreviations are enzymes. Carotenoid metabolites are in lowercase boldface. Dashed arrows represent series of enzymatic reactions. Bar-headed lines represent enzyme inhibitors [norflurazon (NF) and 2-(4-chlorophenylthio) triethylamine hydrochloride (CPTA)] or mutant name (yofi). ABA, Abscisic acid; CCDs, carotenoid cleavage dioxygenases; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; GGPP, geranylgeranyl diphosphate; GGPPS, geranylgeranyl pyrophosphate synthase; NCED, 9-cis-epoxycarotenoid dioxygenase; β-OHase, β-carotene hydroxylase; VDE, violaxanthin deepoxidase; ZEP, zeaxanthin epoxidase.
Melon (Cucurbitaceae) is an important crop with an estimated annual yield of more than 32 million tons worldwide (http://faostat3.fao.org). Fruit flesh (mesocarp) color is an important commercial trait and can be categorized into three distinctive phenotypes: white, green, and orange (Chayut et al., 2015). The orange phenotype is a result of massive β-carotene accumulation in the fruit mesocarp governed by the golden single-nucleotide polymorphism (SNP) of the Orange (CmOr) gene (Tzuri et al., 2015). Melon plants carrying a dominant CmOr allele bear orange-fleshed fruits, while homozygous recessive Cmor plants bear white- or green-fleshed fruits. The golden SNP transversion alters Arg to His at position 108; thus, the recessive and dominant alleles will be termed hereafter Cmor-Arg and CmOr-His, respectively. CmOr natural allelic variation has no effect on the transcript levels of carotenogenesis genes (Chayut et al., 2015), and its mechanism of action in melon fruit is still largely unknown.
The Or gene was discovered originally in cauliflower (Brassica oleracea botrytis; BoOr) and shown to enhance sink strength by triggering the biogenesis of chromoplasts in nongreen tissues (Lu et al., 2006). Carotenoid biosynthetic gene expression was not changed by the BoOr mutation (Li et al., 2001). The OR protein is targeted to the plastid and contains a Cys-rich zinc finger domain that is found in DnaJ cochaperones. However, OR lacks the J domain that defines this class of proteins (Giuliano and Diretto, 2007). In Arabidopsis (Arabidopsis thaliana), OR proteins were shown to interact directly and posttranscriptionally regulate PSY in controlling carotenoid biosynthesis (Zhou et al., 2015). Overexpression of the Arabidopsis OR protein mutagenized in the corresponding site of the melon golden SNP (altering Arg to His) resulted in high β-carotene levels (Yuan et al., 2015a). The high β-carotene accumulation in these cases has been associated with chromoplast biogenesis (Lu et al., 2006; Yuan et al., 2015a). However, how OR regulates various processes in ultimately affecting carotenoid accumulation remains elusive.
In this work, we isolated an ethyl methanesulfonate (EMS)-induced nonsense mutation in CmOr (low-β) that dramatically lowered β-carotene level in melon fruit mesocarp. By comparing low-β with its isogenic progenitor, we show that an active CmOr is required to maintain PSY protein level and activity in developing melon fruit mesocarp. However, by examining various melon accessions differing in the golden SNP, we found that regulation of PSY level and activity is not the mechanism by which CmOr natural allelic variation governs fruit β-carotene levels in melon. Our biochemical inhibition and genetic studies show that the golden SNP variation does not change the β-carotene synthesis rate. Chemical inhibition and genetic arrest of the carotenogenic flux resulted in the accumulation of carotenoids, independently of the golden SNP. Thus, we suggest that CmOr-His stabilizes β-carotene and inhibits its turnover, resulting in β-carotene accumulation and chromoplast formation potentially as a metabolite-derived process.
RESULTS
Isolation of an EMS-Induced low-β Mutant and Its Fruit Phenotype Analysis
To further study the molecular role of CmOr in melon fruit, we screened M2 families of the previously described orange-fleshed CEZ-based EMS library for fruit color changes (Tadmor et al., 2007; Galpaz et al., 2013). Twelve plants from each of 1,000 M2 families were grown in the field, and the mature fruit were phenotyped visually. The M2 family cem1697 showed lower flesh color intensity that was inherited recessively monogenic, as concluded from segregation analyses. One pale-fleshed individual of cem1697 was backcrossed to the parent line CEZ and then self-pollinated followed by selection of a green-fruited individual, which was self-pollinated for an additional two generations. The line obtained was isogenic to CEZ but with green-fleshed fruits and was named low-β (Fig. 2A).
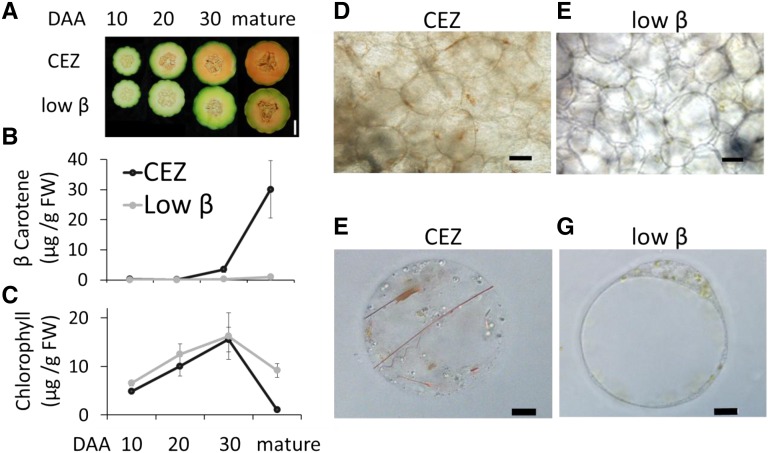
Figure 2.
Phenotypes of low-β mutant and CEZ fruits. A, Cross sections of low-β and CEZ fruits at four developmental stages: 10, 20, and 30 d after anthesis (DAA) and mature stage. Bar = 5 cm. B, Fruit flesh β-carotene levels in the four fruit developmental stages shown in A. Data are means ± sd of three biological replicates (different fruits from different plants). FW, Fresh weight. C, Total fruit chlorophyll a and b levels during fruit development. D and E, Bright visible light microscopy of mature fruit mesocarp cells of CEZ (D) and low-β (E). Bars = 100 μm. F and G, Bright-field light microscopy images of mature CEZ protoplast (F) and mature low-β protoplast (G). Bars = 10 μm.
Comparative visualization of over 100 low-β and CEZ plants under commercial field and greenhouse conditions did not reveal any phenotypic variation except the modified fruit flesh color. The first slightly visible color difference between low-β and CEZ fruit appeared at 20 DAA (Fig. 2A). The color difference became most notable at 30 DAA; CEZ flesh turned orange, while low-β flesh remained green. When the fruit matured (40–42 DAA), CEZ flesh became uniformly deep orange whereas the mutant flesh was green with pale orange tissue adjacent to the seed cavity (Fig. 2A).
We measured flesh carotenoid concentration in CEZ and low-β developing fruit by HPLC. The β-carotene level was 11.7- and 30.1-fold lower at 30 DAA and at the mature stage, respectively, in the flesh of the mutant compared with CEZ (Fig. 2B; Supplemental Fig. S1). Other carotenoids were detected at low levels. Lutein was found only in low-β mature fruits but not in CEZ, while α-carotene was present only in CEZ but not in low-β (Supplemental Fig. S1). During CEZ fruit development from 10 to 30 DAA, total chlorophyll levels increased 3.2-fold and then almost completely disappeared at the mature stage. During early low-β fruit development, the total chlorophyll levels increased similarly until 30 DAA. However, unlike CEZ, low-β chlorophyll levels declined only mildly at the mature stage (Fig. 2C).
Light microscopy of thin cross sections of mature CEZ fruit mesocarp revealed the existence of one or two orange bodies, probably chromoplasts, per cell (Fig. 2D). In contrast, most low-β mesocarp cells were colorless or spotted with a small number of green chloroplasts, which were usually concentrated in a single area (Fig. 2E). In protoplasts isolated from CEZ fruit mesocarp, orange β-carotene structures were sometimes seen inside chromoplasts (Fig. 2F; Supplemental Fig. S2, A and B), but often as large floating crystals with lengths of up to the entire protoplast diameter (Fig. 2F; Supplemental Fig. S2C). In contrast, isolated protoplasts of low-β mesocarp cells contained chloroplasts but did not contain chromoplasts and visible carotenoid crystals (Fig. 2G; Supplemental Fig. S2, E and F). The crystal shapes found in CEZ fruit were diverse, as fine rods (Fig. 2F; Supplemental Fig. S2G), thin plane polygons (Supplemental Fig. S2G), spiral threads (Fig. 2F), or shapeless aggregates (Supplemental Fig. S2, C and H). Most of the low-β protoplasts contained up to 20 chloroplasts (Fig. 2G), seldom up to approximately 100 chloroplasts in one protoplast (Supplemental Fig. S2, E and F). Chlorophyll autofluorescence was never observed in CEZ protoplasts of mature fruit mesocarp cells, indicating the complete absence of chloroplasts in these cells (Supplemental Fig. S2D). Protoplast sizes varied tremendously in both lines, ranging from approximately 25 µm to approximately 85 µm in diameter (Supplemental Fig. S2, B and I). In summary, the microscopic analysis indicated that low-β mature fruit mesocarp cells contained chloroplasts and lacked free β-carotene crystal structures, as opposed to CEZ mature fruit mesocarp cells.
low-β Carries a Mutation in the CmOr Gene
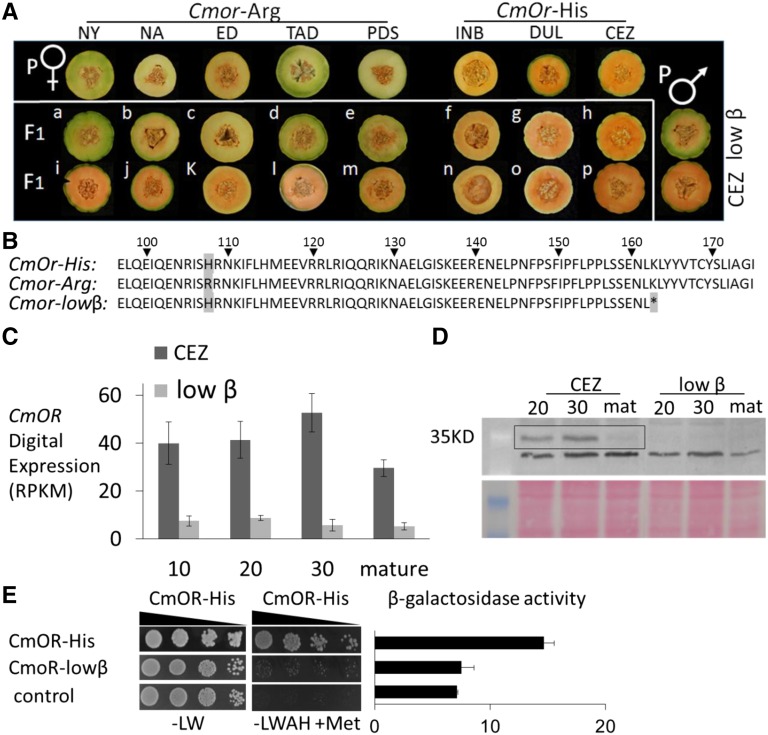
Based on the phenotypic similarity with the recessive homozygous Cmor-Arg melon fruits (Tzuri et al., 2015), we hypothesized that low-β was mutated either in the CmOr gene or alternatively in a structural gene of the carotenoid metabolic pathway. To differentiate between these two options, we conducted a genetic complementation test. We crossed low-β as well as its parental line CEZ (homozygous for CmOr-His) with a panel of nonorange lines (NY, NA, ED, TAD, and PDS, all homozygous for Cmor-Arg; Table I). As a control, crossings also were made with CmOr-His homozygous orange lines (INB, DUL, and CEZ; Table I). We visually phenotyped the color of the hybrid fruit flesh. Complementation of the low-β phenotype through a cross with a nonorange line would indicate that the low-β mutation is not allelic to CmOr. In all crosses with CEZ, the F1 hybrid showed the expected orange-flesh phenotype (Fig. 3A, i–p). Similarly, when low-β was crossed with homozygous dominant CmOr-His lines, the F1 also was orange fleshed (Fig. 3A, f–h). However, in all crosses between low-β and homozygous Cmor-Arg lines, the F1 had a nonorange flesh color, indicating that the low-β mutation was not complemented (Fig. 3A, a–e). These results strongly suggested that the low-β phenotype was caused by a mutation at the CmOr locus, a presumably third CmOr allele hereby designated Cmor-lowβ.
Table I. Accession full names and genetic backgrounds.
| Variety Full Name | Variety Short Name | Marketing Type | Taxonomic Groupa | CmOr Allele | Mature Fruit Flesh Color |
|---|---|---|---|---|---|
| CEZ | CEZ | Charentais | Cantalupensis | CmOr-His | Orange |
| Low β-Carotene | low-β | Charentais | Cantalupensis | CmOr-lowβ | Green |
| Dulce | DUL | Cantaloupe | Reticulatus | CmOr-His | Orange |
| Tam-Dew | TAD | Honey Dew | Inodorus | Cmor-Arg | Green |
| Noy Amid | NA | Yellow Canary | Inodorus | Cmor-Arg | Green-white |
| Noy Yizre'el | NY | Ha’Ogen | Cantalupensis | Cmor-Arg | Green |
| Vedrantais | VEP | Charentais | Cantalupensis | CmOr-His | Orange |
| Indian Best | INB | –b | Chandalc | CmOr-His | Light orange |
| Ein Dor | ED | Ananas | Reticulatus | Cmor-Arg | Cream-white |
| Piel De Sapo | PDS | Piel de Sapo | Inodorus | Cmor-Arg | Green-white |
| Yellow Orange Flesh | yofi | Charentais | Cantalupensis | CmOr-His | Yellow |
After Pitrat (2000).
INB has no marketing type.
Figure 3.
Comparisons of low-β with CEZ. A, Complementation test of low-β and CEZ with a panel of Cmor-Arg homozygous lines: NY, NA, ED, TAD, and PDS. The CmOr-His homozygous lines INB, DUL, and CEZ served as controls. Images of the female parent fruits are in the top row. The male parent was either low-β or CEZ (fruits shown at right). F1 hybrid fruits of the crosses with low-β (a–h) and with CEZ (i–p) are shown. In a to e, nonorange fruits in hybrids indicated that low-β is allelic to CmOr. B, Aligned partial protein sequences of CmOR-His (CEZ) and CmOR-Arg (TAD) and the low-β OR protein with its premature stop codon (asterisk) at position 163. The golden SNP (His to Arg) at position 108 is highlighted. C, CmOr mRNA digital expression in CEZ and low-β developing fruit mesocarp. Columns represent average reads per kilobase of exon model per million mapped reads (RPKM) ± se of three independent biological repeats. D, CmOR protein levels in CEZ and low-β mutant at 20 and 30 DAA and in mature melon fruit. CmOR protein was detected by immunoblot analysis. A Ponceau S-stained membrane is shown below. E, Yeast-split ubiquitin analysis (-LW = selective medium lacking Leu and Trp; -LWAH+Met = selective medium lacking Leu, Trp, adenine, and His and supplemented with 150 μm Met).
The low-β Mutation Leads to Reduced CmOr Transcript and CmOR Protein Levels
Sequence analysis of the CmOr cDNA in low-β revealed a transversion of Ala-487 to Thr, which altered CmOR Lys-163 to a stop codon (AAG to TAG; Fig. 3B; the full coding sequence is shown in Supplemental Fig. S3). CmOr mRNA levels were found to be significantly lower in low-β compared with CEZ at all the tested fruit developmental stages (Fig. 3C). We compared CmOR protein levels by western-blot analysis and used high-density (15%) separating gels in order to include the predicted truncated CmOR with an expected size of 18 kD (Supplemental Fig. S4). CmOR was not detected in any fruit developmental stage of low-β, while in CEZ, CmOR-His was more abundant at 20 and 30 DAA than at the mature stage (Fig. 3D), correlating with the CmOr expression pattern (Fig. 3C).
The N-terminal region of OR is required for interaction with PSY, while the C-terminal region of OR is crucial for dimerization (Zhou et al., 2015). Therefore, we analyzed whether the CmOR truncation in low-β affected the capacity to form dimers and to interact with PSY. For these purposes, we used the yeast split-ubiquitin system and coexpressed proteins fused to either the N-terminal or C-terminal ubiquitin moiety. An interaction-growth test and quantification of the reporter gene lacZ activity confirmed that CEZ CmOR-His formed homodimers, consistent with previous findings (Zhou et al., 2015; Fig. 3E). However, CmOR-lowβ lost its ability to form homodimers (Fig. 3E), while the N-terminal 162 amino acids were sufficient to interact with PSY similarly to CmOR-His (Supplemental Fig. S5).
The low-β Mutation Dramatically Reduces PSY Protein Level
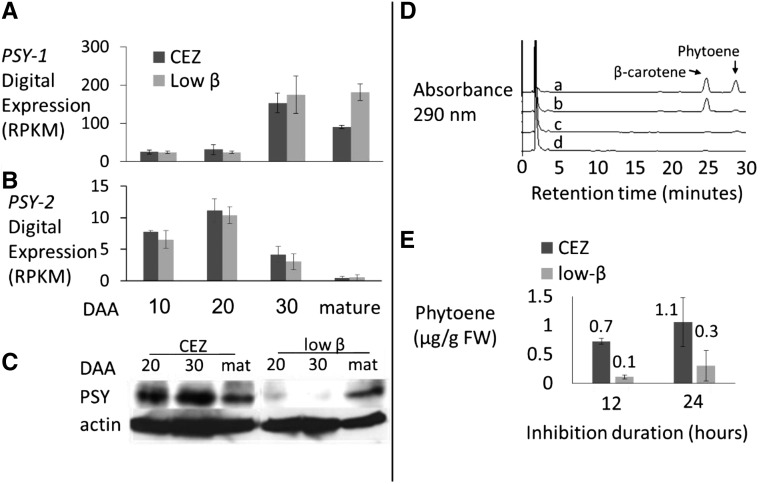
While β-carotene levels were significantly lower in low-β fruits compared with CEZ, our fruit transcriptome analysis revealed that the carotenoid biosynthetic pathway genes were expressed similarly at 10, 20, and 30 DAA (Supplemental Table S1). However, in the mature low-β fruit, transcripts of several genes of the methyl-erythritol phosphate and carotenoid pathways were up-regulated compared with CEZ (Supplemental Fig. S6). These genes included 1-deoxy-d-xylulose-5-phosphate synthase, geranylgeranyl diphosphate synthase, and PSY-1. PSY-1 is one of the three PSY paralogs in the melon genome, which was shown to correlate with fruit carotenoid accumulation (Qin et al., 2011). This was confirmed by our transcriptome analysis (Fig. 4A). PSY-2 expression was much lower than PSY-1 and showed a different pattern during fruit ripening (Fig. 4B).
Figure 4.
PSY mRNA expression, protein levels, and enzymatic activity analyses in CEZ and low-β fruits. A and B, Digital expression of PSY-1 (A) and PSY-2 (B) in CEZ and low-β developing fruits. Average RPKM values ± se of three biological repeats are shown. C, Western blot showing PSY protein levels at 20 and 30 DAA and in the mature fruit of CEZ and low-β; proteins were extracted from pools of three fruits. D, Representative HPLC results at 290 nm. Carotenoids were extracted from 30-DAA fruit flesh discs of CEZ (a and b) and low-β (c and d) treated with NF (a and c) or with water (b and d) for 12 h in the dark. E, Phytoene accumulated in 30-DAA fruit discs upon NF treatment. Phytoene is expressed in µg g−1 fresh weight (FW) of treated fruit flesh at 12 and 24 h of treatment. Values are means ± sd of three biological repeats.
To test if the strong reduction of CmOR protein in low-β fruit mesocarp affects PSY protein levels, we performed western-blot analysis of the fruit PSY-1 in low-β and in its isogenic progenitor line (CEZ) during fruit development. In CEZ, the PSY protein maintained high levels at 20 and 30 DAA with a slight reduction in mature fruit. In contrast, PSY was almost absent in low-β fruit flesh at 20 and 30 DAA but increased at the mature stage to levels similar to those found in CEZ (Fig. 4C). This correlates with fruit colors, showing maximal difference between low-β and CEZ at 30 DAA and the appearance of orange coloration around the low-β mature fruit seed cavity (Fig. 2A). Since AtOr-like (AT5G06130.2) partially compensates AtOR in maintaining PSY protein level in Arabidopsis (Zhou et al., 2015), we examined the expression of CmOr-like (MELO3C024554). CmOr-like expression was stable from 10 to 30 DAA but rose about 2-fold in the mature fruit, providing a possible explanation for the increased PSY levels found in the mature fruits of low-β in the absence of CmOr (Supplemental Fig. S7, A and B).
Biochemical Inhibition of Carotenogenesis Shows Lower Metabolic Flux in low-β
Apparently, the low PSY protein level in low-β was responsible for the reduced accumulation of β-carotene in the low-β fruits. To confirm this, we determined the metabolic flux following chemical inhibition. Phytoene levels upon NF inhibition are considered to be correlated directly with PSY activity (Beisel et al., 2011; Lätari et al., 2015). We treated developing fruit flesh discs of low-β and CEZ with NF and determined phytoene accumulation by HPLC at 12 and 24 h after treatment. At 30 DAA, when fruit carotenoid metabolic flux peaks (Supplemental Fig. S8A), 12 h of NF treatment resulted in a substantial elevation of phytoene in CEZ fruit discs (Fig. 4D, a) but only a minor elevation in low-β (Fig. 4D, c). Trace levels of phytoene also were detected in water-treated control fruit discs of CEZ but not in low-β (Fig. 4D, b and d). On average, CEZ accumulated 6.7- and 3.6-fold more phytoene than low-β during 12 and 24 h of treatment, respectively (Fig. 4E). In 20-DAA fruit discs, CEZ accumulated 2.8- and 1.8-fold more phytoene than low-β during 12 and 24 h of inhibition, respectively (Supplemental Fig. S8B).
Flux differences between low-β and CEZ were further confirmed by the use of CPTA, an inhibitor of lycopene cyclase (Fig. 1), followed by lycopene quantification by HPLC. CPTA-treated CEZ fruit discs accumulated about 2-fold more lycopene than low-β after 24 h of treatment in 20- and 30-DAA fruit discs (Supplemental Fig. S8, C and D). Clearly, carotenogenesis inhibition by both inhibitors suggested a lower carotenoid biosynthesis rate in fruits of the low-β mutant than in CEZ, which was likely due to the reduced PSY protein levels resulting from the dysfunctional CmOR in the low-β mutant.
Cmor-lowβ Reduces the Metabolic Flux in Developing Fruits of the crtiso Mutant
To confirm the Cmor-lowβ effect on the metabolic flux in vivo, we designed a double mutant experiment using the melon crtiso mutant called yofi. This line exhibits a dysfunctional crtiso due to a premature stop codon, which prevents carotenoid isomerization in the fruit flesh and causes the accumulation of light yellow prolycopene (Galpaz et al., 2013). The prolycopene amounts in a CRTISO-deficient background would indicate pathway flux without the impact of further metabolism of β-carotene, similar to the chemical inhibition of carotenoid biosynthesis performed so far.
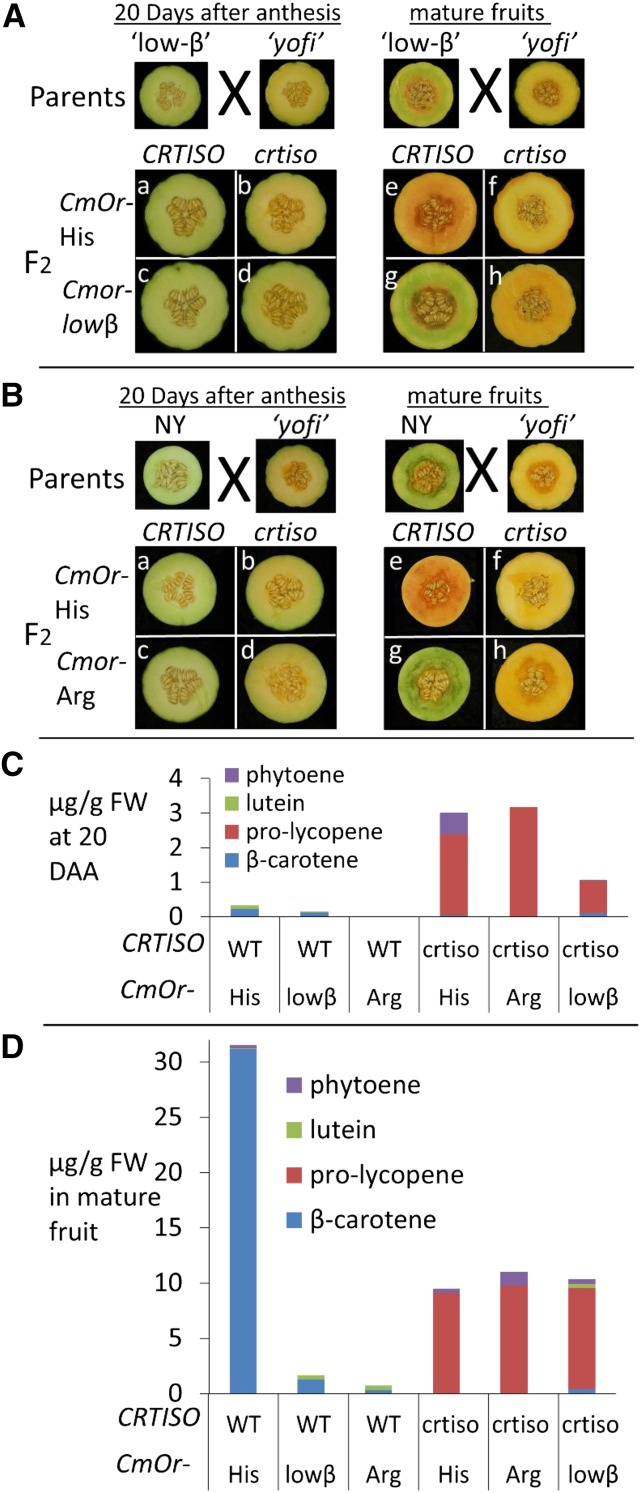
yofi carries the CmOr-His allele, as CEZ is also its wild-type progenitor. We crossed low-β with yofi. The F1 hybrid fruit flesh color was orange, as expected, since they were heterozygous at both the CRTISO as well as the CmOr loci. The F2 offspring segregated as expected for two independent recessive genes. We used physiological and molecular markers to select four combinations of CRTISO and Or as follows: the double wild-type alleles, the two single gene mutants, and the double mutant (Fig. 5A). Representative fruits of these genotypes, at 20 DAA and at the mature stage, were analyzed for carotenoid composition by HPLC (Fig. 5C; Supplemental Table S2). Since yofi accumulates prolycopene in the female flower ovules (Galpaz et al., 2013), we also examined the color of transected female flowers at the day of anthesis.
Figure 5.
Fruit flesh color and carotenoid levels of various melon genotypes. Transected fruit at 20 DAA (left) and mature stage (right) are shown. A, Transected fruits of low-β and yofi parental lines (top) and the four possible allelic combinations (bottom). CmOr genotypes (CmOr-His and Cmor-lowβ) are indicated on the left, and CRTISO genotypes (the wild type and nonsense mutated crtiso) are indicated on the top of each image. B, Transected fruits of yofi and NY parental lines (top) and the four possible allelic combinations (bottom). C and D, Average amounts (μg g−1 fresh weight [FW]) of phytoene, lutein, prolycopene, and β-carotene accumulated at 20 DAA (C) and in mature fruit (D) of F2 defined segregants derived from yofi × low-β (CmOr-His and Cmor-lowβ) and from yofi × NY (Cmor-Arg) crosses. WT, Wild type. Exact numbers and se values are given in Supplemental Table S2.
At 20 DAA, yofi mutant fruit exhibited pale yellow color (Fig. 5A) due to substantial amounts of prolycopene and phytoene and a very low level of β-carotene (Fig. 5C; Supplemental Table S2). At the same fruit development stage, the F2 progeny of the low-β × yofi cross that carried a wild-type functional CRTISO allele exhibited light green color similar to the low-β parental line (Fig. 5A, a and c). However, progeny that carried mutated crtiso was either pale yellow in fruits that carried CmOr-His or light green in fruits that carried the Cmor-lowβ allele (Fig. 5A, b and d). Accordingly, at 20 DAA, prolycopene levels in crtiso/Cmor-lowβ fruit flesh were 2.46-fold lower than in crtiso/CmOr-His fruit (Fig. 5C; Supplemental Table S2). Phytoene levels also were reduced; fruit flesh of F2 progeny carrying crtiso/CmOr-His contained similar amount of phytoene as the yofi parental line, while fruit flesh of progeny carrying crtiso/Cmor-lowβ contained only a trace amount of phytoene (Fig. 5C; Supplemental Table S2). Thus, the Cmor-lowβ mutant allele conferred lower prolycopene and phytoene levels at 20 DAA under the metabolic inhibition caused by CRTISO deficiency. This confirms that Cmor-lowβ had a reduced carotenoid biosynthetic metabolic flux in developing fruits compared with its wild-type progenitor. The reduction of carotenogenesis in the double mutant also was evident on the day of flower anthesis; the crtiso yellow ovule phenotype was completely masked in the background of Cmor-lowβ, which exhibited a green ovule phenotype (Supplemental Fig. S9).
Cmor-lowβ Retained the Chloroplastic Carotenoids in Ripe crtiso Mutant Fruit
The mature fruit flesh of yofi exhibited yellow color due to an increased accumulation of prolycopene (Fig. 5A) and contained high phytoene levels (Fig. 5D; Supplemental Table S2). Mature F2 fruits of the low-β × yofi cross, which carried wild-type functional CRTISO, exhibited the expected phenotypes, depending on the CmOr allele present in low-β or CEZ progenitor lines (Figs. 2A and 5A, e and g). However, while the mature fruit flesh of crtiso/CmOr-His was light yellow, similar to yofi, as expected (Fig. 5A, f), the mature fruit of the double mutant crtiso/Cmor-lowβ was, surprisingly, neither green nor yellow. Its hue was between yellow and orange (Fig. 5A, h). Accordingly, the carotenoid profile of F2 progeny carrying homozygous crtiso and CmOr-His was, as expected, similar to that of yofi, with trace levels of β-carotene, high levels of prolycopene and phytoene, and no detectable lutein (Fig. 5D; Supplemental Table S2). The double mutant crtiso/Cmor-lowβ mature fruit carotenoid profile was a mixture of the parental phenotypes, with lutein and β-carotene levels similar to low-β alongside high prolycopene levels similar to yofi (Fig. 5D; Supplemental Table S2). This indicates that an active CmOr-His is necessary for chloroplast disintegration or the chloroplast-to-chromoplast transition but not for prolycopene accumulation, which happens in a crtiso background, independently of the presence or absence of CmOr.
The Golden SNP Does Not Govern PSY Protein Level and Activity
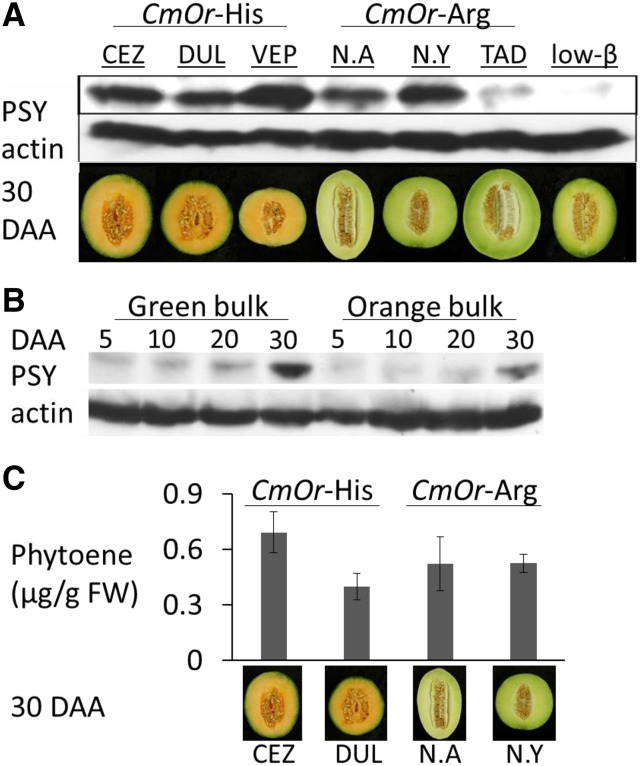
The results described so far indicated that the low PSY protein levels in low-β reduced carotenoid levels in fruits by lowering the metabolic flux. The similarity in fruit flesh color between low-β and homozygous Cmor-Arg melon lines suggested a similar decrease of PSY protein abundance in nonorange melon fruits as compared with orange fruits. Therefore, we analyzed PSY protein levels in fruits of six melon accessions: three orange-fruited accessions harboring CmOr-His and three Cmor-Arg nonorange fruit flesh accessions (Table I). We also included low-β in the analysis. Fruits were sampled at 30 DAA, a time point when carotenoid synthesis rate and PSY-1 transcript level peak (Fig. 4A; Supplemental Fig. S8A). PSY levels in low-β were much lower than in the six analyzed inbred lines, in which the highest PSY protein level was measured in a CmOr-His orange-fruited line (VEP) and the lowest was measured in a Cmor-Arg green-fruited line (TAD). However, PSY protein levels were similar in the remaining four lines independent from the CmOr natural allelic variation (Fig. 6A). This suggested a lack of association between PSY protein levels and the natural allelic variation of CmOr in different melon accessions.
Figure 6.
PSY protein and phytoene levels in melon lines. A, Western blot showing PSY protein levels at 30 DAA in six melon inbred lines and low-β with actin signals used as a loading control; the fruit flesh phenotypes at 30 DAA are shown below. Each sample is a pool of six fruits from six different plants. B, PSY protein levels at 5, 10, 20, and 30 DAA of green flesh and orange flesh bulked segregants originating from a cross between DUL and TAD and sharing the same fruit flesh color. Each sample is a bulk of 75 fruits originating from 25 different F3 families. C, Phytoene accumulated in 30-DAA fruit discs upon NF treatment. Phytoene is expressed in µg g−1 fresh weight (FW) of treated fruit flesh at 24 h of treatment. Values are means ± sd of three biological repeats. Although PSY protein and phytoene levels upon NF treatment are quite similar, β-carotene content in CmOR-His is much higher than in Cmor-Arg melons.
To determine the effect of the golden SNP on PSY protein levels while minimizing the possible effects of additional genes, we crossed a CmOr-His line (DUL) with a Cmor-Arg line (TAD) and analyzed PSY protein levels in green-fleshed (genotype, homozygous Cmor-Arg) and orange-fleshed (genotype, homozygous CmOr-His) bulks of 75 fruits equally representing 25 F3 families. The bulked green- or orange-fruited plants inherited their respective fruit flesh color from the CmOr alleles, but all their other CmOr-unlinked traits segregated arbitrarily (Chayut et al., 2015). In all analyzed fruit developmental stages, the bulks of the green and orange fruit flesh exhibited similar levels of PSY protein (Fig. 6B), confirming that PSY level was indeed inherited independently of CmOr natural allelic variation in developing fruit.
We treated fruit flesh discs of CmOr-His accessions (CEZ and DUL) and of Cmor-Arg accessions (NA and NY) with NF and determined phytoene accumulation 24 h after NF treatment by HPLC. Phytoene levels were not significantly different between CmOr-His and Cmor-Arg lines (Fig. 6C). Thus, high and low β-carotene levels in fruit mesocarp did not correlate with PSY protein abundance or PSY enzymatic activity.
Carotenoid Metabolic Flux in Developing Fruits Is Independent of CmOr-His/Cmor-Arg Natural Allelic Variation
PSY protein levels and activity across melon germplasm suggested that the CmOr natural allelic variation does not govern carotenogenesis metabolic flux (Fig. 6). To confirm this in planta, we crossed yofi (crtiso/crtiso; CmOr-His/CmOr-His) with the green-fleshed accession NY (CRTISO/CRTISO; Cmor-Arg/Cmor-Arg) and generated F2 segregants. We selected again the four genotypes for CRTISO and CmOr (Fig. 5B) and analyzed carotenoids in representative 20-DAA fruitlets and in mature fruit (Supplemental Table S2).
At 20 DAA, the NY parent exhibited light green color (Fig. 5B) and contained lutein that was not detectable in yofi and 8-fold more β-carotene than yofi (Supplemental Table S2). The F2 segregants of the NY × yofi cross had light green fruit flesh if they contained a functional CRTISO allele and pale yellow flesh in a homozygous mutated crtiso allele background, regardless of the CmOr allele (Fig. 5B, a–d). Accordingly, prolycopene levels in the homozygous recessive crtiso F2 segregants were similar, regardless of the CmOr allele (Supplemental Table S2). This confirmed that pathway flux at this early fruit developmental stage was not affected by CmOr-His/Cmor-Arg natural allelic variation.
crtiso Is Epistatic to CmOr-His/Cmor-Arg Natural Allelic Variation in Regulating Ripe Fruit Color and Carotenoid Composition
The mature fruit flesh of NY had its characteristic distinctive intense green color (Fig. 5B). Mature fruit flesh of NY × yofi F2 segregants carrying a functional CRTISO was intense green or orange depending on the CmOr allele (Fig. 5B, e and g). However, similar to the results from 20-DAA fruits, mature fruit flesh color and carotenoid levels in mature fruits of CRTISO-deficient crosses were similar and independent from the CmOr allele (Fig. 5B, f and h; Supplemental Table S2). Prolycopene levels in these fruit were similar, regardless of the CmOr allele (Supplemental Table S2). Thus, the fruit flesh color phenotype of the F2 segregants derived from a cross between NY and yofi revealed an epistatic interaction between CmOr and CRTISO genes. CRTISO regulated carotenoid levels and composition regardless of CmOr natural allelic variation. In other words, these results indicated that in planta CmOr natural allelic variation did not affect the carotenoid synthesis rate. Instead, it affected β-carotene accumulation by inhibiting metabolism downstream of β-carotene.
DISCUSSION
The natural occurrence of a large copia-like retrotransposon insertion in the cauliflower BoOr gene revealed the ability of the highly conserved Or gene to induce carotenoid accumulation in noncolored plant tissues (Lu et al., 2006). However, the mechanism by which the cauliflower mutated Or gene operates remains elusive. The pleiotropic effects of BoOr and the multiple alternatively sliced transcripts of the naturally occurring BoOr active allele make it difficult to resolve the Or mechanism of action. We recently reported that β-carotene accumulation in melon fruit flesh is governed by a single SNP in CmOr, providing a precise biological tool with which to study OR function (Tzuri et al., 2015) and to utilize it in biotechnology (Yuan et al., 2015a). The identification of the novel low-β mutant provides an invaluable tool for further deciphering the Or gene mechanism of action in the regulation of carotenoid accumulation. Studies reported here show that carotenoid biosynthesis is fully active and functional in nonorange melon fruit and that the specific inhibition of carotenoid metabolism leads to carotenoid accumulation and chromoplast formation. In addition, we show that CRTISO, whose deficiency arrests carotenoid metabolism at prolycopene, is epistatic to CmOr. Thus, a CRTISO-deficient background was used to show that carotenoids are synthesized in melon fruit regardless of the golden SNP allelic variation (Fig. 7).
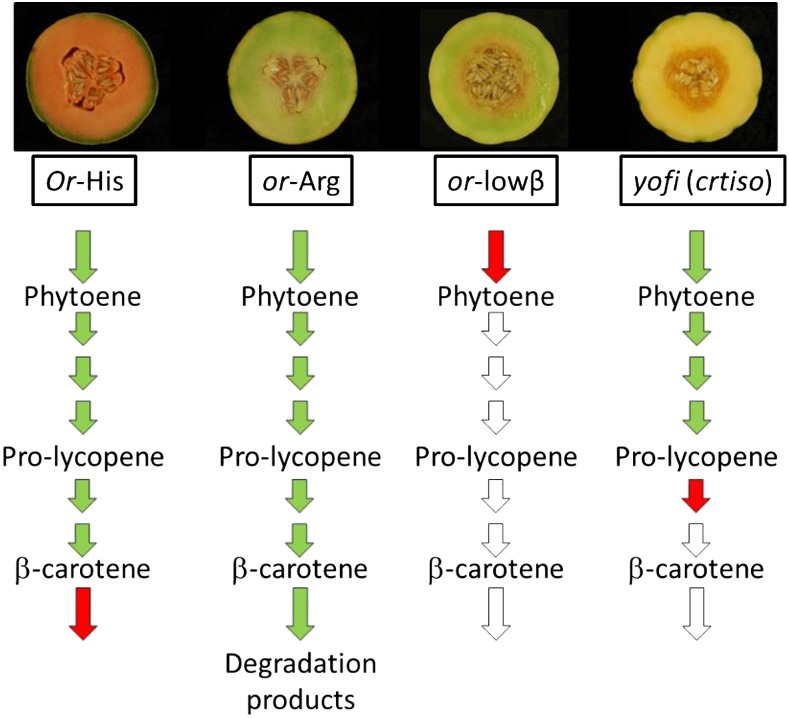
Figure 7.
A model of carotenogenesis under the effect of three alleles of CmOr and of a dysfunctional CRTISO. In the presence of both OR-His and OR-Arg, carotenoid pathway flux occurs at high levels (green arrows) but only OR-His prevents further metabolism downstream of β-carotene (red arrow), resulting in strongly increased β-carotene accumulation in OR-His melon fruits but not in OR-Arg. In contrast, low PSY protein levels, caused by a truncated OR-lowβ, results in a weak pathway flux (red arrow) and, hence, low carotenoid levels in OR-lowβ fruit. Dysfunctional CRTISO in yofi melons results in metabolic arrest downstream of prolycopene (red arrow). Accordingly, prolycopene is accumulated in yofi genotypes regardless of the golden SNP, which operates downstream in the pathway.
The Roles of OR
Recent studies of OR proteins of melon (Chayut et al., 2015; Tzuri et al., 2015), Arabidopsis (Yuan et al., 2015a; Zhou et al., 2015), and sweet potato (Park et al., 2015) and previous works with cauliflower (Li et al., 2006, 2012) and potato (Solanum tuberosum; Lopez et al., 2008; Li et al., 2012) suggest two complementary roles of OR in carotenoid accumulation: posttranscriptional regulation of PSY protein and biogenesis of chromoplasts. Stabilization of PSY protein in controlling carotenoid pathway flux has been shown in Arabidopsis (Zhou et al., 2015) and transgenic potato (Lopez et al., 2008). Our analyses of the nonsense mutation of Cmor-lowβ reaffirm that OR is required to maintain PSY level and suggest that OR is involved in the chloroplast-to-chromoplast transition during fruit development. Moreover, our results provide solid evidence that PSY levels are not the cause of OR-mediated high carotenoid accumulation in melon fruit, therefore reopening the question of the exact mechanism by which Or operates. The results of our experiments with chemical (NF and CPTA) and genetic (yofi, a CRTISO mutant) metabolic flux inhibitors indicate that OR facilitates β-carotene accumulation by inhibiting carotenoid metabolism downstream of β-carotene (hydroxylation or degradation). This is also shown by the presence of only traces of lutein while other xanthophylls are not detected in orange melon fruit (Chayut et al., 2015; Supplemental Table S2). We suggest that, in melon and probably in other fruit, chromoplast biogenesis is triggered in a metabolite-dependent mechanism, similar to what has been suggested in transgenic tomato overexpressing PSY (Fraser et al., 2007).
Facilitation of the Active PSY Enzyme
Immunoblot analysis showed that the nonsense mutation of Cmor-lowβ dramatically reduced fruit mesocarp PSY protein level but not Psy gene expression at 20 and 30 DAA (Fig. 4, A–C; Supplemental Fig. S8A). The Cmor-lowβ allele encoded a truncated CmOR protein and also was associated with significantly suppressed Cmor-lowβ transcript levels (Fig. 3C), probably by a nonsense-mediated mRNA decay mechanism (Kerényi et al., 2008). The truncated CmOR-lowβ protein was unable to dimerize (Fig. 4E), which is expected to negatively affect its stability (Zhou et al., 2015). The inability to dimerize and the significantly reduced Cmor-lowβ mRNA levels lead to the absence of CmOR protein in low-β melon fruits (Fig. 3D; Supplemental Fig. S4).
As OR was shown to be a major posttranslational regulator of PSY, we concluded that the absence of CmOR in low-β melons results in low levels of PSY protein, similar to OR-deficient Arabidopsis lines (Zhou et al., 2015). As PSY catalyzes the rate-limiting step in carotenoid biosynthesis (Arango et al., 2014), the reduction of PSY protein could account for the reduced carotenoid accumulation in low-β fruit. This was confirmed by the determination of phytoene accumulation in detached developing melon fruit tissues upon the inhibition of PDS activity by NF, an indicative assay for PSY activity (Beisel et al., 2011; Lätari et al., 2015). The low-β mutant accumulated significantly less phytoene than CEZ (Fig. 4, D and E), confirming that OR mediated PSY protein activity. The lower PSY activity in low-β fruit reduced the metabolic flux toward β-carotene synthesis, indicating that CmOr promoted the carotenoid metabolic flux toward β-carotene synthesis in wild-type melon fruits.
However, the golden SNP, which governs fruit flesh β-carotene levels across a wide spectrum of melon germplasm (Tzuri et al., 2015), was not associated with a reduction in PSY protein level. Green and orange 30-DAA melon fruits showed similar levels of PSY protein independently of the golden SNP genotype and accumulated a similar amount of phytoene 24 h after NF inhibition (Fig. 6). PSY localization was shown to be a major mode of regulation of PSY activity (Shumskaya et al., 2012); it has been shown previously that the golden SNP did not alter PSY localization (Yuan et al., 2015a). Our study confirms that the golden SNP-induced carotenoid accumulation acts independently of PSY levels and activity.
In the malfunctioning crtiso genetic background, CmOr-lowβ lowered prolycopene levels in female flowers (Supplemental Fig. S9) and in developing fruit but not in the mature fruit (Fig. 5D; Supplemental Table S2). The latter phenomenon is not surprising, since PSY protein level, the carotenogenic flux determinant, was not significantly affected in low-β mature fruit although it was severely lowered at 20 and 30 DAA (Fig. 5C). Increased expression of PSY at the mature fruit stage also was observed by Galpaz et al. (2013) when they compared yofi and CEZ at the mature stage. CRTISO has been suggested as a regulatory node in the carotenoid pathway (Cazzonelli and Pogson, 2010); however, the exact mechanism by which it operates is still unclear. The mature low-β fruit accumulated orange β-carotene in its inner part close to the seed cavity (Fig. 2A), showing the incomplete nature of the low-β phenotype. While β-carotene accumulated at trace amounts in the mature low-β fruit (Fig. 2B), prolycopene accumulated significantly in mature fruit regardless of OR (Fig. 5D). This can probably be best explained by the efficient inhibition of the carotenoid biosynthesis metabolic flux by crtiso in chromoplast as opposed to chloroplast, in which photoisomerization may occur (Isaacson et al., 2002).
Regulation of Plastid Fate
In orange cauliflower, it was suggested that the BoOr mutant allele triggers the biogenesis of chromoplasts, the specialized plastids that synthesize and store carotenoids (Lu et al., 2006; Li and Yuan, 2013). Chromoplasts also were formed in potato tubers overexpressing BoOr (Lopez et al., 2008) and in Arabidopsis overexpressing engineered AtOr-His (Yuan et al., 2015a). We show here that low-β carries a loss-of-function allele of CmOr, resulting in the absence of orange chromoplasts, which were observed in fruit mesocarp cells of CEZ, the isogenic progenitor of low-β (Fig. 3, D–G; Supplemental Fig. S2). This supports the suggested role of OR as a molecular switch of chromoplast biogenesis.
While cauliflower curd, potato tubers, and Arabidopsis calli grown in the dark are nongreen tissues, containing noncolored proplastids and amyloplasts, melon fruit mesocarp contains chloroplasts during the first stages of fruit development, as evidenced by the presence of chlorophylls in young fruit (Chayut et al., 2015). Here, we show that both CEZ and low-β similarly accumulated chlorophylls until 30 DAA (Fig. 2C). However, while CEZ fruits degraded all fruit flesh chlorophylls from 30 DAA until maturation, low-β mature fruit maintained high chlorophyll levels. Accordingly, our microscopy study of mature fruit mesocarp cells revealed chloroplasts in low-β but not in CEZ and chromoplasts in CEZ but not in low-β (Fig. 2, D–G; Supplemental Fig. S2). Our biochemical analysis identified lutein, the major chloroplastic carotenoid (Jahns and Holzwarth, 2012), as present in low-β but absent in CEZ (Supplemental Fig. S1; Supplemental Table S2). This observation further supports that Cmor-lowβ affected chloroplast occurrence in mature fruit mesocarp. These findings suggest that the Cmor-lowβ nonsense mutation inhibited chromoplast formation and chloroplast disintegration from 30 DAA until fruit ripening. While our results cannot exclude the possibility that chromoplast formation and chloroplast disintegration are independent, they strongly suggest that melon plastids follow the chloroplast-to-chromoplast transition under the control of CmOr. Such a transition has been well characterized in pepper (Capsicum annuum) and tomato fruit (Egea et al., 2010) but was never associated previously with the Or gene.
Furthermore, Cmor-lowβ in a CRTISO-deficient background (double mutant segregant of a yofi × low-β cross) maintained its chloroplast-specific carotenoid pattern (lutein and low levels of β-carotene) in the mature fruit accompanied by a high accumulation of prolycopene, a chromoplast-specific carotenoid (Fig. 5B; Supplemental Table S2). Moreover, both Cmor-lowβ and Cmor-Arg increased chlorophyll levels in mature fruits compared with CmOr-His (Fig. 2C; Chayut et al., 2015). Therefore, the mature double mutant phenotype suggests that the CmOr-His allele is needed for chloroplast disintegration in the late fruit-ripening stage. However, the induction of high carotenoid accumulation in fruit by OR can be replaced by carotenogenic flux inhibition resulting from a mutated CRTISO. The accumulation of carotenoids in a yofi/low-β double mutant fruit despite low carotenoid metabolism (Fig. 5B; Supplemental Table S2) and the low PSY protein levels at early fruit development stages (Fig. 4C) are probably caused by the efficient inhibition of carotenoid metabolism by yofi. The presence of carotenoids downstream of prolycopene in the absence of a functional CRTISO in these segregants is probably due to the extended presence of photosynthetic organelles where light-induced photoisomerization of cis-configured carotenes could occur (Isaacson et al., 2002).
Inhibition of β-Carotene Turnover
Does the biogenesis of chromoplasts trigger carotenoid buildup or does carotenoid buildup trigger the plastid rearrangement leading to chromoplasts? Plastid development in white- and red-fleshed grapefruit (Citrus paradisi) indicates that the differentiation of distinctive plastids could be correlated with a characteristic carotenoid content (Lado et al., 2015). While the red-fruited cv Star Ruby accumulates high carotenoid levels in the ripe fruit (predominantly phytoene, phytofluene, and lycopene), the white-fruited cv Marsh accumulates very low carotenoid levels (phytoene and violaxanthin). A comparative transcriptional study of carotenoid biosynthesis genes suggests that low expression of β-LCY causes the high lycopene accumulation in the red-fruited cultivar (Alquezar et al., 2013). Microscopic analysis indicates that the white grapefruit pulp exhibited fewer chromoplasts per cell and that carotenoid crystalloid structures were absent, while in the red grapefruit pulp, chromoplasts were more abundant and frequently contained crystals (Lado et al., 2015). In this example, the differences in carotenoid composition and accumulation were reflected by the development of specific chromoplast types. Therefore, it seems that, in grapefruit, lycopene accumulation, caused by a specific inhibition of carotenoid metabolism flux, resulted from low β-LCY transcription, which triggers this characteristic chromoplast biogenesis.
In the early developmental stage of tomato fruit overexpressing PSY-1, alteration of plastid type was found along with higher carotenoid levels and different carotenoid composition. The authors suggested a metabolite-induced chloroplast-to-chromoplast transition (Fraser et al., 2007). The induced chromoplasts lacked the distinctive crystalline lycopene structures found in ripe tomato fruits probably as a result of high β-carotene, which was more abundant than lycopene in the studied fruitlets of the mature green stage. In nongreen tissues (roots and callus) of Arabidopsis, an increase of carotenogenic flux by overexpression of PSY caused carotenoid accumulation in crystalline-type chromoplasts (Maass et al., 2009). Thus, chromoplast could be formed from high carotenoid abundance, which can result from flux elevation or from an interruption within an existing carotenoid metabolism flux.
It was shown previously that the natural allelic variation of CmOr does not change the expression of carotenogenesis genes in melon fruit (Chayut et al., 2015). In this work, we show that fruit PSY protein levels were similarly unchanged by the golden SNP. For example, we found high PSY levels in NY, a green-fruited melon containing the Cmor-Arg allele, as well as in CEZ, an orange-fruited CmOr-His variety (Fig. 6A). In a CRTISO-deficient background, both naturally occurring CmOr alleles allowed high prolycopene levels, which are thus independent of CmOr natural allelic variation (Fig. 6C; Supplemental Table S2). However, if pathway flux is allowed to proceed beyond prolycopene (as in the functional CRTISO background), β-carotene accumulates only if CmOR-His is present. This indicates that CmOR-His did not affect carotenoid pathway flux upstream of β-carotene but had a major impact on the stability and further metabolism of β-carotene, correlated with the formation of chromoplasts. Thus, the surprising epistasis of CRTISO over Or arises because, in the carotenoid metabolic pathway, the production of prolycopene precedes the production of β-carotene (Figs. 1 and 7).
Our results suggest that naturally regulated expression of CmOR-His suppresses the downstream metabolism of β-carotene in melon fruits and show that, in mature CmOR-His melon fruit, only traces of lutein could be found while other xanthophylls were not detected (Supplemental Table S2; Chayut et al., 2015). However, previous studies have shown that the overexpression of CmOr-His or AtOr-His induces high accumulation of β-carotene, together with significant amounts of lutein, in nongreen tissue (calli) of Arabidopsis (Tzuri et al., 2015; Yuan et al., 2015a). Lutein also was amassed in potato tubers overexpressing BoOr (Lu et al., 2006) and in sweet potato calli overexpressing engineered IbOr (Kim et al., 2013). These results suggest that the inhibition of carotenoid metabolism flux by OR also could affect lutein fate and is not limited to β-carotene. The golden SNP-specific stabilization of β-carotene and possibly lutein, depending on plant species, may imply its yet to be elucidated molecular mechanism.
CONCLUSION
Global food security demands provitamin A-biofortified staple crops (Graham et al., 2001; Fraser and Bramley, 2004). The Or gene, a molecular switch that induces carotenoid accumulation, is a promising tool for this purpose (Lu et al., 2006). The discovery of the golden SNP in the melon CmOr gene (Tzuri et al., 2015) and the confirmation of this SNP potency (Yuan et al., 2015a) precisely define the site for DNA editing as a possible new path toward β-carotene biofortification of crops by a nontransgenic approach. This study sheds new light on the mechanism of action of the Or gene in melon fruit, with its possible applications in other crop systems.
We showed that the induced nonsense mutation in CmOr in low-β melons reduced the abundance of functional CmOR protein and, in consequence, of PSY protein, which dramatically affected both carotenoid content and plastid fate. By contrast, the natural allelic variations of CmOr in melon fruits, CmOr-His and Cmor-Arg, similarly enhanced carotenoid biosynthesis by posttranscriptionally regulating PSY protein amounts. However, only the golden SNP allele CmOR-His prevents the further metabolism of β-carotene, resulting in its accumulation in chromoplasts. This suggests metabolic flux analysis as a necessary criterion when selecting potential superior germplasm for DNA editing to introduce the Or golden SNP into other crops.
MATERIALS AND METHODS
Plant Material
CEZ is a Charentais-type melon (Cucumis melo melo variety Cantalupensis Naudin). The low-β and yofi (CRTISO) mutants are EMS-mutated CEZ isogenic lines produced as described (Tadmor et al., 2007). All melon varieties used in this work are listed and described in Table I.
We also used 50 F3 families selected from a segregating population originating from a cross between DUL and TAD. Each family resulted from the self-pollination of a single F2 plant. We used the cleaved-amplified polymorphic sequence (CAPS) molecular marker and fruit color visualization of F3 families to select 25 orange-fruited families that are homozygous CmOr-His and 25 green-fruited families that are homozygous CmOr-Arg as described (Chayut et al., 2015).
Double Mutant Production
To generate double mutant plants, we crossed the crtiso mutant yofi (Galpaz et al., 2013) with low-β and with NY. Light yellow instead of green cotyledons is the first visualized phenotypic marker for the homozygous crtiso (yofi) mutation (Galpaz et al., 2013). A total of 600 F2 seeds of each cross were germinated in a light- and temperature-controlled growth room (12 h of artificial light, 27°C/12 h of darkness, 20°C). After emergence, yofi plants were selected by their yellow cotyledon color. A total of 148 and 157 plantlets resembled yofi in the cross with low-β and with NY, respectively. After 5 to 8 d, all seedlings looked alike due to the photoisomerization of prolycopene and grew alike under the experimental conditions. For both genotypes (crtiso/crtiso and CRTISO/−), we genotyped 100 seedlings for the CmOr allele using the CAPS marker of the CmOr gene. In the analysis of segregants derived from the cross of yofi with NY, we defined the natural allelic variation genotype with the previously described CAPS marker using the forward primer 5′-GTATCATAGAGGGACCGG-3′ and the reverse primer 5′-CCCAATGCAGCTAAGTCAA-3′ (Tzuri et al., 2015). The PCR-resulting amplicon was subjected to enzymatic digestion with HinfI, which cuts only the dominant allele. For the genotyping of segregants derived from the cross of yofi with low-β, we used the forward primer 5′-ATGTAGGTTCGTCGGCTGAG-3′ and the reverse primer 5′-ACCTTAGCTGCATTGGGAGA-3′, which amplified a novel polymorphic fragment. The amplicon was subjected to enzymatic digestion with AcuI, which cleaves the wild-type allele and does not cleave the low-β allele. For each cross, eight to 10 plants of each of the four possible genotypes were grown in a semicontrolled greenhouse. We manually self-pollinated two female flowers on each trellised plant and marked the anthesis date. Three fruits (the first successfully pollinated) from three plants of each genotype, at 20 DAA and upon fruit maturation, were visually phenotyped, sampled, and stored at −80°C until use. For each developmental stage, the three fruits were sampled on the same day. Nonpollinated female flowers, at the day of anthesis, were picked from all plants for ovule color phenotyping.
Carotenoid and Chlorophyll Analyses
A hexane:acetone:ethanol (2:1:1, v/v/v) mixture was used to extract carotenoid pigments as described (Tadmor et al., 2005). Separation of carotenoids was carried out on a C18 Nova-Pak (Waters) column (250 × 4.6 mm i.d., 60 A, 4 mm) and a Nova-Pak Sentry Guard cartridge (Waters) using a Waters 2695 HPLC apparatus equipped with a Waters 996 photodiode array detector as described (Tadmor et al., 2000). Carotenoids were identified by their characteristic absorption spectra, distinctive retention times, and comparison with authentic standards (ExtraSynthase). Quantification of specific carotenoids was performed using Millennium chromatography software (Waters) that integrates the peak areas of the HPLC results and calculates concentration using authentic standard curves. To distinguish undetected from unquantifiable levels, we defined as trace amount any carotenoids with integrated peak area lower than 20,000 absorbance units min−1.
Total chlorophylls were quantified by recording the absorbance at 661.6 and 644.8 nm of 10× diluted acetone extracts as described (Lichtenthaler, 1987). The following equation was used to quantify chlorophyll: Chl a + Chl b (µg mL−1 acetone) = (11.24 × A661.6 – 2.04 × A644.8) + (20.13 × A644.8 – 4.19 × A661.6).
Microscopy
Protoplasts were isolated by gentle shaking of 1 g of mature fruit mesocarp in 1.5 mL of solution of 18% (w/v) mannitol, 1% (w/v) cellulose, 0.3% (w/v) macerozyme, and 0.05% (w/v) pectolyase at 28°C overnight (enzymes were manufactured by Duchefa Biochemie). Protoplasts and manually sliced mature fruit mesocarp tissues were observed with a phase contrast and direct interference contrast compound microscope (BX61; Olympus) equipped with a high-resolution digital camera (DP73; Olympus) and image-analysis software (Soft Imaging System; Olympus).
RNA Sequencing Transcriptome Analysis
CEZ and low-β plants were field grown in summer 2012 at a Newe Ya’ar Research Center plot. Female flowers were marked at the day of anthesis, and three typical fruits were sampled at 10, 20, and 30 DAA and upon ripening (40–45 d, but it varies between fruits). Each fruit served as a biological replicate and was picked from a different plant. All fruits were picked in the morning to diminish circadian effects. The fruit flesh representative portion was immediately frozen in liquid nitrogen and kept at −80°C until extraction. Total RNA from 24 developing fruit mesocarp tissue samples (two genotypes × four developmental stages × three biological replicates) was extracted following the protocol described (Portnoy et al., 2008) and used to construct the strand-specific RNA sequencing library as described (Zhong et al., 2011). The barcoded libraries were pooled and sequenced on a single lane of the Illumina HiSeq 2000 sequencing system at the Cornell University core facility.
RNA Sequencing Data Analysis
RNA sequencing reads were first aligned to the ribosomal RNA database (Zhong et al., 2011) using Bowtie (Langmead et al., 2009), and those that were aligned were discarded. The resulting reads were aligned to the melon genome (Garcia-Mas et al., 2012) using TopHat (Trapnell et al., 2009). Following the alignments, raw counts for each melon gene were derived and normalized to RPKM. The raw counts of melon genes were fed to edgeR (Robinson et al., 2010) to identify differentially expressed genes between CEZ and the low-β mutant at each of the four developmental stages.
Protein Extraction
Proteins of CEZ and low-β fruit were extracted from the same fruit tissue samples that were used for RNA extraction (field-grown plants from summer 2012). Three samples of 20- and 30-DAA and of the mature fruit developmental stage were bulked. Proteins from 30-DAA fruit of CEZ and low-β and additional inbred lines were extracted from six bulked fruits grown on six different trellised plants in semicontrolled greenhouse conditions during the summer of 2013. Proteins from developing bulked F3 families were extracted from 75 pooled developing or mature fruits, grown on 75 different plants, three from each family originating from a cross between DUL and TAD inbred lines. All proteins were extracted with phenol according to the protocol as described (Maass et al., 2009).
Western-Blot Analysis
Protein concentration was determined with the Bio-Rad protein assay. Proteins were resolved electrophoretically on a 12% SDS-polyacrylamide gel and then blotted to either nitrocellulose or polyvinylidene difluoride membranes (pore size, 0.45 µm; Carl Roth). Ponceau S staining was used to show equal loading and transfer quality. Membranes were blocked by soaking in 5% milk powder in Tris-buffered saline plus 0.1% Tween 20 for 1 h at 20°C to 25°C and then incubated with antibodies against BoOr (Lu et al., 2006) or PSY (Maass et al., 2009) for 1 to 2 h at room temperature. After washing three times for 10 min each with Tris-buffered saline plus 0.1% Tween 20, the blot was incubated with the horseradish peroxidase-conjugated antibody solution for 1 h at room temperature. The signal was detected with enhanced chemiluminescence (GE Healthcare). Blots were stripped (Sennepin et al., 2009) and reprobed with anti-actin antibodies (Sigma).
Protein Interaction Analysis
The split-ubiquitin system was used (Obrdlik et al., 2004). OR dimerization and OR-PSY interaction were analyzed as described previously (Zhou et al., 2015). Basically, the cDNA sequences were cloned to make either N-terminal or C-terminal fusions. Overnight cultures from yeast strains that coexpressed two fused proteins were spotted on nonselective and selective media lacking Leu and Trp (-LW) or lacking Leu, Trp, adenine, and His (-LWAH), supplemented with Met in a series of 1:10 dilutions, and grown at 29°C. β-Galactosidase activity was determined by assaying enzyme velocity using the substrate ortho-nitrophenyl-β-d-galactopyranoside as described (Zhou et al., 2015). Amounts of ortho-nitrophenol were determined photometrically at 420 nm using the molar extinction coefficient of ortho-nitrophenol (3,300 g L−1 mol−1). β-Galactosidase activity was calculated as nmol ortho-nitrophenol min−1 optical density at 600 nm−1. All β-galactosidase assays were performed in triplicate.
In Vivo Measurement of Carotenogenic Metabolic Flux
Trellised plants of CEZ and low-β were grown in semicontrolled greenhouse conditions. Each plant was self-pollinated once, and the flower was marked at the pollination day. Bearing a single fruit per plant eliminated competition between fruits within plants and lowered the variance in fruit maturation pace between plants. Fruitlets were collected at 20 and 30 DAA, and each served as an independent biological repeat. Discs 10 mm in diameter and 5 mm in height were cut with a cork borer from fruitlet mesocarp. Each disc was treated with 12 µL of 1 mm NF or 0.1 mm CPTA aqueous solution. Controls were treated with 12 µL of water. One control sample was frozen immediately after water treatment. Six discs from each treatment were sampled at 12 and 24 h. Water-treated controls were sampled as well. Carotenoids were quantified by HPLC (Tadmor et al., 2005).
Accession Numbers
The raw sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession number SRP071059.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HPLC results.
Supplemental Figure S2. Protoplast microscopy.
Supplemental Figure S3. CmOr coding sequences of CEZ and low-β.
Supplemental Figure S4. Immunoblotting of CmOR using a high-density (15%) fractionating gel.
Supplemental Figure S5. Yeast split-ubiquitin analysis.
Supplemental Figure S6. Different digital gene expression (RPKM) of metabolic genes.
Supplemental Figure S7. CmOr-like digital expression.
Supplemental Figure S8. In planta metabolic flux measurements.
Supplemental Figure S9. Transected female flowers of low-β × yofi F2 segregants at the day of anthesis exposing the ovule color.
Supplemental Table S1. Mean carotenogenesis gene digital expression during CEZ and low-β fruit development.
Supplemental Table S2. Fruit carotenoid content at 20 DAA and at mature fruit of various inbred lines and F2 segregants.
Supplementary Material
Acknowledgments
We thank Joseph Hirschberg of the Department of Genetics at the Hebrew University of Jerusalem for supplying CPTA inhibitor and advising on the inhibition experiments and on carotenoid quantification; Alexander Vainstein of the Faculty of Agriculture, Food, and Environment at the Hebrew University of Jerusalem for kindly providing the NF inhibitor; Shira Gal from the Newe Ya’ar Research Center for microscopy assistance; and Harry Paris for critically editing the article.
Glossary
- SNP
single-nucleotide polymorphism
- EMS
ethyl methanesulfonate
- DAA
days after anthesis
- NF
norflurazon
- CAPS
cleaved-amplified polymorphic sequence
- RPKM
reads per kilobase of exon model per million mapped reads
Footnotes
This work was supported by the United States-Israel Binational Agricultural Research and Development Fund (grant nos. US–4423–11 and US–4918–16CR), by the Israeli Ministry of Agriculture Chief Scientist (grant no. 256–1103–15), by the Center for the Improvement of Cucurbit Fruit Quality, Agricultural Research Organization, Israel, and by the Harvestplus Research Consortium (to R.W.).
Articles can be viewed without a subscription.
References
- Alquezar B, Rodrigo MJ, Lado J, Zacarias L (2013) A comparative physiological and transcriptional study of carotenoid biosynthesis in white and red grapefruit (Citrus paradisi Macf.). Tree Genet Genomes 9: 1257–1269 [Google Scholar]
- Arango J, Jourdan M, Geoffriau E, Beyer P, Welsch R (2014) Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in Orange carrots. Plant Cell 26: 2223–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel KG, Schurr U, Matsubara S (2011) Altered turnover of β-carotene and Chl a in Arabidopsis leaves treated with lincomycin or norflurazon. Plant Cell Physiol 52: 1193–1203 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Chayut N, Yuan H, Ohali S, Meir A, Yeselson Y, Portnoy V, Zheng Y, Fei Z, Lewinsohn E, Katzir N, et al. (2015) A bulk segregant transcriptome analysis reveals metabolic and cellular processes associated with Orange allelic variation and fruit β-carotene accumulation in melon fruit. BMC Plant Biol 15: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li F, Wurtzel ET (2010) Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol 153: 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech JC (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43: 228–265 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EMA, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM (2007) Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19: 3194–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Burger Y, Lavee T, Tzuri G, Sherman A, Melamed T, Eshed R, Meir A, Portnoy V, Bar E, et al. (2013) Genetic and chemical characterization of an EMS induced mutation in Cucumis melo CRTISO gene. Arch Biochem Biophys 539: 117–125 [DOI] [PubMed] [Google Scholar]
- Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Hénaff E, Câmara F, Cozzuto L, Lowy E, et al. (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci USA 109: 11872–11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Diretto G (2007) Of chromoplasts and chaperones. Trends Plant Sci 12: 529–531 [DOI] [PubMed] [Google Scholar]
- Graham RD, Welch RM, Bouis HE (2001) Addressing micronutrient malnutrition through enhancing the nutritional quality of staple foods: principles, perspectives and knowledge gaps. Adv Agron 70: 77–142 [Google Scholar]
- Howitt CA, Pogson BJ (2006) Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ 29: 435–445 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns P, Holzwarth AR (2012) The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim Biophys Acta 1817: 182–193 [DOI] [PubMed] [Google Scholar]
- Kerényi Z, Mérai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D (2008) Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J 27: 1585–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ahn YO, Ahn MJ, Jeong JC, Lee HS, Kwak SS (2013) Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol Biochem 70: 445–454 [DOI] [PubMed] [Google Scholar]
- Lado J, Zacarías L, Gurrea A, Page A, Stead A, Rodrigo MJ (2015) Exploring the diversity in Citrus fruit colouration to decipher the relationship between plastid ultrastructure and carotenoid composition. Planta 242: 645–661 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lätari K, Wüst F, Hübner M, Schaub P, Beisel KG, Matsubara S, Beyer P, Welsch R (2015) Tissue-specific apocarotenoid glycosylation contributes to carotenoid homeostasis in Arabidopsis leaves. Plant Physiol 168: 1550–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lu S, Cosman KM, Earle ED, Garvin DF, O’Neill J (2006) β-Carotene accumulation induced by the cauliflower Or gene is not due to an increased capacity of biosynthesis. Phytochemistry 67: 1177–1184 [DOI] [PubMed] [Google Scholar]
- Li L, Paolillo DJ, Parthasarathy MV, Dimuzio EM, Garvin DF, Plant US, Road T (2001) A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J 26: 59–67 [DOI] [PubMed] [Google Scholar]
- Li L, Yang Y, Xu Q, Owsiany K, Welsch R, Chitchumroonchokchai C, Lu S, Van Eck J, Deng XX, Failla M, et al. (2012) The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol Plant 5: 339–352 [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H (2013) Chromoplast biogenesis and carotenoid accumulation. Arch Biochem Biophys 539: 102–109 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophyll fluorescence signatures of leaves during the autumnal chlorophyll breakdown. J Plant Physiol 131: 101–110 [Google Scholar]
- Lopez AB, Van Eck J, Conlin BJ, Paolillo DJ, O’Neill J, Li L (2008) Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. J Exp Bot 59: 213–223 [DOI] [PubMed] [Google Scholar]
- Lu S, Van Eck J, Zhou X, Lopez AB, O’Halloran DM, Cosman KM, Conlin BJ, Paolillo DJ, Garvin DF, Vrebalov J, et al. (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass D, Arango J, Wüst F, Beyer P, Welsch R (2009) Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE 4: e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiani G, Castón MJP, Catasta G, Toti E, Cambrodón IG, Bysted A, Granado-Lorencio F, Olmedilla-Alonso B, Knuthsen P, Valoti M, et al. (2009) Carotenoids: actual knowledge on food sources, intakes, stability and bioavailability and their protective role in humans. Mol Nutr Food Res (Suppl 2) 53: S194–S218 [DOI] [PubMed] [Google Scholar]
- Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8: 68–82 [DOI] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SC, Kim SH, Park S, Lee HU, Lee JS, Park WS, Ahn MJ, Kim YH, Jeong JC, Lee HS, et al. (2015) Enhanced accumulation of carotenoids in sweetpotato plants overexpressing IbOr-Ins gene in purple-fleshed sweetpotato cultivar. Plant Physiol Biochem 86: 82–90 [DOI] [PubMed] [Google Scholar]
- Pitrat M. (2000) Some comments on infraspecific classification of cultivars of melon. Acta Hortic 510: 29–36 [Google Scholar]
- Portnoy V, Benyamini Y, Bar E, Harel-Beja R, Gepstein S, Giovannoni JJ, Schaffer AA, Burger J, Tadmor Y, Lewinsohn E, et al. (2008) The molecular and biochemical basis for varietal variation in sesquiterpene content in melon (Cucumis melo L.) rinds. Plant Mol Biol 66: 647–661 [DOI] [PubMed] [Google Scholar]
- Qin X, Coku A, Inoue K, Tian L (2011) Expression, subcellular localization, and cis-regulatory structure of duplicated phytoene synthase genes in melon (Cucumis melo L.). Planta 234: 737–748 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennepin AD, Charpentier S, Normand T, Sarré C, Legrand A, Mollet LM (2009) Multiple reprobing of western blots after inactivation of peroxidase activity by its substrate, hydrogen peroxide. Anal Biochem 393: 129–131 [DOI] [PubMed] [Google Scholar]
- Shumskaya M, Bradbury LMT, Monaco RR, Wurtzel ET (2012) Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell 24: 3725–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor Y, Katzir N, Meir A, Yaniv-Yaakov A, Sa’ar U, Baumkoler F, Lavee T, Lewinsohn E, Schaffer A, Burger J (2007) Induced mutagenesis to augment the natural genetic variability of melon (Cucumis melo L.). Isr J Plant Sci 55: 159–169 [Google Scholar]
- Tadmor Y, King S, Levi A, Davis A, Meir A, Wasserman B, Hirschberg J, Lewinsohn E (2005) Comparative fruit colouration in watermelon and tomato. Food Res Int 38: 837–841 [Google Scholar]
- Tadmor Y, Larkov O, Meir A, Minkoff M, Lastochkin E, Edelstein M, Levin S, Wong J, Rocheford T, Lewinsohn E (2000) Reversed-phase high performance liquid chromatographic determination of vitamin E components in maize kernels. Phytochem Anal 11: 370–374 [Google Scholar]
- Telfer A. (2002) What is beta-carotene doing in the photosystem II reaction centre? Philos Trans R Soc Lond B Biol Sci 357: 1431–1439; discussion 1439–1440, 1469–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzuri G, Zhou X, Chayut N, Yuan H, Portnoy V, Meir A, Sa’ar U, Baumkoler F, Mazourek M, Lewinsohn E, et al. (2015) A ‘golden’ SNP in CmOr governs the fruit flesh color of melon (Cucumis melo). Plant J 82: 267–279 [DOI] [PubMed] [Google Scholar]
- Walter MH, Strack D (2011) Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep 28: 663–692 [DOI] [PubMed] [Google Scholar]
- Yuan H, Owsiany K, Sheeja T, Zhou X, Rodriguez C, Li Y, Chayut N, Yang Y, Welsch R, Thannhauser T, et al. (2015a) A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis. Plant Physiol 169: 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zhang J, Nageswaran D, Li L (2015b) Carotenoid metabolism and regulation in horticultural crops. Hortic Res 2: 15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ (2011) High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949 [DOI] [PubMed] [Google Scholar]
- Zhou X, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu J, Thannhauser TW, Li L (2015) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci USA 112: 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.