PHOTOPERIOD1 (Ppd-H1) and VERNALIZATION1 (HvVRN1) interact to control reproductive development under high ambient temperature in barley.
Abstract
Ambient temperature has a large impact on reproductive development and grain yield in temperate cereals. However, little is known about the genetic control of development under different ambient temperatures. Here, we demonstrate that in barley (Hordeum vulgare), high ambient temperatures accelerate or delay reproductive development depending on the photoperiod response gene PHOTOPERIOD1 (Ppd-H1) and its upstream regulator EARLY FLOWERING3 (HvELF3). A natural mutation in Ppd-H1 prevalent in spring barley delayed floral development and reduced the number of florets and seeds per spike, while the wild-type Ppd-H1 or a mutant Hvelf3 allele accelerated floral development and maintained the seed number under high ambient temperatures. High ambient temperature delayed the expression phase and reduced the amplitude of clock genes and repressed the floral integrator gene FLOWERING LOCUS T1 independently of the genotype. Ppd-H1-dependent variation in flowering time under different ambient temperatures correlated with relative expression levels of the BARLEY MADS-box genes VERNALIZATION1 (HvVRN1), HvBM3, and HvBM8 in the leaf. Finally, we show that Ppd-H1 interacts with regulatory variation at HvVRN1. Ppd-H1 only accelerated floral development in the background of a spring HvVRN1 allele with a deletion in the regulatory intron. The full-length winter Hvvrn1 allele was strongly down-regulated, and flowering was delayed by high temperatures irrespective of Ppd-H1. Our findings demonstrate that the photoperiodic and vernalization pathways interact to control flowering time and floret fertility in response to ambient temperature in barley.
Climate models predict that an increase in global average temperature will have large impacts on crop yield (Lobell et al., 2011). High temperatures are particularly critical during plant reproductive development and affect flowering time, flower fertility, and seed set. To sustain high crop yields under changing climatic conditions, it is important to understand the genetic basis of plant development in response to ambient temperature.
Temperature-dependent flowering is regulated by the vernalization and ambient temperature pathways. Whereas vernalization requires long periods of cold during the winter, the ambient temperature pathway modulates flowering in response to short-term temperature changes (Wigge, 2013). Research in the model plant Arabidopsis (Arabidopsis thaliana), a facultative long-day (LD) plant, has demonstrated that the temperature and photoperiod pathways interact to control reproductive development. For example, high temperature accelerates flowering and overcomes the delay in flowering commonly observed under short photoperiods in Arabidopsis (Balasubramanian et al., 2006). Early flowering in response to high temperature was correlated with an increase in the expression of the floral integrator gene FLOWERING LOCUS T (FT) independently of day length (Halliday et al., 2003; Balasubramanian et al., 2006). The FT protein acts as a long-distance signal (florigen) that conveys the information to induce flowering from leaves to the shoot meristem (Corbesier et al., 2007; Jaeger and Wigge, 2007; Kobayashi and Weigel, 2007; Mathieu et al., 2007; Tamaki et al., 2007). In addition, recent studies have identified EARLY FLOWERING3 (ELF3), a repressor of light signals to the circadian clock as an essential component of ambient temperature response (Thines and Harmon, 2010). ELF3 forms together with ELF4 and LUX ARRHYTHMO (LUX), the so-called “evening complex” (EC) that functions as a night-time repressor of gene expression in the circadian clock of Arabidopsis (Nusinow et al., 2011; Herrero et al., 2012). The circadian clock is an autonomous oscillator that produces endogenous biological rhythms with a period of about 24 h and consists of at least three interlocking feedback loops. The core loops comprise (1) the inhibition of EC genes by CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) late at night, (2) the inhibition of PSEUDO RESPONSE REGULATOR (PRR) genes by the EC early at night, and (3) the inhibition of LHY/CCA1 by TIMING OF CAB EXPRESSION1/PRR1 in the morning (Pokhilko et al., 2012).
Several independent studies have recently found that elevated temperatures, specifically during dark periods, inhibit the activity of the EC by an unknown mechanism (Box et al., 2015; Mizuno et al., 2014; Thines et al., 2014; Raschke et al., 2015), leading to increased expression of PHYTOCHROME-INTERACTING FACTOR4 (PIF4; Koini et al., 2009). PIF4 binding to the promoter of FT and consequent transcriptional activation of FT is promoted by an improved chromatin accessibility through temperature-dependent histone modifications at the FT promoter (Kumar and Wigge, 2010; Kumar et al., 2012). However, high temperature also accelerated flowering in pif4 mutants under long photoperiods, suggesting that a PIF4-independent thermoresponsive flowering pathway acts through components of the photoperiod pathway (Koini et al., 2009; Press et al., 2016).
In addition, the MADS-box genes SHORT VEGETATIVE PHASE-like (SVP), FLOWERING LOCUS C (FLC), and FLOWERING LOCUS M (FLM; MAF1) play a role in the thermosensory regulation of flowering in Arabidopsis (Balasubramanian et al., 2006; Lee et al., 2007; Gu et al., 2013). Loss of function of either SVP or FLM results in partial temperature-insensitive early flowering (Balasubramanian et al., 2006; Lee et al., 2007, 2013; Posé et al., 2013). Moreover, FLM is subject to temperature-dependent alternative splicing (Balasubramanian et al., 2006; Sureshkumar et al., 2016) resulting in two major splice forms, that either facilitate or inhibit SVP dependent repression of FT, and the floral homeotic genes SUPPRESSOR OF OVEREXPRESSION1 and SEPALLATA (Posé et al., 2013). Interestingly, a structural polymorphism in the first intron of FLM affects its expression, splicing, and also regulates flowering predominantly at lower ambient temperatures (Lutz et al., 2015). Such structural polymorphisms within the first intron are typical within the family of MADS-box transcription factor genes and play an important role for expression variation and possibly adaptation to different environments across different species (Hong et al., 2003; Distelfeld et al., 2009; Schauer et al., 2009; Yoo et al., 2011).
While flowering-time control in response to temperature is well described in Arabidopsis, little is known about the genetic determinants of ambient temperature response in cereal grasses (Bullrich et al., 2002; Appendino and Slafer, 2003; Lewis et al., 2008; Hemming et al., 2012). In barley (Hordeum vulgare), a complex interplay between day length and temperature in the regulation of flowering has been reported. Under LD conditions, barley plants accelerated reproductive development at 25°C compared with 15°C, whereas the opposite was the case under short days (SDs) (Hemming et al., 2012). In contrast to Arabidopsis, the transcript level of the barley homolog of FT was not influenced by temperature, and no clear candidate genes for the integration of thermal signals into the flowering-time pathways have been identified so far (Hemming et al., 2012). Barley is a facultative LD plant and is characterized by winter and spring growth habits as determined by natural variation at the two vernalization genes, VERNALIZATION1 (HvVRN1; HvBM5a) and HvVRN2 (Yan et al., 2003, 2004; Trevaskis et al., 2006). Winter types accelerate flowering after a prolonged period of cold (vernalization), whereas spring barley does not respond to vernalization. The MADS-box gene HvVRN1 is characterized by a series of different deletions and insertions in the first regulatory intron, which has been linked to differences in vernalization response and flowering behavior (Hemming et al., 2009). Photoperiod response, rapid flowering under LDs, is determined by natural variation at the PHOTOPERIOD-H1 (Ppd-H1) gene, which is homologous to the PRR genes of the circadian clock in Arabidopsis (Turner et al., 2005). The wild-type allele is prevalent in winter barley, while a natural mutation in the conserved CCT domain of Ppd-H1 causes a delay in flowering under LDs and is predominant in spring barley from cultivation areas with long growing seasons (Turner et al., 2005; von Korff et al., 2006, 2010; Wang et al., 2010). Ppd-H1 induces flowering under LDs by up-regulating HvFT1, the barley homolog of FT in Arabidopsis (Turner et al., 2005; Campoli et al., 2012b). Ppd-H1 is repressed during the night by HvELF3, HvLUX1, and PHYTOCHROME C, and mutations in these genes result in a day-neutral up-regulation of HvFT1 and early flowering (Faure et al., 2012; Zakhrabekova et al., 2012; Campoli et al., 2013; Pankin et al., 2014). Consequently, the major vernalization and photoperiod response genes are known in barley, whether these also play a role for thermoresponsive flowering is not known.
In Arabidopsis, commonly used macroscopic indicators of reproductive phase change or floral transition are time to bolting or rosette leaf number under the first open floral bud (Pouteau and Albertini, 2009). Under optimal conditions, floral transition, bolting, and flowering are well correlated in Arabidopsis. In barley, most stages of reproductive development, including flowering, occur within the leaf sheath and can therefore only be scored upon dissection of the shoot. Waddington et al. (1983) developed a quantitative scale for barley and wheat development based on the morphogenesis of the shoot apex and carpels. This scale is based on the progression of the most advanced floret primordium and carpel of the inflorescence. The enlargement of the apical dome at Waddington stage (W) 1.0 represents an apex that is transitioning to a reproductive state and indicates the end of the vegetative phase. The emergence of the first floret primordia on the shoot apex at the double ridge stage (W2.0) specifies a reproductive shoot apical meristem (SAM). At the stamen primordium stage (W3.5), the first floral organ primordia differentiate, and stem elongation initiates. In barley, the induction of floret primordia on the inflorescence continues until the awn primordium stage (W5.0). Anthesis and pollination of the most advanced floret occurs at the last stage of the Waddington scale (W10.0). This last step can be scored macroscopically because it is marked by the emergence of awn tips from the top of the leaf sheath (heading). Most commonly, flowering is scored as heading in barley. However, the different phases of shoot apex development differ in their sensitivity to environmental cues and are controlled by different genetic factors, so that floral transition and flowering may not be correlated and separated in time by many weeks (Digel et al., 2015). Variation in the timing of different developmental phases in turn affects the number of floret primordia, fertile flowers, and seeds per spike (Digel et al., 2015). To better understand the effects of temperature on development, it is therefore important to investigate the effects of environmental and genetic variation on individual phases of shoot apex development.
The objective of present study was to elucidate the genetic control of reproductive development under high ambient temperature in barley. We show that high ambient temperature delays the phase and reduces the amplitude of clock gene expression. Further, we demonstrate that under high ambient temperature, flowering time and seed number are controlled by interactions between Ppd-H1 and HvVRN1 and correlate with expression levels of the BARLEY MADS-box genes HvBM3 and HvBM8 in the leaf. These findings provide new insights into the genetic and molecular control of flowering time and inflorescence development under high ambient temperature in barley.
RESULTS
High Ambient Temperature Delays Reproductive Development and Reduces Seed Set in Spring Barley
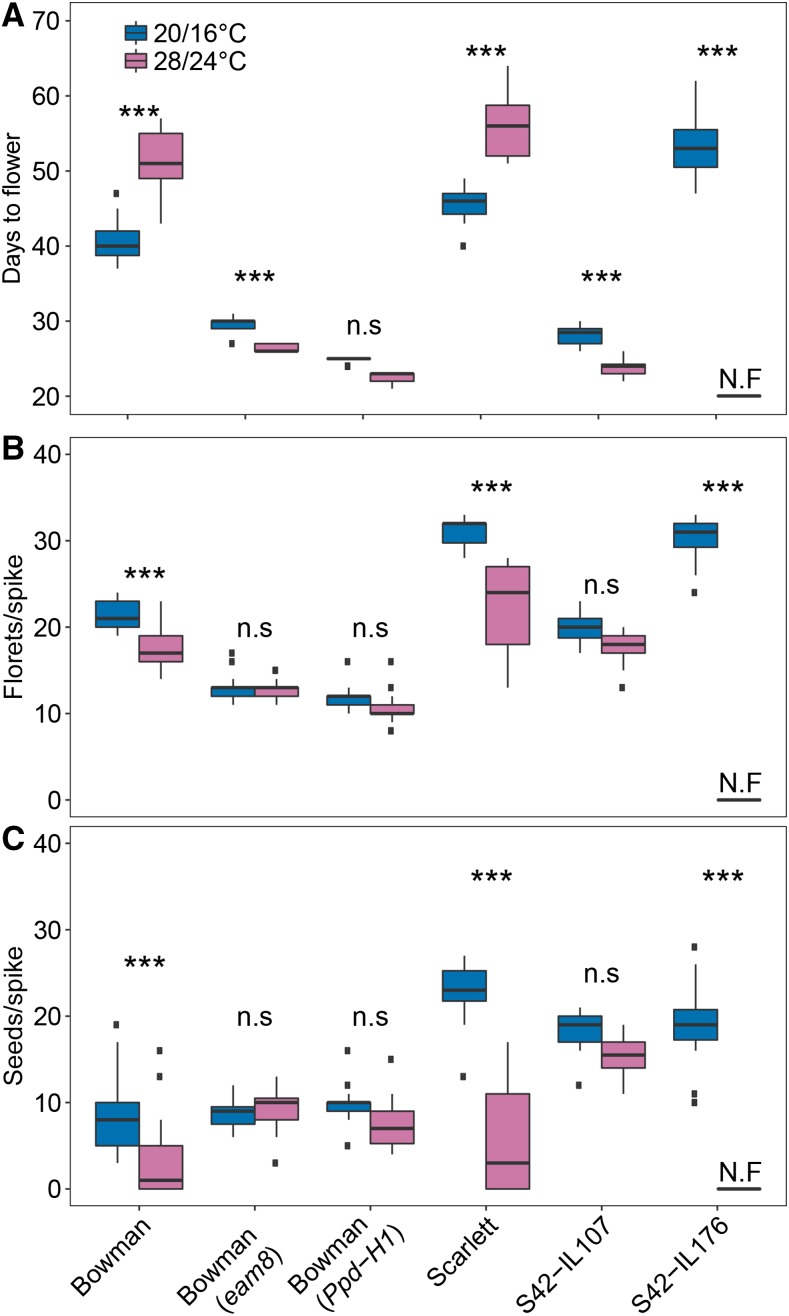
To examine the effect of high ambient temperature on flowering in barley, we scored the spring barley genotypes Bowman and Scarlett for days to flowering under control (20°C/16°C) and high temperatures (28°C/24°C) in LDs. These genotypes carry a mutated ppd-H1 allele, a functional HvELF3 allele, and a spring HvVRN1 allele and therefore do not respond to vernalization and are late flowering under LDs. Flowering was significantly delayed in both Bowman and Scarlett under high as compared to control temperatures (Fig. 1A). In addition, high temperature reduced floret and seed number per spike in both genotypes (Fig. 1, B and C). The total number of florets and seeds per spike were reduced in Bowman by 19% and 34% and in Scarlett by 30% and 74%, respectively, at high compared to control temperatures (Fig. 1, B and C). Under short-day condition (8 h light/16 h dark), Bowman and Scarlett plants never flowered neither under control nor under high ambient temperature conditions (data not shown).
Figure 1.
High ambient temperature affects flowering time, floret, and seed number per main spike in barley. Days to flower (A), the number of florets (B), and the number of seeds per main spike (C) under control (blue; 20°C/16°C, day/night) and high ambient temperatures (pink; 28°C/24°C, day/night) in the spring barley varieties Bowman and Scarlett and the derived ILs Bowman(eam8) (Hvelf3), Bowman(Ppd-H1), S42-IL107 (Ppd-H1), and S42-IL176 (Hvvrn1). Flowering time, floret, and seed number were recorded for 20 plants per genotype and treatment under LDs (16 h light/8 h night). N.F indicates nonflowering plants. Statistical differences were calculated by a two-factorial ANOVA and a post-hoc Tukey’s multiple comparison test: *P < 0.05, **P < 0.01, ***P < 0.001; n.s = nonsignificant.
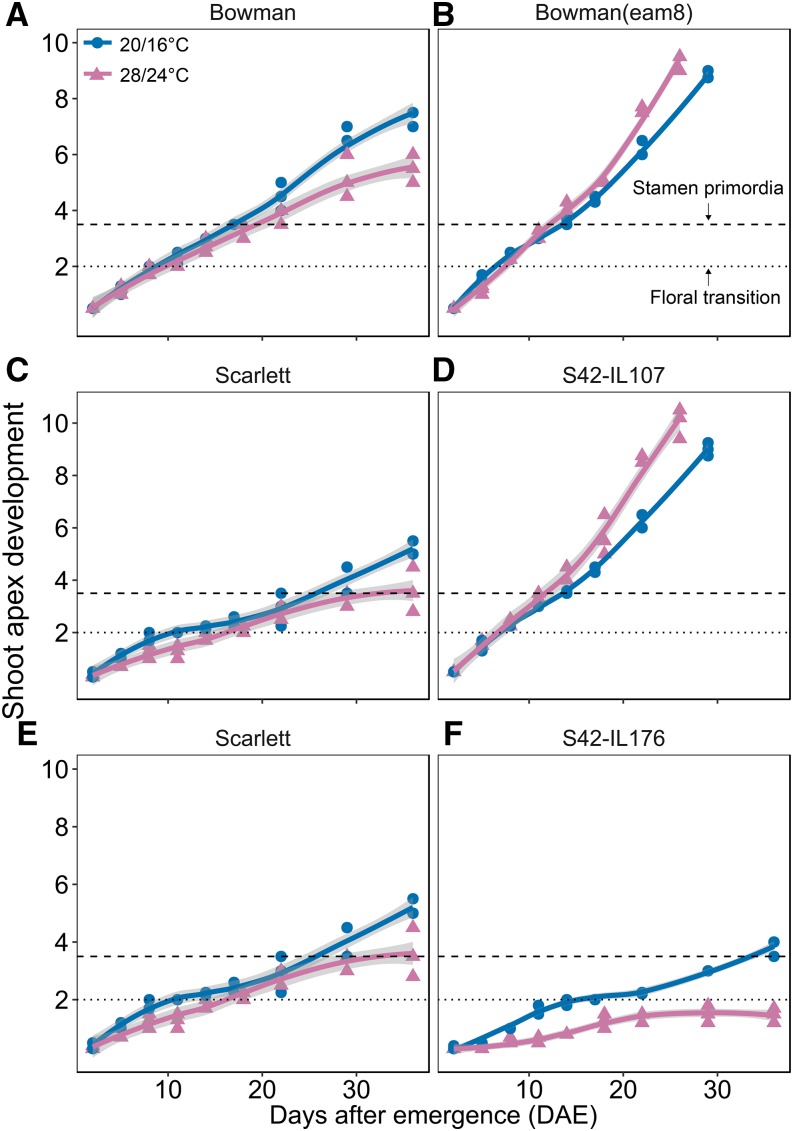
The effect of increased temperature on floral development was evaluated by monitoring the progression of the main shoot apex (MSA) in Bowman and Scarlett plants grown at 20°C/16°C and 28°C/24°C according to the Waddington scale (Waddington et al., 1983). Microscopic dissection of the MSA revealed that high temperature did not have a strong effect on floral transition (W2.0), but greatly delayed the late reproductive phase of inflorescence development (after W3.5) both in Bowman and Scarlett (Fig. 2, A and C). In summary, high ambient temperature primarily delayed inflorescence development and reduced the number of seeds per spike in the spring barley genotypes Bowman and Scarlett.
Figure 2.
High ambient temperature affects MSA development in barley. Microscopic development of the MSA was scored under control (blue; 20°C/16°C, day/night) and high ambient (pink; 28°C/24°C, day/night) temperatures every 3 d according to the Waddington scale (Waddington et al., 1983). MSA development was delayed under high compared to control temperature in Bowman (A) and Scarlett (C and E), accelerated in Bowman(eam8) (B) and S42-IL107 (D), and further delayed floral transition in S42-IL176 (F). Three or four plants per genotype were dissected at each time point in each treatment under LDs (16 h light/8 h night). Statistical differences (P < 0.05) were calculated using a polynomial regression model at 95% confidence interval (Loess smooth line).
High Ambient Temperature Accelerates Flowering Time in Genotypes with a Nonfunctional Hvelf3 Allele and a Dominant Ppd-H1 Allele
In Arabidopsis, the circadian clock and photoperiod pathways modulate ambient temperature responses to regulate flowering. Therefore, we further characterized reproductive development in introgression lines (ILs) with a nonfunctional Hvelf3 or dominant Ppd-H1 alleles under control and high ambient temperatures. HvELF3 is a component of the EC in Arabidopsis and represses Ppd-H1 expression in the night in barley (Faure et al., 2012). Therefore, the barley Hvelf3 mutant genotype is characterized by high expression of Ppd-H1 during the night (Faure et al., 2012). The IL Bowman(eam8) carrying a nonfunctional Hvelf3 allele in the background of Bowman and the ILs S42-IL107 and Bowman(Ppd-H1) with the wild-type Ppd-H1 gene in the background of Scarlett and Bowman were analyzed along with the parental genotypes for flowering time, floret fertility, and seed set. In addition, the microscopic development of the MSA was evaluated in Scarlett, Bowman, S42-IL107, and Bowman(eam8) under control and high ambient temperatures.
Microscopic dissection of the MSA revealed that in contrast to Bowman with a delayed development under high temperatures, Bowman(eam8) showed an accelerated MSA development at 28°C/24°C compared to 20°C/16°C (Fig. 2B). As a result, Bowman(eam8) plants flowered on average 5 d earlier at high compared to control temperatures (Fig. 1A).
Since HvELF3 might control flowering time through its downstream target Ppd-H1, we evaluated if variation at Ppd-H1 mediated the flowering response under high ambient temperature. In contrast to the parental lines, S42-IL107 and Bowman(Ppd-H1) plants flowered on average 7 and 2 d earlier under high ambient compared to control temperatures (Fig. 2D). The dissection of the MSA in Scarlett and S42-IL107 revealed that high ambient temperature accelerated in particular the phase of stem elongation and inflorescence development (Fig. 2D). In addition, the ANOVA for floret and seed number revealed a significant interaction between Ppd-H1 and HvELF3 with temperature (Supplemental Table S1). High ambient temperatures caused a larger reduction in floret and seed number in Bowman and Scarlett than in the ILs Bowman(eam8), S42-IL107, and Bowman(Ppd-H1) (Fig. 1, B and C; Supplemental Table S1).
Taken together, high ambient temperature affected inflorescence development and flowering time in a HvELF3- and Ppd-H1-dependent manner. Quantitative variation in the reduction of seed number under high ambient temperatures was dependent on HvELF3 and Ppd-H1.
Variation at HvVRN1 Affects Reproductive Development under High Ambient Temperature
Natural variation in the length of the first regulatory intron of HvVRN1 has a strong effect on vernalization response in barley. Therefore, we examined whether this variation also affected the response to ambient temperature variation in barley. For this purpose, we compared the development of Scarlett with that of S42-IL176. Scarlett carries a spring HvVRN1 allele with a deletion in the first regulatory intron, whereas S42-IL176 carries an introgression of the full-length winter Hvvrn1 allele. Although high ambient temperature delayed reproductive development in both genotypes, the effect was more pronounced in S42-IL176, which did not undergo floral transition and did not flower until 160 d after emergence (DAE) when the experiment was stopped (Fig. 2F, 1A). Consequently, the full-length intron of Hvvrn1 was correlated with a strong delay in floral transition under high ambient temperatures. In order to assess if variation at HvVRN1 was also associated with inflorescence development in response to ambient temperature, we shifted Scarlett and S42-IL176 plants from 20°C/16°C to 28°C/24°C only after floral transition (W2.0). Under these conditions, the IL with the winter Hvvrn1 allele also showed a strong delay in inflorescence development under high ambient temperatures compared to control conditions (Supplemental Fig. S1B). Flowering was delayed by about 2 weeks under 28°C/24°C compared to 20°C/16°C. However, S42-IL176 plants were able to produce flowers and seeds, when the temperature treatment was started after floral transition (Supplemental Fig. S1C).
High Ambient Temperature Affects the Expression of Clock Genes
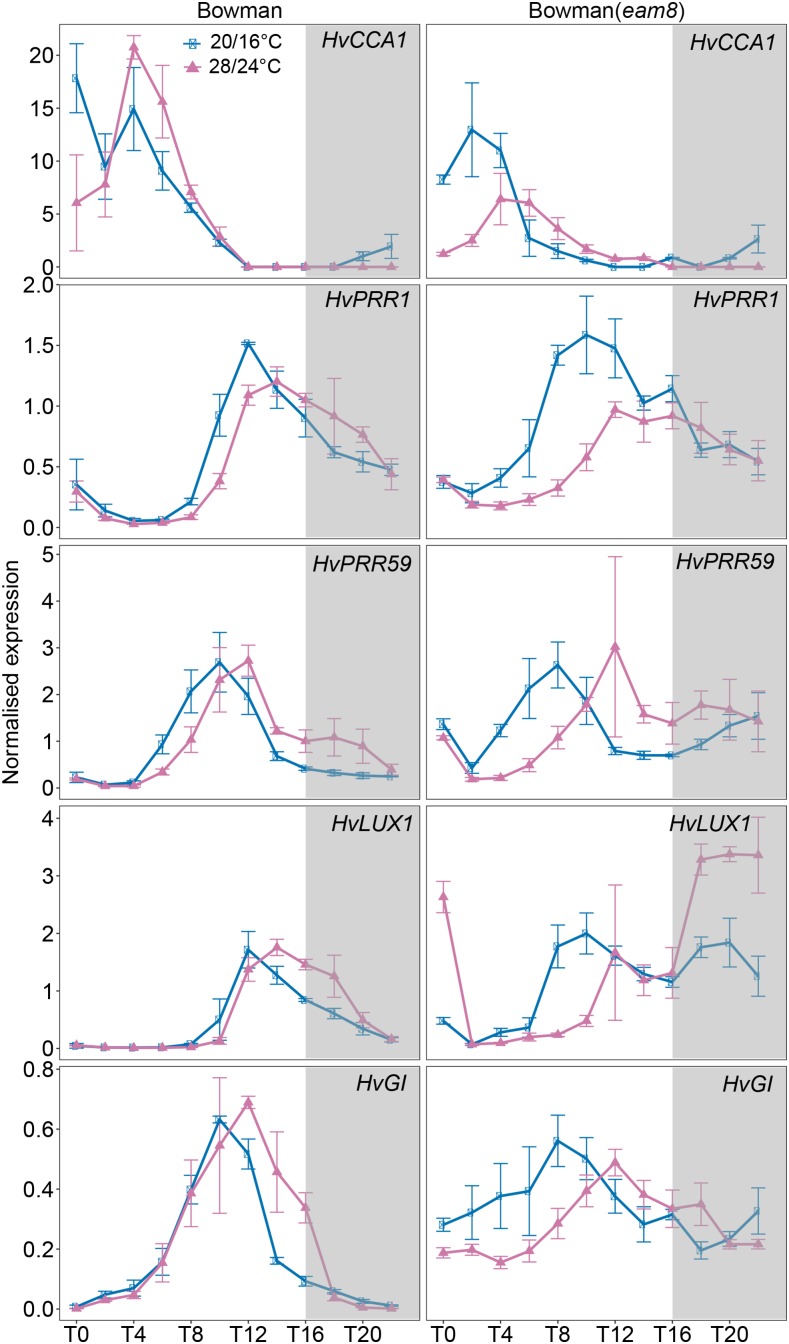
To further characterize the Ppd-H1-, HvELF3-, and HvVRN1-dependent effects of high temperature on barley development, we analyzed the expression of barley genes from the circadian clock, photoperiod, and vernalization response pathways in the parental and ILs. Because the barley clock is plastic under abiotic stresses, we first tested the effects of high ambient temperature on variation in the diurnal pattern of clock gene expression. Under control conditions, the circadian clock genes showed a diurnal pattern of expression with clock genes peaking at different times of the day, corroborating previous results (Campoli et al., 2012b; Habte et al., 2014). The expression phase of clock genes did not differ between the parental lines Scarlett, and the ILs S42-IL107 and S42-IL176, suggesting that Ppd-H1 and HvVRN1 did not affect diurnal clock oscillations. By contrast, the expression phase and shape of clock genes were significantly different between Bowman and Bowman(eam8). The expression phase of the clock genes in Bowman(eam8) was advanced by 2 h. The expression peaks were less defined and broader in Bowman(eam8) than in Bowman. Moreover, Bowman(eam8) exhibited higher levels of Ppd-H1 expression at most time points during the day compared to Bowman. Consequently, the loss-of-function mutation in HvELF3 affected the diurnal pattern of clock gene expression and caused a strong increase in Ppd-H1 expression independent of the ambient temperature.
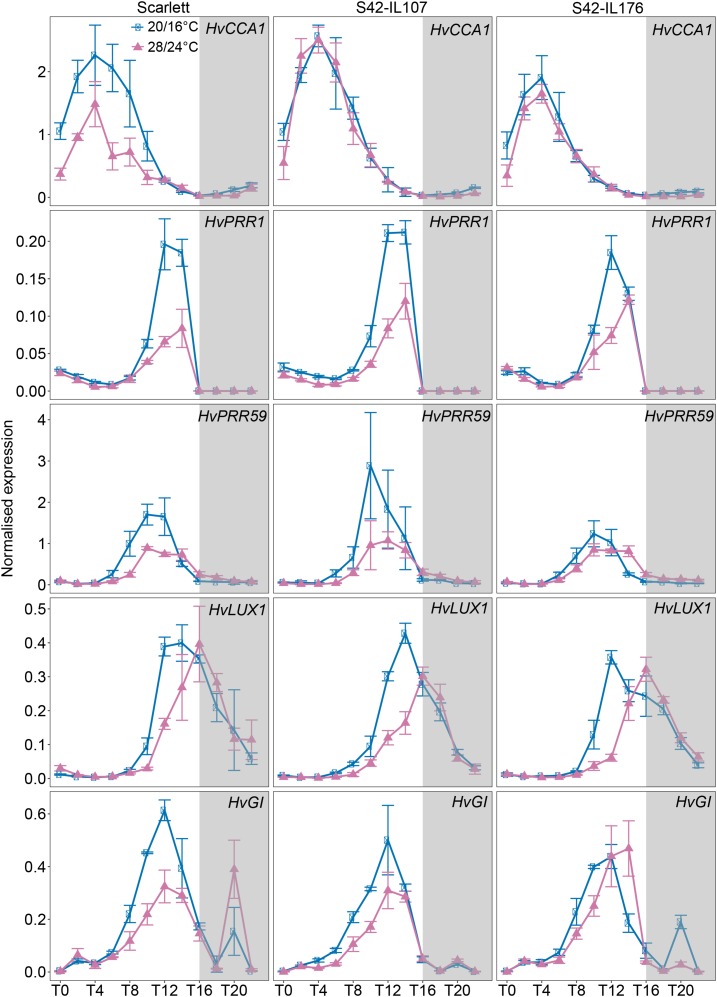
High ambient temperatures caused a decrease in the expression of clock genes, as seen for most clock genes in Scarlett and for HvCCA1 and HvPRR1 in Bowman (Figs. 3 and 4). Furthermore, the expression phase of clock genes was delayed by 4 h under high ambient temperature compared to control conditions in Scarlett and Bowman. This reduction in expression amplitude and the shift in the expression phase were also observed in all ILs, suggesting that temperature affected the phase of clock gene expression independently of the genotype.
Figure 3.
Diurnal expression patterns of circadian clock genes in Scarlett, S42-IL107, and S42-IL176 under control and high ambient temperatures. Diurnal expression of circadian clock genes was assayed every 2 h for 24 h under control (blue; 20°C/16°C, day/night) and high ambient (pink; 28°C/24°C, day/night) temperatures under LDs (16 h light/8 h night). Gray boxes indicate nights. Error bars indicate ± sd of three biological replicates.
Figure 4.
Diurnal expression of circadian clock genes in Bowman and Bowman(eam8) under control and high ambient temperatures. Diurnal expression of circadian clock genes was assayed every 2 h for 24 h under control (blue; 20°C/16°C, day/night) and high ambient (pink; 28°C/24°C, day/night) temperatures under LDs (16 h light/8 h night) are shown. Gray boxes indicate nights. Error bars indicate ± sd of three biological replicates.
High Ambient Temperature Reduces Expression of Flowering Time Genes
As the clock genes are putative upstream regulators of flowering-time genes, we investigated whether the temperature-dependent changes in clock gene expression correlated with changes in the expression of flowering-time genes. As observed for the clock genes, most flowering-time regulators showed a significantly lower expression under high ambient temperature. Ppd-H1 exhibited a reduction in expression in Scarlett, Scarlett-derived ILs, and Bowman(eam8), but not in Bowman under high ambient temperature. The expression levels of HvCO1, the barley homolog of the major Arabidopsis photoperiod response gene CONSTANS, were reduced, and the peak expression was delayed by approximately 4 h under high ambient temperature in Scarlett and Scarlett-derived ILs (Supplemental Fig. S4). While in Bowman HvCO1 expression peaked at dusk (T16) under control temperature, it showed an expression peak in the night at T20 under high ambient temperature (Supplemental Fig. S5). This suggested that HvCO1 expression was controlled by the clock and a temperature-dependent phase shift of clock genes. However, no consistent changes in the level and peak time of HvCO1 expression were observed in Bowman and Bowman(eam8) (Supplemental Fig. S5).
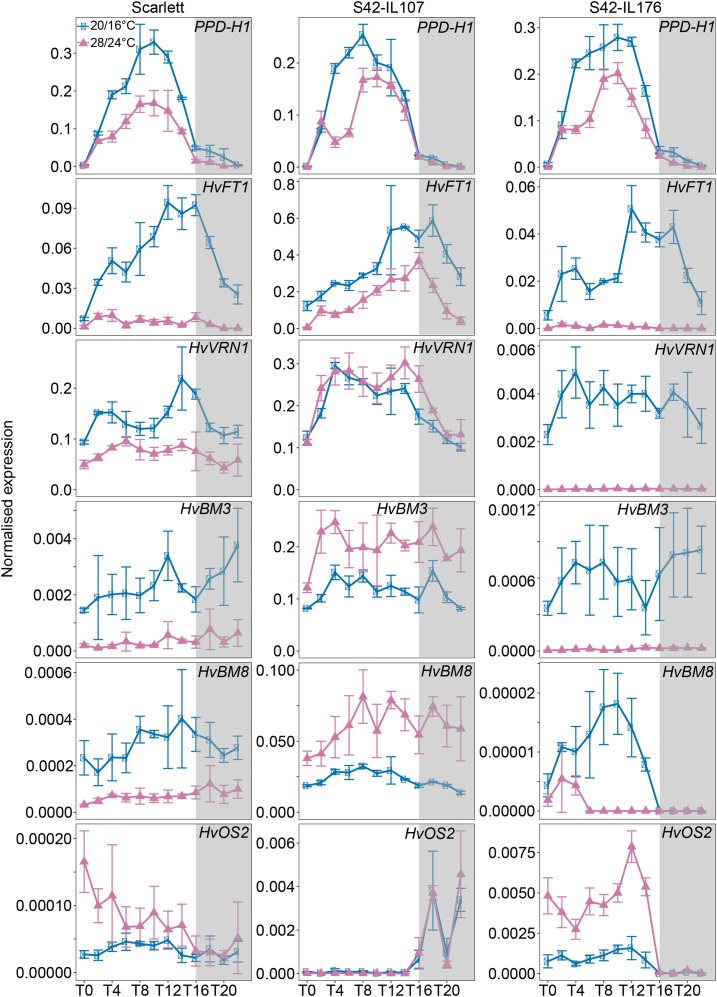
The expression levels of HvFT1, a putative target of Ppd-H1, were significantly down-regulated under high temperature in all genotypes. In addition, HvFT1 expression levels were overall significantly different between genotypes with higher transcript abundance in S42-IL107 and Bowman(eam8) and lower transcript levels in S42-IL176 compared to the parental lines (Figs. 5 and 6).
Figure 5.
High ambient temperature affects the expression of flowering-time genes in Scarlett, S42-IL107, and S42-IL176 under control and high ambient temperatures. Diurnal expression of flowering-time genes was assayed every 2 h for 24 h under control (blue; 20°C/16°C, day/night), and high ambient (pink; 28°C/24°C, day/night) temperatures under LDs (16 h light/8 h night) are shown. Gray boxes indicate nights. Error bars indicate ± sd of three biological replicates.
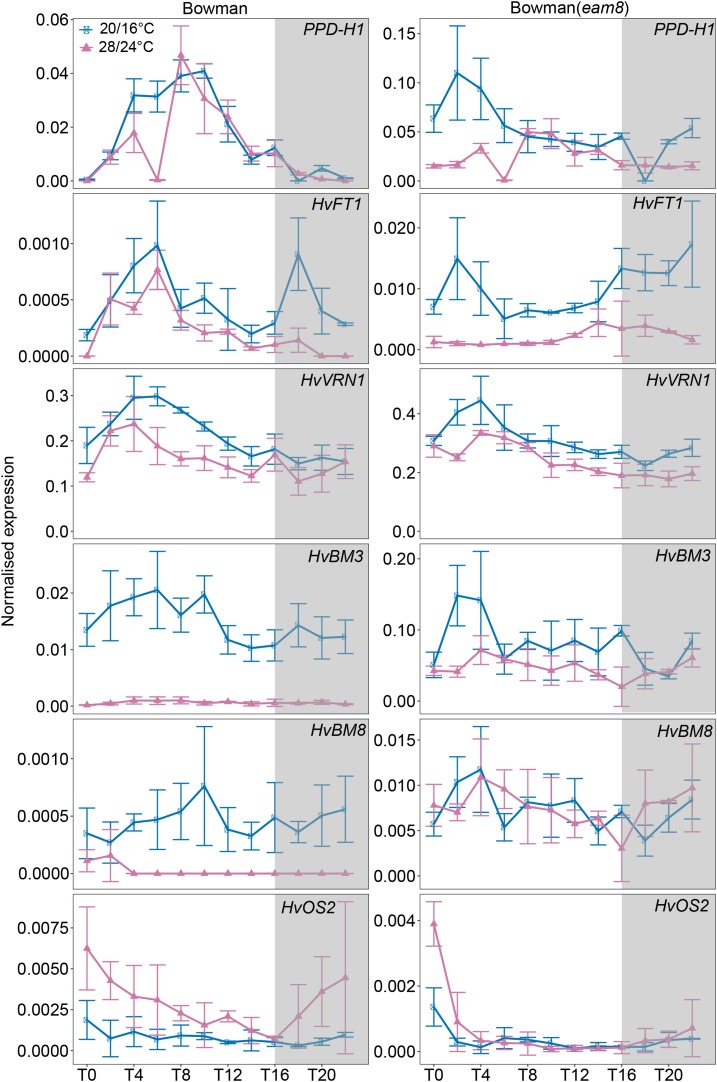
Figure 6.
Diurnal expression of flowering-time genes in Bowman and Bowman(eam8) under control and high ambient temperatures. Diurnal expression of flowering-time genes sampled every 2 h for 24 h under control (blue; 20°C/16°C, day/night) and high ambient (pink; 28°C/24°C, day/night) temperatures under LDs (16 h light/8 h night) are shown. Gray boxes indicate nights. Error bars indicate ± sd of three biological replicates.
The MADS-box genes HvVRN1, HvBM3, and HvBM8 were also strongly down-regulated under high versus control temperatures. In S42-IL176, the expression levels of the winter Hvvrn1 allele were 90-fold lower, while the expression levels of the spring HvVRN1 allele in Scarlett were only 2-fold lower under high ambient compared to control temperature (Fig. 5). This suggested that the winter allele of HvVRN1 was repressed by high ambient temperatures (Fig. 5). The expression patterns of HvBM3 and HvBM8 were comparable to those of HvVRN1 with a stronger temperature-dependent down-regulation in S42-IL176 compared to Scarlett. In contrast, S42-IL107 with a dominant Ppd-H1 allele exhibited an up-regulation of HvVRN1, HvBM3, and HvBM8 under high compared to control temperatures (Fig. 5). In Bowman(eam8), expression levels of the HvBM genes were approximately 10-fold higher compared to Bowman under control and high ambient temperature conditions. In addition, HvVRN1 and HvBM3 were only slightly down-regulated under high versus control temperatures, while expression of HvBM8 was not significantly different between control and high-temperature conditions (Fig. 6). HvOS2, a repressor of flowering and homolog of the major Arabidopsis vernalization gene FLC (Greenup et al., 2010, Ruelens et al., 2013), was up-regulated under high versus control temperatures and was controlled by Ppd-H1, HvELF3, and HvVRN1. Expression levels of HvOS2 were up-regulated under high ambient temperature in Scarlett and Bowman, but very low during the day in S42-IL107 and Bowman(eam8) under both temperatures. HvOS2 expression levels were further increased under high temperatures in S42-IL176 with the winter Hvvrn1 allele and no detectable expression of Hvvrn1. HvOS2 expression levels were consequently negatively correlated with HvVRN1 expression and controlled by ambient temperature.
Variation at Ppd-H1 and HvELF3, therefore, correlated with the temperature-dependent regulation of the MADS-box transcription factor genes. It is interesting to note, that in S42-IL107, the expression patterns of HvFT1 and the HvBM genes were not correlated under the different temperature regimes, as the HvBM genes were up-regulated, but HvFT1 was down-regulated under high compared to control temperatures. The expression patterns of the HvBM genes, but not of HvFT1, correlated with the differential flowering time in response to high ambient temperatures. Low expression of the HvBM genes under high temperatures in Scarlett and Bowman coincided with a delay in reproductive development, while accelerated inflorescence development in S42-IL107 correlated with an up-regulation of the HvBM genes under high ambient versus control temperatures. In Bowman(eam8) with accelerated development under high temperatures, the expression of HvBM3 and HvBM8 was strongly increased compared to Bowman and not very different between temperature regimes. In S42-IL176 with a winter Hvvrn1 allele, a complete down-regulation of HvFT1 and HvBM genes correlated with a strong delay in reproductive development, as this genotype did not undergo floral transition under high temperatures.
Taken together, the wild-type Ppd-H1 and a loss-of-function Hvelf3 allele correlated with an accelerated development under high compared to control temperatures and a higher expression of HvBM genes under high compared to control temperatures. In addition, variation in the regulatory region of the first intron in HvVRN1 controlled the expression of HvVRN1 itself, of the related HvBM genes and reproductive development under high ambient temperatures.
Ppd-H1 and HvVRN1 Interact to Control Inflorescence Development under High Ambient Temperatures
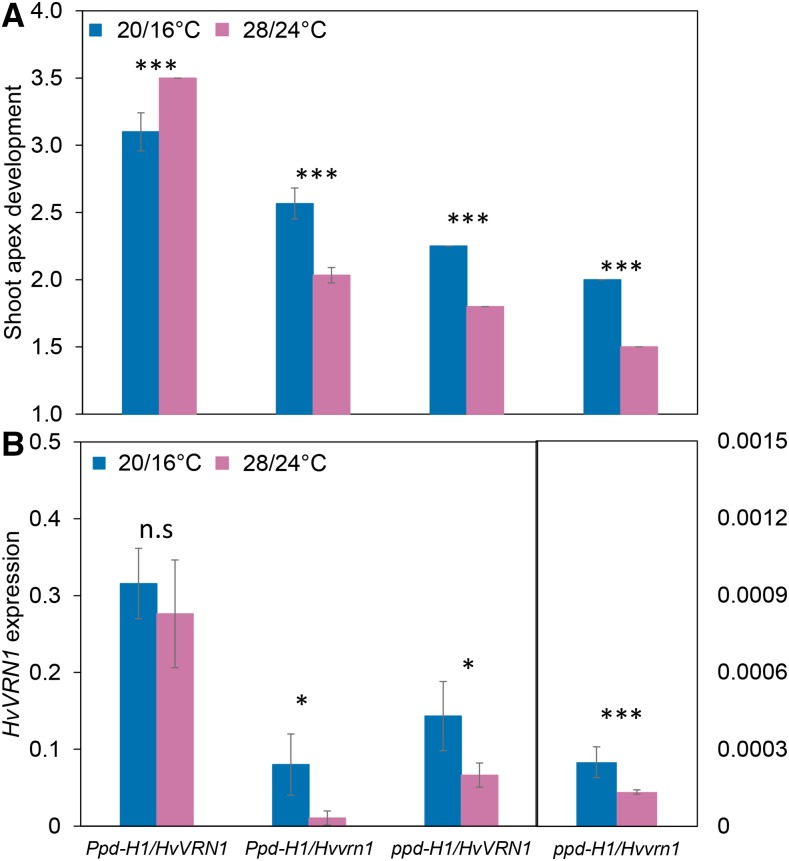
Our results showed that variation at Ppd-H1 was correlated with the expression of HvBM genes including HvVRN1 under high compared to control temperatures. Therefore, we examined if Ppd-H1 and HvVRN1 interacted to control reproductive development under different ambient temperatures. For this purpose, we analyzed MSA development and gene expression of HvVRN1 in F3 families selected from a cross between the winter barley variety Igri and the spring barley variety Golden Promise. The F3 families were segregating for variation at Ppd-H1 and HvVRN1 but were fixed for the spring alleles at the other major flowering loci HvVRN2 and HvFT1. Reproductive development was delayed under high ambient temperatures in F3 plants with a spring ppd-H1 allele irrespective of the HvVRN1 allele as seen for Scarlett and S42-IL176. In addition, under high ambient temperature, the dominant Ppd-H1 allele accelerated development in the background of a spring HvVRN1 allele, as observed for S42-IL107. F3 plants carrying a winter Hvvrn1 allele and a wild-type Ppd-H1 allele exhibited a delay in MSA development under high ambient temperature compared to control conditions (Fig. 7A). Consequently, Ppd-H1 interacted with HvVRN1 to control the development under high temperatures, where only plants with a dominant Ppd-H1 and a spring HvVRN1 allele showed an accelerated development under high versus control temperatures. The expression of the spring HvVRN1 allele was not affected in the presence of a dominant Ppd-H1 allele under high ambient versus control temperatures. However, the winter Hvvrn1 allele was down-regulated in the Ppd-H1 and ppd-H1 backgrounds under high compared to control temperature (Fig. 7B). The winter Hvvrn1 allele was stronger down-regulated than the spring HvVRN1 allele as shown for Scarlett and S42-IL176. These results indicated that Ppd-H1 interacts with HvVRN1, where a dominant Ppd-H1 allele only accelerated floral development under high ambient temperature in the background of a spring HvVRN1 allele.
Figure 7.
Ppd-H1 and HvVRN1 interact to control the development of the MSA under different ambient temperatures. Microscopic changes in MSA development were scored under control (blue; 20°C/16°C, day/night) and high ambient (pink; 28°C/24°C, day/night) temperatures under LDs (16 h light/8 h night) in F3 families derived from a cross between the winter barley Igri and the spring barley Golden Promise. Selected F3 families segregated for Ppd-H1 and HvVRN1 and were fixed for the spring alleles at HvVRN2 (deleted) and HvFT1. Early MSA development was accelerated under high temperature in Ppd-H1/HvVRN1 and delayed in Ppd-H1/Hvvrn1, ppd-H1/HvVRN1, and ppd-H1/Hvvrn1. Significant differences were determined by a two-way ANOVA and a Tukey HSD pairwise comparison test: *P < 0.05, **P < 0.01, ***P < 0.001., n.s = nonsignificant.
DISCUSSION
Understanding how ambient temperature controls plant development and eventually grain yield in crop plants is gaining importance in the light of a predicted increase in average global temperatures. The circadian clock influences plant adaptation to different abiotic stresses and controls many different output traits, including plant development. Furthermore, the circadian clock itself is altered in response to changing environmental conditions. For example, osmotic stress increased the amplitude and advanced the expression phase of clock genes in barley, and high salinity resulted in a lengthening of the circadian period in wheat (Erdei et al., 1998; Habte et al., 2014). We found that an increase in ambient temperature from 20°C/16°C to 28°C/24°C decreased expression levels and delayed the phase of clock gene expression. Although the clock is temperature-compensated and maintains an approximately 24 h period over a range of ambient temperatures (Pittendrigh, 1954; Gould et al., 2006; Salomé et al., 2010), previous studies have reported changes in the expression phase and amplitude of oscillator components under different temperatures. For example, in Arabidopsis peak expression levels of CCA1 and LHY RNA rhythms increased in amplitude as temperatures decreased from 17°C to 12°C (Gould et al., 2006; Mizuno et al., 2014). Temperatures of above 30°C are considered as heat stress for temperate cereals (Barnabás et al., 2008). However, an induction of a stress response when increasing the temperature from 20°C to 28°C cannot be excluded. Therefore, the observed alterations in clock oscillations in this work may be related to changes in the level of stress-response hormones. In Arabidopsis, application of the stress hormone abscisic acid lengthened the period of the Arabidopsis clock (Hanano et al., 2006), probably through evolutionary conserved ABA-responsive elements (ABREs) ABREs present in the promoters of TIMING OF CAB EXPRESSION1, LHY, and CCA1 (Bieniawska et al., 2008; Spensley et al., 2009; Picot et al., 2010; Habte et al., 2014). In addition, the heat shock transcription factor HsfB2b repressed transcription of PRR7 at high temperatures and in response to drought (Kolmos et al., 2014). Salomé et al. (2010) and Kolmos et al. (2014) found that the PRR genes are important for the temperature compensation of the clock in Arabidopsis, as high temperature led to overcompensation and lengthening of the period in a HsfB2b overexpression line or double prr7/9 mutant. In our study, the changes in clock gene expression under high ambient temperature were also observed in S42-IL107 and Bowman(eam8), suggesting that these temperature-mediated changes of the clock were not controlled by the PRR homolog Ppd-H1 or its upstream regulator HvELF3. In addition, the down-regulation of all PRR genes under high ambient temperature suggested that the repressive EC consisting of HvELF3, HvELF4, and HvLUX1 was not reduced in its activity under high temperature in barley as demonstrated for Arabidopsis (Mizuno et al., 2014).
Although the function of clock plasticity under different environmental conditions is not well understood, it may affect the expression of different clock output genes and related traits. We observed that the altered clock expression patterns correlated with changes in the diurnal expression patterns of flowering-time genes. Similar to the reduction in the expression amplitudes of clock genes, the expression levels of the majority of flowering-time genes including Ppd-H1 and its downstream target HvFT1 were strongly reduced under high ambient temperatures. However, in contrast to the clock genes, temperature-dependent changes in the expression of flowering-time genes were controlled by Ppd-H1, HvELF3, and HvVRN1. Ppd-H1 and HvFT1 transcripts were reduced under high compared to the control temperatures in all genotypes. In contrast, relative expression patterns of BARLEY MADS-box (BM) genes HvBM3, HvVRN1 (HvBM5a), and HvBM8 were genotype and condition specific. While in Scarlett HvVRN1, HvBM3, and HvBM8 were down-regulated, they were not down-regulated or even up-regulated under high versus control temperature in S42-IL107 and Bowman(eam8). This indicated that Ppd-H1 and HvELF3 controlled the relative expression levels of BM genes under different ambient temperature conditions. HvBM3 and HvBM8 are known targets of Ppd-H1 under LD conditions and their expression patterns correlate with the development of the inflorescence (Digel et al., 2015, 2016). In this study, we show that the effect of Ppd-H1 on HvBM3 and HvBM8 expression and flowering time was temperature dependent. Scarlett and Bowman with a mutated ppd-H1 allele showed a relatively lower expression of HvBM3, HvVRN1, and HvBM8 and a delay in floral development under high versus control temperatures. S42-IL107, with a wild type Ppd-H1 allele, exhibited a relatively higher expression of HvBM3, HvVRN1, and HvBM8 and was characterized by a faster inflorescence development under high versus control temperatures. Interestingly, functional variation at Ppd-H1 also had a strong effect on the number of florets and seeds per main spike under high ambient temperatures. While in Scarlett and Bowman the mutated ppd-H1 allele was correlated with a strong reduction of the number of seeds per main spike, S42-IL107 did not show a significant reduction in seed number under high temperatures. This suggested that Ppd-H1 affected floret fertility and seed set under high ambient temperatures, possibly by controlling the rate of development of the inflorescence.
A previous study found that high ambient temperatures accelerated flowering time under LDs, but delayed development under SDs in a winter barley with a wild-type Ppd-H1 allele (Hemming et al., 2012). The authors suggested an interaction between the photoperiod and thermosensitive pathway. Our study demonstrates that this interaction is mediated by Ppd-H1, which is functional under LDs, but not under SDs (Digel et al., 2015). Furthermore, we show that the effect of Ppd-H1 on early reproductive development under high temperatures is dependent on HvVRN1. Only in the background of a spring HvVRN1 allele or after up-regulation of Hvvrn1 by vernalization, the wild-type Ppd-H1 allele is capable of accelerating early reproductive development under high ambient temperatures.
Among the BM genes, HvVRN1 has been extensively characterized for its role in vernalization response. The winter HvVRN1 allele is up-regulated by a prolonged exposure to cold to allow flowering after winter. Our results suggest that HvVRN1 expression is negatively regulated by high ambient temperature, and this down-regulation of the winter Hvvrn1 allele correlated with a strong delay in reproductive development. The full-length winter Hvvrn1 allele in S42-IL176 was more strongly down-regulated by high ambient temperature compared to the spring HvVRN1 allele with a deletion in the first intron. Interestingly, a recent study has revealed that natural variation in the first intron of the MADS-box gene FLM was responsible for differential temperature response in Arabidopsis (Lutz et al., 2015). Consequently, structural variation in related MADS-box transcription factors may play a role in temperature adaptation across different species. In Arabidopsis, high ambient temperature accelerates plant development and growth. However, different Arabidopsis ecotypes show substantial variation in the thermosensitive response mediated by natural variation at the vernalization gene and floral repressor FLC. High expression levels of FLC in autonomous pathway mutants functioned as a potent suppressor of thermal induction (Balasubramanian et al., 2006). HvOS2, the putative barley homolog of FLC, was up-regulated under high ambient temperature in a HvVRN1-dependent manner. The barley vernalization gene and floral inducer HvVRN1, in turn, was down-regulated by high temperature, and this correlated with a down-regulation of HvBM3 and HvBM8 and a delay in floral development. Different vernalization genes might, therefore, mediate thermosensitive flowering across different species.
CONCLUSION
Our study demonstrates that an interaction of Ppd-H1 and HvVRN1 controls reproductive development and the number of seeds per spike under high ambient temperatures. These genetic interactions between Ppd-H1 and HvVRN1 are important to consider for breeding barley better adapted to climate change.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Phenotyping
Flowering time, development of the SAM, flower fertility, and seed set were scored in the spring cultivars Bowman and Scarlett and four derived ILs. Bowman and Scarlett are characterized by a mutation in the CCT domain of Ppd-H1 and the spring allele (HvVRN1-1; Hemming et al., 2009) at the vernalization response gene HvVRN1. The IL Bowman(eam8.w) carries a base pair mutation leading to a premature stop codon in HvELF3, orthologous to ELF3 in Arabidopsis (Arabidopsis thaliana; Faure et al., 2012). Bowman(Ppd-H1) carries an introgression of the dominant Ppd-H1 allele from wild barley (Hordeum vulgare; Druka et al., 2011). The Scarlett derived ILs S42-IL107 and S42-IL176 carry a dominant allele of Ppd-H1 and a recessive winter Hvvrn1 allele, respectively, both derived from wild barley (von Korff et al., 2006; Schmalenbach et al., 2008; Wang et al., 2010). In addition, development of the MSA and expression of HvVRN1 were analyzed in selected F3 families derived from a cross between the winter barley Igri and the spring barley Golden Promise. These F3 families segregated for natural variation at Ppd-H1 and HvVRN1 and were fixed for the spring alleles at HvFT1 and VRN-H2 (locus deleted, Mulki et al., 2016).
For scoring of shoot apex development, flowering time, floret number, and seed number per spike seeds were stratified at 4°C for 5 d for even germination followed by a transfer to controlled growth chambers with day/night temperatures of 20°C/16°C or 24°C/28°C, a light intensity of ∼300 µm and long photoperiods (LD; 16 h light/8 h dark). Light and temperature were monitored throughout the experiments using WatchDog series 1000 light sensors. Plants were fertilized once per week, and trays were shuffled twice a week to normalize for position effects. Plants were either shifted to high ambient temperatures after stratification or after floral transition as determined by the formation of a double ridge SAM (Waddington et al., 1983). Experiments were replicated two or three times using different randomizations to minimize the environmental effects.
The MSA of three to four representative plants per genotype (Bowman, Bowman(eam8), Scarlett, S42-IL107, and S42-IL176) and treatment were dissected every 3 to 7 d starting from the third DAE until 36 DAE. At each time point, the developmental stage of the MSA was determined according to the quantitative scale of Waddington et al. (1983), which rates the development of the most advanced floret primordium. Images of apices were obtained using the Nikon imaging software and a stereo microscope (Nikon SMZ18) equipped with a digital camera (Nikon digital sight DS-U3). The apex was dissected with a microsurgical stab knife (5 mm blade at 15° [SSC#72-1551], Sharepoint, Surgical Specialties) under the stereo microscope to confirm the developmental stage of each harvested MSA. In addition, morphological phenotypes of the main shoot, i.e. heading date (at Z49; Zadoks et al., 1974), the number of florets per spike, and the number of grains per spike, were recorded during development and at plant maturity for 20 plants per genotype.
Leaf Sampling, RNA Extraction, and Gene Expression Analysis
For the analysis of diurnal expression variation in clock and flowering-time genes in Scarlett, S42-IL107, S42-IL176, Bowman, and Bowman(eam8.w), plants were grown in 96-well trays (Einheitserde) under day/night temperatures of 20°C/16°C or 24°C/28°C, a light intensity of ∼300 µm and long photoperiods (LD; 16 h light/8 h dark). Leaf samples were harvested 21 DAE at 2 h intervals starting from the onset of light (ZT0) to the end of the night (T22). For all genotypes and treatment conditions, three biological replicates of two pooled plants were sampled per time point. Total RNA extraction, cDNA synthesis, and quantitative real-time-PCRs using gene-specific primers as detailed in Supplemental Table S2 were performed as explained in Campoli et al. (2012a, 2012b). The expression of target genes was normalized against the geometric mean of the three internal controls HvACTIN, HvGAPDH, and HvβTUBULIN (Supplemental Table S1). Two technical replicates were used for each sample, and each data point was quantified based on the titration curve for each target gene and normalized against the geometric mean of the three housekeeping genes using the LightCycler 480 Software (Roche; version 1.5).
Statistical Analysis
Significant differences in flowering time, floret, and seed number were calculated with a two-factorial ANOVA with the factors genotype and temperature treatment. In addition, least square means for each gene by temperature combination were calculated followed by a Tukey’s multiple comparison test. Significant differences in HvVRN1 expression were calculated with an ANOVA including temperature treatment, HvVRN1, and Ppd-H1 genotype and all possible interaction effects. Statistical differences in the MSA development between temperature regimes were calculated using a polynomial regression model at 95% confidence interval (Loess smooth line).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HvVRN1 affects reproductive development in response to ambient temperature after floral transition.
Supplemental Figure S2. Diurnal expression of circadian clock genes HvPRR73 and HvPRR95 in Scarlett, S42-IL107, and S42-IL176 under control and high ambient temperatures.
Supplemental Figure S3. Diurnal expression of circadian clock genes HvPRR73 and HvPRR95 in Bowman and Bowman(eam8) under control and high ambient temperatures.
Supplemental Figure S4. High ambient temperature down-regulates the expression of the flowering-time gene HvCO1 in Scarlett, S42-IL107, and S42-IL176.
Supplemental Figure S5. Effect of high ambient temperature on diurnal expression of the flowering-time gene HvCO1 in Bowman and Bowman(eam8).
Supplemental Table S1. Two-factorial ANOVA and least square means for heading date, floret, and seed number.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Kerstin Luxa, Caren Dawidson, and Andrea Lossow for excellent technical assistance.
Glossary
- LD
long day
- EC
evening complex
- SAM
shoot apical meristem
- MSA
main shoot apex
- IL
introgression line
Footnotes
This work was supported by the Max Planck Society and by DFG Priority Programme 1530 (“Flowering time control: from natural variation to crop improvement”) and the Excellence Cluster EXC1028. M.E. received a fellowship from the DAAD (German Academic Exchange Service).
References
- Appendino M, Slafer GA (2003) Earliness per se and its dependence upon temperature in diploid wheat lines differing in the allelic constitution of a major gene Eps-Am1 alleles. J Agric Sci 141: 149–154 [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabás B, Jäger K, Fehér A (2008) The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ 31: 11–38 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA (2008) Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AA, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Bullrich L, Appendino L, Tranquilli G, Lewis S, Dubcovsky J (2002) Mapping of a thermo-sensitive earliness per se gene on Triticum monococcum chromosome 1A(m). Theor Appl Genet 105: 585–593 [DOI] [PubMed] [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M (2012a) Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J 69: 868–880 [DOI] [PubMed] [Google Scholar]
- Campoli C, Pankin A, Drosse B, Casao CM, Davis SJ, von Korff M (2013) HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol 199: 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Shtaya M, Davis SJ, von Korff M (2012b) Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol 12: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Digel B, Pankin A, von Korff M (2015) Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 27: 2318–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digel B, Tavakol E, Verderio G, Tondelli A, Xu X, Cattivelli L, Rossini L, von Korff M (2016) Photoperiod-H1 (Ppd-H1) controls leaf size. Plant Physiol 172: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12: 178–184 [DOI] [PubMed] [Google Scholar]
- Druka A, Sato K, Muehlbauer GJ (2011) Genome Analysis: The state of knowledge of barley genes. In SE Ulrich, ed, Barley: Production, Improvement, and Uses, John Wiley and Sons, New York, pp. 85–111 [Google Scholar]
- Erdei L, Szegletes Z, Barabás KN, Pestenácz A, Fülöp K, Kalmár L, Kovács A, Tóth B, Dér A (1998) Environmental stress and the biological clock in plants: changes of rhythmic behavior of carbohydrates, antioxidant enzymes and stomatal resistance by salinity. J Plant Physiol 152: 265–271 [Google Scholar]
- Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, Laurie DA (2012) Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA 109: 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al. (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenup AG, Sasani S, Oliver SN, Talbot MJ, Dennis ES, Hemming MN, Trevaskis B (2010) ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol, 153: 1062–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, He Y (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4: 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habte E, Müller LM, Shtaya M, Davis SJ, von Korff M (2014) Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ 37: 1321–1327 [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC (2003) Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 33: 875–885 [DOI] [PubMed] [Google Scholar]
- Hanano S, Domagalska MA, Nagy F, Davis SJ (2006) Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B (2009) Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics 282: 107–117 [DOI] [PubMed] [Google Scholar]
- Hemming MN, Walford SA, Fieg S, Dennis ES, Trevaskis B (2012) Identification of high-temperature-responsive genes in cereals. Plant Physiol 158: 1439–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong RL, Hamaguchi L, Busch MA, Weigel D (2003) Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA (2007) FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Karsai I, Igartua E, Casas A, Kiss T, Soós V, Balla K, Bedő Z, Veisz O (2013) Developmental patterns of a large set of barley (Hordeum vulgare) cultivars in response to ambient temperature. Ann Appl Biol 162: 309–323 [Google Scholar]
- Kobayashi Y, Weigel D (2007) Move on up, it’s time for change—mobile signals controlling photoperiod-dependent flowering. Genes Dev 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kolmos E, Chow BY, Pruneda-Paz JL, Kay SA (2014) HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc Natl Acad Sci USA 111: 16172–16177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lee JH, Ryu H-S, Chung KS, Posé D, Kim S, Schmid M, Ahn JH (2013) Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342: 628–632 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH (2007) Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S, Faricelli ME, Appendino ML, Valárik M, Dubcovsky J (2008) The chromosome region including the earliness per se locus Eps-Am1 affects the duration of early developmental phases and spikelet number in diploid wheat. J Exp Bot 59: 3595–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell DB, Schlenker W, Costa-Roberts J (2011) Climate trends and global crop production since 1980. Science 333: 616–620 [DOI] [PubMed] [Google Scholar]
- Lutz U, Posé D, Pfeifer M, Gundlach H, Hagmann J, Wang C, Weigel D, Mayer KF, Schmid M, Schwechheimer C (2015) Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of FLOWERING LOCUS M. PLoS Genet 11: e1005588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55: 958–976 [DOI] [PubMed] [Google Scholar]
- Mulki MA, von Korff M (2016) CONSTANS controls floral repression by up-regulating VERNALIZATION2 (VRN-H2) in barley. Plant Physiol 170: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankin A, Campoli C, Dong X, Kilian B, Sharma R, Himmelbach A, Saini R, Davis SJ, Stein N, Schneeberger K, et al. (2014) Mapping-by-sequencing identifies HvPHYTOCHROME C as a candidate gene for the early maturity 5 locus modulating the circadian clock and photoperiodic flowering in barley. Genetics 198: 383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot E, Krusche P, Tiskin A, Carré I, Ott S (2010) Evolutionary analysis of regulatory sequences (EARS) in plants. Plant J 64: 165–176 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS. (1954) On temperature independence in the clock system controlling emergence time in Drosophila. Proc Natl Acad Sci USA 40: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol Syst Biol 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M (2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- Pouteau S, Albertini C (2009) The significance of bolting and floral transitions as indicators of reproductive phase change in Arabidopsis. J Exp Bot 60: 3367–3377 [DOI] [PubMed] [Google Scholar]
- Press MO, Lanctot A, Queitsch C (2016) PIF4 and ELF3 act independently in Arabidopsis thaliana thermoresponsive flowering. PLoS ONE 11: e0161791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke A, Ibañez C, Ullrich KK, Anwer MU, Becker S, Glöckner A, Trenner J, Denk K, Saal B, Sun X, et al. (2015) Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biol 15: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K (2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4: doi/10.1038/ncomms3280 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Weigel D, McClung CR (2010) The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolaños A, Curtis MD, Grossniklaus U (2009) Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J 59: 987–1000 [DOI] [PubMed] [Google Scholar]
- Schmalenbach I, Körber N, Pillen K (2008) Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor Appl Genet 117: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Spensley M, Kim J-Y, Picot E, Reid J, Ott S, Helliwell C, Carré IA (2009) Evolutionarily conserved regulatory motifs in the promoter of the Arabidopsis clock gene LATE ELONGATED HYPOCOTYL. Plant Cell 21: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S, Dent C, Seleznev A, Tasset C, Balasubramanian S (2016) Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat Plants 2: 16055. [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Thines B, Harmon FG (2010) Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc Natl Acad Sci USA 107: 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines BC, Youn Y, Duarte MI, Harmon FG (2014) The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J Exp Bot 65: 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140: 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- von Korff M, Léon J, Pillen K (2010) Detection of epistatic interactions between exotic alleles introgressed from wild barley (H. vulgare ssp. spontaneum). Theor Appl Genet 121: 1455–1464 [DOI] [PubMed] [Google Scholar]
- von Korff M, Wang H, Léon J, Pillen K (2006) AB-QTL analysis in spring barley: II. Detection of favourable exotic alleles for agronomic traits introgressed from wild barley (H. vulgare ssp. spontaneum). Theor Appl Genet 112: 1221–1231 [DOI] [PubMed] [Google Scholar]
- Waddington S, Cartwright P, Wall P (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot 51: 119–130 [Google Scholar]
- Wang G, Schmalenbach I, von Korff M, Léon J, Kilian B, Rode J, Pillen K (2010) Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor Appl Genet 120: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA. (2013) Ambient temperature signalling in plants. Curr Opin Plant Biol 16: 661–666 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Wu X, Lee JS, Ahn JH (2011) AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J 65: 62–76 [DOI] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14: 415–421 [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, Müller AH, Lundqvist J, Ahmann K, Dockter C, Matyszczak I, Kurowska M, Druka A, et al. (2012) Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA 109: 4326–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.