Hybridization impacts the evolution of lineages through many mechanisms, including adaptive introgression, transgressive segregation, and hybrid speciation.
Abstract
Hybridization has played an important role in the evolution of many lineages. With the growing availability of genomic tools and advancements in genomic analyses, it is becoming increasingly clear that gene flow between divergent taxa can generate new phenotypic diversity, allow for adaptation to novel environments, and contribute to speciation. Hybridization can have immediate phenotypic consequences through the expression of hybrid vigor. On longer evolutionary time scales, hybridization can lead to local adaption through the introgression of novel alleles and transgressive segregation and, in some cases, result in the formation of new hybrid species. Studying both the abundance and the evolutionary consequences of hybridization has deep historical roots in plant biology. Many of the hypotheses concerning how and why hybridization contributes to biological diversity currently being investigated were first proposed tens and even hundreds of years ago. In this Update, we discuss how new advancements in genomic and genetic tools are revolutionizing our ability to document the occurrence of and investigate the outcomes of hybridization in plants.
In natural populations, hybridization can act in opposition to divergence, introduce adaptive variation into a population, drive the evolution of stronger reproductive barriers, or generate new lineages. Hybridization is purposefully employed in the breeding of domesticated plants to take advantage of transient hybrid vigor, move desirable variation among lineages, and generate novel phenotypes. With the advent of next-generation sequencing and the availability of genomic data sets has come a tide of interest in hybridization and introgression. This includes the development of methods for detecting gene flow and a steadily growing set of empirical studies of natural hybridization (for review, see Payseur and Rieseberg, 2016) as well as a shift toward thinking of phylogenies as reticulate webs rather than strictly bifurcating trees (Mallet et al., 2016). One reason for this trend is that genomic data are particularly well suited to address the problem of detecting gene flow. Another is the growing recognition that hybridization is widespread and may have significant evolutionary consequences, a long-held belief about plants that is increasingly extended to animals (Mallet, 2005; Arnold, 2006; Abbott et al., 2013; Vallejo-Marín and Hiscock, 2016).
The study of hybridization in plants has a rich history. Verne Grant (1981) noted that much of the historical work on hybridization in plants could be partitioned into cataloging the frequency of hybridization and exploring the evolutionary consequences of hybridization. To this day, our research on hybridization still focuses on these two themes. In plants, scientific identification of hybrids is thought to have begun in 1716, when Cotton Mather described corn/maize (Zea mays) and squash (Cucurbita spp.) plants as being of hybrid origin (Zirkle, 1934). Around the same time Thomas Fairchild produced what was likely the first intentional wild plant hybrid between two Dianthus species (Zirkle, 1934). Over the next 300 years, botanists including J.E. Smith (1804), Wilhelm Olbers Focke (1881), and Leonard Cockayne (1923) made notable efforts to catalog natural hybridization (Anderson and Stebbins, 1954; Stebbins, 1959). Until the advent of molecular data, hybrids had to be identified by phenotypic comparisons, a practice that was eventually formalized into the hybrid index (Anderson, 1949).
Joseph Gottlieb Kölreuter (1766) is credited with the first rigorous investigations of the consequences of hybridization, showing, for instance, that early-generation hybrids tend to be phenotypically intermediate between parents but may be more luxuriant, while later-generation hybrids more closely resemble parental forms. Following Kölreuter (1766), many botanists have introduced or developed major hypotheses regarding the consequences of hybridization, including work on heterosis (Jones, 1917; East, 1936), transgressive segregation and adaptive introgression (Lotsy, 1916), and hybrid speciation (Winge, 1917; Müntzing, 1930). Finally, Edgar Anderson (1949) and G. Ledyard Stebbins (1950) both synthesized and developed many of these ideas, making major botanical contributions to the modern synthesis.
Our goal is to draw connections between the conception and development of ideas in plant hybridization and the recent and future work in these areas. This Update is not meant to be an exhaustive review of the literature; rather, we hope to present a handful of research areas that combine rich histories of botanical and evolutionary thought with exciting recent advancements. In particular, we consider the ways in which genomic data have changed how we think about hybridization in plants and highlight areas that we believe are especially accessible to genomic study. We also recognize that, while genomic data provide previously inaccessible insight into the evolutionary history of plant populations, they are most powerful when combined with classical experiments (i.e. to determine the strength of selection in the field or the molecular function of a particular allele).
IDENTIFYING HYBRIDIZATION
One of the greatest achievements of genomics is revealing the fundamental role of hybridization in shaping the history of life on earth. In spite of some disagreement regarding the definition of hybridization (Box 1), it is clear that a significant proportion of plant and animal taxa have experienced hybridization and introgression (Mallet, 2005). The concept of genetic introgression, defined as the movement of genetic material between parental types through the production of and mating with hybrids (Grant, 1981), predates the genomic era and was founded upon observations of increased phenotypic variation in areas of contact between plant species (Du Rietz, 1930; Marsden-Jones, 1930). Introgression was formerly inferred by using hybrid indices and pictorialized scatter diagrams, which scored individuals from putative hybrid populations based on the similarity to phenotypes of parental forms (Anderson, 1949; Grant, 1981). These indices are based on the idea that parental phenotypes are recombined in hybrids and that the proportion and distribution of these phenotypes will reflect the amount and nature of introgression. However, Anderson (1948) lamented that “Gene flow from one species to another may go far beyond any point which could be detected by ordinary morphological techniques. We shall not be able to assess the real importance of introgression until we can study genetically analyzed species in the field and determine the actual spread of certain marker genes.”
As predicted by Anderson (1948), analyses of sequence divergence, haplotype structure, and allele frequency distributions in genomic data have fundamentally improved our ability to detect hybridization and even identify introgressed loci (Rieseberg et al., 1993; Payseur and Rieseberg, 2016).
The evolutionary history of a population is reflected in the genetic variation of its genomes. Model-based methods are widely used to infer global (genome-average) and local (locus-specific) ancestry from population variation data (Gompert and Buerkle, 2013; Liu et al., 2013). For example, the program STRUCTURE uses a hierarchical Bayesian model to identify subpopulations and estimate global ancestry for each sampled individual based on allele frequency data (Pritchard et al., 2000; Porras-Hurtado et al., 2013) and has been extended to estimate locus-specific ancestry (Falush et al., 2003). Maximum likelihood-based programs, like ADMIXTURE (Alexander et al., 2009), allow for less computationally intensive estimates of genetic ancestry. Model-based methods that infer locus-specific ancestry (Falush et al., 2003; Sankararaman et al., 2008; Paşaniuc et al., 2009; Price et al., 2009) are particularly useful for detecting hybridization and introgression without requiring a priori assignment of samples into different populations and can be used on taxa without a reference genome (Vähä and Primmer, 2006; Porras-Hurtado et al., 2013). For instance, such analyses have been used to identify crop-wild introgression in chicory (Cichorium intybus) and maize (Kiær et al., 2009; Hufford et al., 2013). However, many of these model-based analyses may have difficulty distinguishing between different evolutionary histories, as they do not account for incomplete lineage sorting (ILS) or estimate the timing of introgression (Falush et al., 2016).
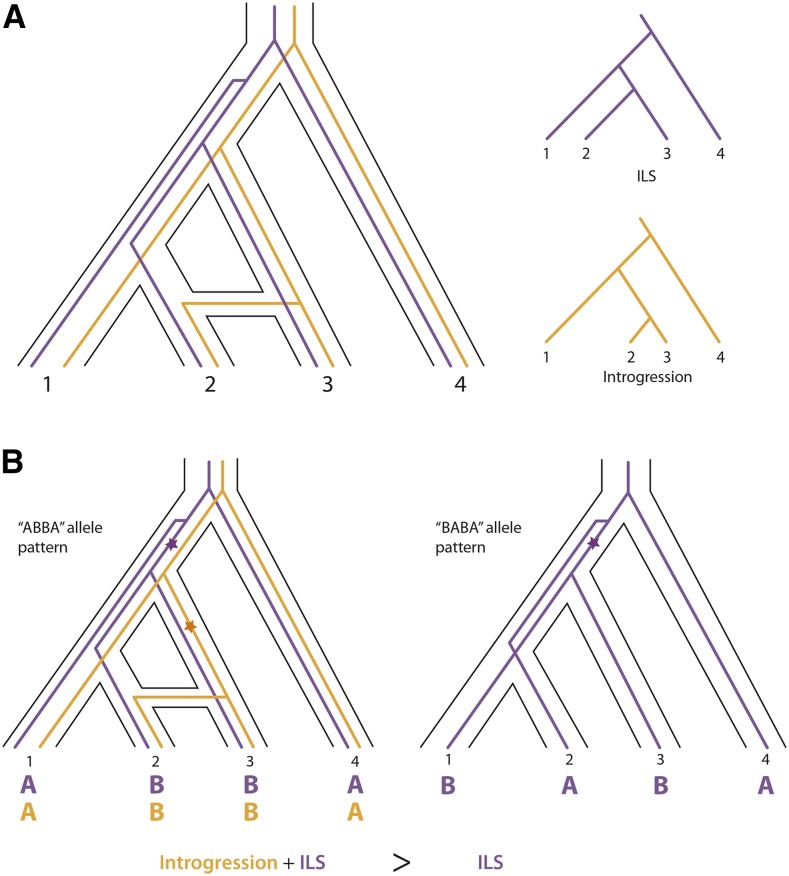
Independent mutations accumulate in the genomes of reproductively isolated taxa; therefore, the amount and pattern of genetic differences between species reveal the relative time of divergence between them. Phylogenetics-based analyses utilize this property of genetic variation to infer hybridization and introgression based on gene tree discordance and relative divergence patterns. Specifically, a sequence that is introgressed is expected to show less divergence than is expected based on the phylogenetic relationship of two lineages. A phylogenetic analysis of such loci will be discordant with the species tree (Fig. 1A). But introgression is not the only phenomenon that can cause discrepancies between gene trees. The persistence of ancestral polymorphism after the divergence of two species can produce phylogenetic signals that differ from the species tree. This phenomenon, known as ILS, produces a signal of incongruence that, in some ways, mimics introgression (Fig. 1A).
Figure 1.
Differentiating between introgression and ILS. A, Individual gene trees may be incongruent with the species tree (outlined in black) due to either ILS (purple) or introgression (orange). Genetic divergence, as indicated by total branch length, between taxa 2 and 3 is predicted to be shorter under introgression than ILS. B, The ABBA-BABA test is used to detect an excess of one pattern of discordance relative to the other in four taxon phylogenies (three ingroup taxa and an outgroup) by comparing counts of allele patterns at polymorphic sites that differ from the species tree (outlined in black). If the star symbol represents mutation from ancestral A alleles to derived B alleles, then in this example, incongruent ABBA allele patterns are due to either introgression (orange) or ILS (purple). BABA allele patterns are due to ILS alone. An equal number of incongruent ABBA and BABA allele patterns are expected under ILS alone; therefore, a significant excess of ABBA allele patterns is consistent with a history of introgression.
Several phylogenomic analyses have been developed to infer introgression in spite of ILS. The ABBA-BABA test is currently the most widely used and is based on counts of ancestral (A) and derived (B) alleles in sets of four samples with known phylogenetic relationships (i.e. three ingroups and an outgroup). Two allele patterns, ABBA and BABA, are incongruent with the species tree BBAA and can be used to infer introgression (Green et al., 2010). Under ILS, the two patterns should be equally frequent; therefore, a significant excess of one pattern over the other (as evaluated with Patterson’s D statistic) is indicative of introgression (Fig. 1B). These analyses have been used to successfully detect ancient and recent introgression in spite of high levels of ILS (Pease et al., 2016; Ru et al., 2016).
Another approach to infer reticulate evolutionary histories is to model phylogenetic networks in which introgression is represented by nodes connecting hybridizing species in a phylogenetic tree (Bapteste et al., 2013; Hahn and Nakhleh, 2016; Mallet et al., 2016). These methods have proven particularly useful for inferring the timing, magnitude, and direction of gene flow (Than et al., 2008; Solís-Lemus and Ané, 2016).
Because recombination breaks apart haplotypes over time, recent introgression is expected to generate long-shared haplotype blocks between hybridizing species, a pattern that is not predicted under ILS. Therefore, the distribution of haplotype block sizes can be used to infer introgression (Pool and Nielsen, 2009; Gravel, 2012; Mailund et al., 2012; Harris and Nielsen, 2013). These methods are less widely used because they require haplotype data from multiple individuals as well as a null distribution of expected haplotype sizes, which is not attainable in many systems.
Although tests to detect hybridization do not require the identification of exchanged genes, similar analyses have been adapted to detect the targets of introgression (Rosenzweig et al., 2016). For instance the f statistic, an expansion of Patterson’s D, is used to search for genomic regions with increased proportions of shared derived variants, likely exchanged by recent gene flow (Green et al., 2010; Durand et al., 2011). Methods to detect long-shared haplotypes also have been used to identify genes involved in adaptive introgression (Pardo-Diaz et al., 2012; Racimo et al., 2015; Dannemann et al., 2016). Finally, because introgressed loci will share a more recent common ancestor than the most recent common ancestor of hybridizing taxa, they should have a lower genetic distance in hybridizing taxa than nonintrogressed loci (Fig. 1A).
Genomic methods have dramatically improved our ability to detect introgression and have expanded the number of taxa amenable to a detailed study of hybridization. However, there are still limits to what we can learn from genomic data. For instance, the timing, direction, and magnitude of gene flow define the biological implications of hybridization. Calculating these parameters is challenging and has traditionally been conducted by modeling population divergence using theoretical frameworks such as the isolation with migration model (Nielsen and Wakeley, 2001; Hey and Nielsen, 2004). These methods are computationally demanding and make controversial evolutionary assumptions (Sousa and Hey, 2013; Payseur and Rieseberg, 2016). Models of phylogenetic networks (Than et al., 2008; Solís-Lemus and Ané, 2016; Wen et al., 2016) and the five-taxa extension of the ABBA-BABA test (Eaton and Ree, 2013; Pease and Hahn, 2015) have made progress toward evaluating the direction and magnitude of introgression, and future efforts should continue to develop such methods.
EVOLUTIONARY CONSEQUENCES OF HYBRIDIZATION
Identifying a history of hybridization still leaves the question of how hybridization affects the evolutionary trajectory of lineages. Although Kölreuter (1766) observed hybrid vigor, he more generally concluded that interspecific hybrids are usually difficult to produce and are frequently sterile. Hybrids are often inviable, sterile, or exceedingly rare, such that genetic exchange between species is not possible. Hybridization without gene flow has fewer evolutionary consequences and, therefore, is not addressed here. Instead, we focus primarily on how hybridization with gene flow affects the genetic and phenotypic composition of populations immediately and over longer evolutionary time scales. Our discussion starts with phenomena in F1 hybrids (heterosis), continues to population-level processes (transgressive segregation and adaptive introgression), and concludes with hybrid speciation and reinforcement.
Heterosis
It has long been observed that crossing two plant species or genotypes can create a hybrid with faster growth rate, more biomass at maturity, and/or greater reproductive output than its parents. This counterintuitive phenomenon is called hybrid vigor or heterosis. Both Kölreuter (1766) and Darwin (1876) described the phenomenon of heterosis in their experimental crosses of plants, but neither offered explanations to the underlying mechanism causing the pattern (Mayr, 1986; Chen, 2013). Following Shull’s (1908, 1911) pioneering experiments in maize, determining the genetic mechanism causing heterosis became one of the earliest problems in the new field of genetics. How does a hybrid that has an allele from each parent perform so much better than either of the parental sources of the alleles?
Early research on heterosis yielded two competing hypotheses that we are still investigating today: dominance (Jones, 1917) and overdominance (East, 1936). The dominance model posits that recessive deleterious alleles accumulated at different loci in each parental taxon and that, in F1 hybrids, these deleterious alleles are masked by beneficial alleles from the other parent. The overdominance hypothesis posits that, at loci contributing to heterosis, the heterozygous genotype is superior to both homozygous genotypes. Recent advances in genetic and genomic methods have allowed for more thorough characterization of the mechanisms causing heterosis and also have implicated epistatic interactions among alleles at multiple loci, epigenetic modifications to the genome, and the activity of small RNAs (Chen, 2013). Despite more than a century of research, the genetic basis of heterosis remains an open question. Early work tended to assume a single, common cause of heterosis (Crow, 1948), but it has become clear that multiple causal mechanisms contribute to heterosis (Grant, 1975; Kaeppler, 2012).
Quantitative trait locus (QTL) mapping experiments have been used to identify and then characterize loci contributing to heterotic phenotypes. Such studies are limited by the density and genomic coverage of genetic markers, so the most convincing genomic characterizations of heterosis come from genetic model systems including rice (Oryza sativa), maize, cotton (Gossypium hirsutum), and Arabidopsis (Arabidopsis thaliana). These genomic studies paint heterosis as the cumulative result of many loci that have a mixture of dominant, overdominant, and epistatic effects (Tang et al., 2010; Zhou et al., 2012; Shen et al., 2014; Shang et al., 2015). There is one notable exception to this pattern, a single locus controlling heterosis for yield in tomato (Solanum lycopersicum). Krieger et al. (2010) show that tomato plants heterozygous for a wild-type and a nonfunctional allele at SINGLE FLOWER TRUSS have significantly greater yield than either homozygote genotype. Thus, heterosis for yield is driven by overdominance at a single locus.
Recent genetic and genomic studies also have revealed that interactions between divergent epigenetic regulatory systems contribute to heterosis in F1 hybrids (Groszmann et al., 2013; Greaves et al., 2015). In Arabidopsis, Wang et al. (2015) demonstrate that F1 hybrids show gene expression levels outside of the parental range for defense, abiotic stress, and hormone response pathways, due in part to epigenetic regulation. In many cases, these pathways are down-regulated, consistent with the idea that there are tradeoffs between growth and defense or abiotic stress response. There is also emerging evidence from Arabidopsis and rice that small RNAs, including microRNAs and small interfering RNAs, may be involved in heterosis, as F1 hybrids often show small RNA expression levels outside of the parental range (Ng et al., 2012). Compellingly, Shen et al. (2012) were able to eliminate heterosis in F1 hybrids of Arabidopsis by treatment with a DNA demethylating agent and by introducing a mutation that compromises gene regulation by small RNAs.
While the fundamental task of explaining the genetic basis of heterosis has persisted for over 100 years, recent genomic studies have made progress toward its solution. In most cases, multiple genetic mechanisms, including dominance, overdominance, epistasis, and epigenetics, act simultaneously in F1 hybrids to produce heterotic phenotypes. The implication of multiple mechanisms and many loci is consistent with the finding that levels of heterosis for different traits are not strongly correlated, suggesting that the basis of heterosis is largely trait specific (Flint-Garcia et al., 2009). Future studies should follow-up genome-wide surveys with molecular studies of individual loci to confirm that heterosis is the result of multiple genetic models acting in concert and to further our mechanistic understanding of how these different genetic models cause heterotic trait values. Although genetic and genomic studies have the potential to improve our understanding of heterosis, it is also important to continue to assess the relative importance of heterosis in natural plant systems. Phenotypic assessments of hybrid vigor versus hybrid sterility and inviability in natural and controlled environments are key to determining the contribution of heterosis in plant evolution more generally.
Transgressive Segregation
Similar to heterosis, transgressive segregation occurs when phenotypic trait values in hybrid populations fall outside the range of parental variation. Transgressive segregation demonstrates how hybridization can produce novel phenotypes and thus enable adaptation to new ecological niches, found new lineages, and play a significant creative role in evolution. Transgressive segregation is distinct from heterosis because it manifests predominantly in the F2 generation and later and may persist indefinitely once established (Rieseberg et al., 1999). This difference suggests possible distinct genetic mechanisms for the two phenomena.
Transgressive segregation is common in hybrid plant populations. Rieseberg et al. (1999) found that 97% (110 of 113) of studies reporting parental and hybrid trait values include at least one transgressive trait. Stebbins (1950) cites early observations of transgressive segregation by Lotsy (1916) and Hagedoorn and Hagedoorn-Vorstheuvel La Brand (1921) and notes the potential of transgressive traits to allow adaptation to a new ecological niche; however, he does not offer hypotheses regarding the genetic mechanism underlying this phenomenon. In the 1970s, it was assumed that the genetic mechanism was understood (Grant, 1975), and yet now we realize that, like heterosis, there are multiple possible causes of transgressive segregation requiring continued investigation.
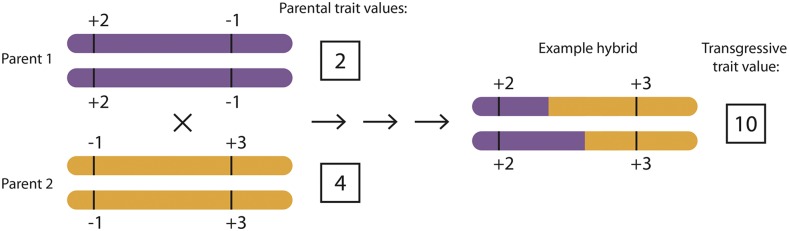
While a number of hypotheses have been proposed (Rieseberg et al., 1999), the best-supported genetic mechanisms causing transgressive segregation are complementary gene action and epistasis (Rieseberg et al., 1999; Dittrich-Reed and Fitzpatrick, 2013). The complementary gene action model requires that both parents harbor additive alleles of opposing sign at different loci affecting a multilocus trait (some + and some −), which then sort in favor of one direction in the segregating hybrids. For example, a late-generation hybrid may acquire + alleles for a trait from both parents across different loci (Fig. 2). Grant (1975) called this an oppositional multiple gene system and credits Nilsson-Ehle (1911) with one of the earliest explicit proposals of the phenomenon in wheat (Triticum aestivum). The epistasis model predicts that nonadditive interactions between loci from different parents can cause extreme trait values in hybrids. Recent advancements in genomic analyses have suggested additional mechanisms underlying transgressive segregation, including a role for small interfering RNAs (Shivaprasad et al., 2012).
Figure 2.
Complementary gene action causes transgressive segregation. Complementary gene action occurs when additive alleles for a multilocus trait act in opposition to one another in both parent lineages but sort in favor of one direction of effect in segregating hybrids. Individual loci contributing to a trait are indicated along a chromosome with their additive contribution to the trait value. The total trait value for each genotype is indicated by the boxed number. One possible hybrid genotype is depicted that has acquired all + alleles and, therefore, has a transgressive trait value.
Many QTL studies of transgressive traits find support for complementary gene action, epistasis, or both (deVicente and Tanksley, 1993; Hagiwara et al., 2006; Mao et al., 2011). For an example in Helianthus spp., see Box 2. Interestingly, oppositional QTLs are more common for morphological traits than physiological traits (Rieseberg et al., 2003b), suggesting that physiological traits are less likely to be transgressive in hybrids. It is unclear why this would be the case, but it may reflect weaker selection on morphological traits that is more permissive to the accumulation of antagonistic alleles.
Can we predict the plausibility of transgressive segregation between two populations? It has been hypothesized that genetic distance is positively correlated with the frequency of transgressive segregation (Rieseberg et al., 1999). Stelkens and Seehausen (2009) found significant evidence in favor of a positive correlation in eudicots; however, this correlation disappears when monocots are included in the analysis. Also, Rieseberg et al. (1999) found transgressive segregation to be significantly more common in intraspecific crosses than in interspecific crosses. One possible explanation for the somewhat ambiguous support for this hypothesis is that the accumulation of fixed differences causing transgressive segregation is masked by the simultaneous accumulation of detrimental genetic incompatibilities (Dittrich-Reed and Fitzpatrick, 2013). Additionally, assuming that complementary gene action is the most common mechanism, the frequency of transgressive segregation should depend not only on divergence but also on the history of selection on traits (Rieseberg et al., 2003b). Drift or stabilizing selection increases the likelihood of fixing antagonistic alleles for a polygenic trait, whereas directional selection will tend to fix alleles with effects in the same direction. Observations of transgressive segregation for agriculturally important traits in domestic plants (Hagiwara et al., 2006; Mao et al., 2011) further complicate this hypothesis, because crops tend to be under strong directional selection yet demonstrate extensive transgressive segregation. Finally, hybrids between inbred plants are much more likely to show transgressive trait values than hybrids between outbred populations (Rieseberg et al., 1999). Thus, describing the likelihood of transgressive segregation is complex and depends on divergence, history of selection, and breeding system.
Transgressive segregation appears to have multiple underlying genetic causes, which partially overlap with those found for heterosis. Complementary gene action and epistasis have amassed the most empirical support, and recent work suggests that epigenetic regulation and small RNA activity also may be important contributors. Future work should continue to investigate the genetic and molecular basis of transgressive segregation, particularly in wild populations. Additionally, experiments in the field demonstrating that transgressive trait values facilitate the adaptation of a hybrid lineage to a new ecological niche will bolster the case for a creative role of hybridization in evolution and speciation. Finally, more research is needed on the factors determining the likelihood of transgressive segregation. Are there patterns in dominance effects of loci underlying transgressive segregation? What is the history of selection on loci implicated in transgressive traits, and have these histories potentiated transgressive segregation in hybrids?
Adaptive Introgression
The production of hybrid offspring generates the potential for gene flow between parent populations. If hybrids are fertile, they may backcross with either or both of the parents, resulting in introgression. Excessive gene flow can lead to genetic swamping and the extinction of rare taxa (Levin et al., 1996; Todesco et al., 2016); however, introgression also may serve as an evolutionarily creative force by introducing new, possibly adaptive, genetic variation into a population. The idea that introgression can move adaptive variation between populations is first credited to the botanist Johannes Lotsy (1916; Stebbins, 1959), but the field’s most influential early thinker was Edgar Anderson, who coined the term “introgressive hybridization” (Anderson and Hubricht, 1938). Anderson (1948) emphasized how open ecological niches that recombined aspects of parental habitats would favor recombinant hybrid offspring that could draw from the genetic variation present in both parents, an idea he called “hybridization of the habitat.” Furthermore, Anderson and Stebbins (1954) proposed that introgression, unlike spontaneous mutation, could introduce large blocks of novel variation into a population, potentially moving an adaptive trait along with its modifiers to allow rapid differentiation into a new ecological niche. Although a number of putative examples of adaptive introgression were proposed in the genera Tradescantia, Melandrium, and Helianthus (Anderson, 1949; Stebbins, 1950), empirical study of adaptive introgression was limited by the difficulty of identifying introgressed loci underlying adaptations.
In order to demonstrate adaptive introgression, it must be shown that a variant in one population is derived from gene flow with a second population and that this variant is adaptive. Demonstrating the latter involves well-established techniques such as reciprocal transplant experiments and common garden experiments to measure selection on traits. However, work on adaptive introgression was historically limited to studying adaptive phenotypes thought to be introgressed without the ability to determine their genetic basis. Genomic analyses can now demonstrate that alleles contributing to an adaptive phenotype in hybrids are in fact introgressed. However, the likelihood of determining the genetic basis and evolutionary history of an adaptive trait will depend on its genetic architecture: adaptive introgressed traits with simple genetic architectures will be easier to detect than traits controlled by many loci.
Much attention has been paid to the exchange of potentially adaptive variation between crop plants and their wild relatives (Kwit et al., 2011; Ellstrand et al., 2013; Warschefsky et al., 2014). For example, adaptive introgression of transgenes conferring herbivore resistance from crop plants to wild relatives has been demonstrated in artificial hybrids of rice (Yang et al., 2011) and sunflower (Helianthus annuus; Snow et al., 2003). Meanwhile, Hufford et al. (2013) found evidence for adaptive introgression from wild teosinte (Zea spp.) into maize crops using a combination of genomic analyses and growth chamber experiments. Adaptive introgression also has been studied extensively in natural hybrid zones or swarms (Grant, 1981). Early researchers observed that hybrid swarms were more common in areas subject to human disturbance, highlighting the importance of habitat in the formation and persistence of hybrids (Wiegand, 1935; Anderson, 1949). Indeed, multiple models proposed to explain the persistence of stable hybrid zones invoke extrinsic selection for hybrid genotypes (Buerkle et al., 2003). Thus, many have advocated the use of natural hybrid zones for studying the process of adaptive introgression (Levin, 1979; Rieseberg and Carney, 1998). For a brief description of research done on adaptive introgression in the genus Helianthus system, see Box 2.
The advancements in genomic sequencing have enabled easier identification of introgressed loci and the detection of genomic signatures of selection in natural hybridizing populations. For example, genome scans for selection in Populus trichocarpa identified a number of candidate loci under strong selection (Geraldes et al., 2014). Further genomic analyses to detect introgressed loci between P. trichocarpa and Populus balsamifera found that at least one of the genomic regions with a history of strong selection also was introgressed from P. balsamifera (Suarez-Gonzalez et al., 2016). Phenotypic data for the sequenced individuals showed that P. trichocarpa individuals with a P. balsamifera haplotype at the region of interest had higher chlorophyll and leaf nitrogen contents than those with native P. trichocarpa haplotypes (Suarez-Gonzalez et al., 2016). Together, these genomic analyses suggest adaptive introgression.
Until recently, researchers interested in adaptive introgression have been limited to investigations of phenotype, without knowledge of the underlying genetic variation. The advent of molecular genetic and genomic tools has allowed for the identification of introgressed loci associated with adaptive traits. Applications of genomic data, such as the examples highlighted above, can be used to test proposed cases of adaptive introgression and identify new cases, enabling an empirical assessment of the idea of Anderson and Stebbins (1954) that introgression is an important mechanism of adaptive evolution. At this time, the most convincing evidence that an introgressed trait is adaptive in wild populations will still come from common garden and reciprocal transplant experiments. However, genomic data complement these classic experiments with more extensive characterization of introgressed genetic variation and its phenotypic effects. Future studies should address questions such as the following. How many traits are typically affected in cases of adaptive introgression, and how is the genetic architecture of those traits distributed across the genome? And what are the frequency and importance of genotype-by-environment interactions in modulating the effects of introgressed alleles?
Reinforcement
Hybridization between diverged lineages often is not adaptive. Hybrids may be inviable, sterile, or maladapted, and this cost to hybridization can generate selection to decrease mating between diverged lineages. The process of increased reproductive isolation due to selection to decrease hybridization is called reinforcement. Reinforcement in plants has been reviewed elsewhere (Hopkins, 2013), but we note here its historical importance in the study of hybridization and recent advancements due to innovative genomic analyses.
The notion that costly hybridization could favor increased isolation is attributed to Alfred R. Wallace (1889), and the process used to be referred to as the Wallace effect (Grant, 1966). During the modern synthesis, Dobzhansky (1940) first clearly articulated how greater assortative mating could be favored by selection to decrease hybridization and maintain coadapted gene complexes. Although much of the formative research on reinforcement was done in animal systems (Dobzhansky and Koller, 1938; Blair, 1955; Dobzhansky et al., 1964; Littlejohn and Loftus-Hills, 1968), there is a long history of botanical research on reinforcement as well (Grant, 1966; Levin and Kerster, 1967; McNeilly and Antonovics, 1968; Paterniani, 1969; Whalen, 1978).
Reinforcement begins with mating between closely related taxa. This hybridization is costly due to low hybrid viability or fertility. Costly hybridization generates selection favoring new traits that increase assortative mating. These novel trait values are selected in sympatric populations because they decrease hybridization, but they are not necessarily favored in allopatry, thus generating a pattern of character displacement (Howard, 1993; Servedio and Noor, 2003). The feasibility of reinforcement is controversial, because gene flow between hybridizing taxa can prevent the evolution of reproductive isolation in sympatry (Felsenstein, 1981; Butlin, 1987). Thus, hybridization is both the source of reinforcing selection and a major hindrance to the success of reinforcement (Kirkpatrick, 2000). In order for new alleles conferring assortative mating to evolve, they must remain genetically associated with alleles causing reduced hybrid viability or fertility (for review, see Servedio, 2009). Extensive theoretical research has illustrated that the feasibility of reinforcement is determined by a balance between the evolutionary forces of selection, gene flow, and recombination (Liou and Price, 1994; Servedio and Kirkpatrick, 1997; Kirkpatrick and Servedio, 1999; Kirkpatrick, 2000).
In light of this controversy, much of the empirical research on reinforcement has focused on demonstrating that the process occurs. Only recently has research expanded to explore how and why reinforcement can occur. What mutations cause reinforcement? How strong is reinforcing selection? How much gene flow occurs during the process of reinforcement? Advancements in genetic and genomic tools have revolutionized our ability to address these questions. We are only just starting to understand the genetic basis of reinforcement (Ortíz-Barrientos and Noor, 2005; Saether et al., 2007), and flower color variation in Phlox spp. is the only case of reinforcement for which the causal genes have been identified (Hopkins and Rausher, 2011). Yet, comparing genome scans for divergence and selection between allopatric and sympatric populations may suggest new candidate genes underlying reinforcement (Smadja et al., 2015). Once causal genes have been identified, future population genomic analyses can be used to infer when and how the mutations evolved (Barbash et al., 2004; Presgraves and Stephan, 2007; Tang and Presgraves, 2009; Sweigart and Flagel, 2015).
Understanding the balance between selection and gene flow during reinforcement requires a complement of genomic and experimental inquires. In Phlox spp., we are beginning to understand the strength of these evolutionary forces. Phlox drummondii evolved divergent flower color in sympatry with the closely related Phlox cuspidata due to reinforcement. Common garden field experiments demonstrate that flower-color divergence decreases hybridization by as much as 50% (Hopkins and Rausher, 2012), and a population genetic model estimates strong selection driving flower-color variation in this system (Hopkins et al., 2014). Genomic analyses also have begun to reveal the extent of gene flow between hybridizing sympatric species that experienced reinforcement (Kulathinal et al., 2009). More analyses investigating the direction, amount, and timing of gene flow between sympatric species that experienced reinforcement are necessary to more fully understand how and why reinforcement occurs.
Hybrid Speciation
Linnaeus (1760) first suggested that new species arose by hybridization in Disquisitio de sexu plantarum, and in so doing, he rejected the notion of immutability. Hybridization is widespread, but the generation of a unique, isolated hybrid lineage is likely very rare. New hybrid lineages must establish reproductive isolation and a unique ecological niche in order to overcome genetic swamping and competition from parental species. A new hybrid lineage may be formed through allopolyploidy or through homoploid hybrid speciation. Allopolyploid lineages may be formed by the fusion of unreduced gametes, genome doubling following hybridization, or via a triploid bridge (Ramsey and Schemske, 1998). Homoploid hybrid speciation describes the formation of a new, reproductively isolated hybrid lineage without a change in ploidy.
Allopolyploid hybrid speciation is the more common and feasible form of hybrid speciation (Soltis and Soltis, 2009). A recent review found that 11% of species across 47 plant genera were likely of allopolyploid origin (Barker et al., 2016). Meanwhile, only a handful of examples of homoploid hybrid speciation have been identified in animals (Mavárez et al., 2006; Lukhtanov et al., 2015), fungi (Leducq et al., 2016), and plants (Rieseberg et al., 2003a). In agreement with the paucity of well-supported examples, simulations suggest that hybridization more likely results in stable hybrid zones or the extinction of a parental species than homoploid hybrid speciation (Buerkle et al., 2003). The apparent difference in frequency also may reflect a bias in the likelihood of detecting polyploid versus homoploid hybrid species. Verifying a case of homoploid hybrid speciation requires both demonstrating the hybrid origin of the lineage and showing that the hybridization is directly accountable for establishing reproductive isolation (Schumer et al., 2014). While homoploid hybrid speciation often is invoked upon the demonstration of hybrid origin and a distinct ecological niche, there are very few examples that draw a compelling mechanistic link between hybridization and the establishment of a new, isolated species (Schumer et al., 2014; Yakimowski and Rieseberg, 2014). Furthermore, there is evidence that genetic divergence affects the relative likelihood of homoploid (more likely between less-diverged species) or polyploid (more likely between more-diverged species) hybrid speciation (Chapman and Burke, 2007). This is consistent with the idea that homologous chromosomes will pair with greater fidelity as the genetic divergence between progenitors of an allopolyploid increases, leading to higher fertility (Stebbins, 1947).
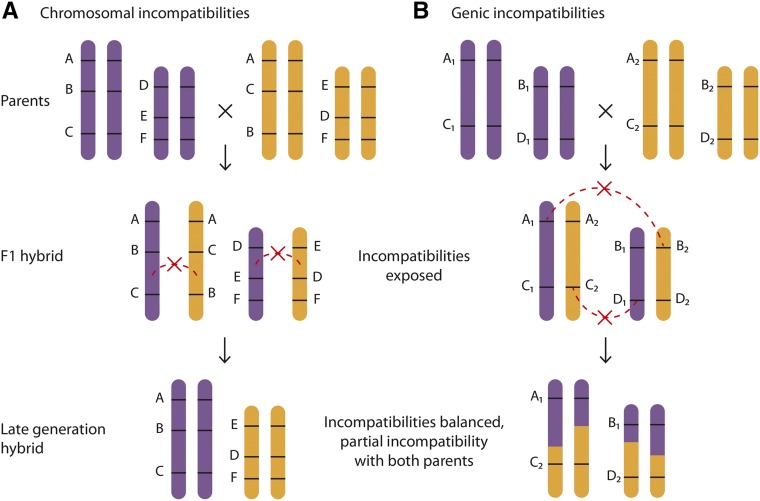
Differences in initial intrinsic reproductive isolation from parental species help explain why these two modes of hybrid speciation differ so dramatically in frequency. The process of genome doubling often produces some degree of immediate reproductive isolation from parental lineages by virtue of the inviability or sterility of interploid progeny. Unlike allopolyploids, homoploid hybrids do not achieve instant reproductive isolation from their parental species. However, homoploid hybrids may evolve partial intrinsic reproductive isolation from both parents relatively quickly. Following the refining of species concepts and Dobzhansky’s (1940) emphasis on reproductive barriers, Müntzing (1930) proposed that homoploid hybrids could become partially reproductively isolated by sorting and fixing genetic incompatibilities with both parents. Grant (1958) later described this process as recombinational speciation. Early work on recombinational speciation focused on describing how chromosomal rearrangements sort in hybrids (Fig. 3A; Grant, 1981), and this framework has since been applied to the sorting of genic incompatibilities as well (Fig. 3B; Schumer et al., 2015). Recombinational speciation has been empirically confirmed in multiple systems (Grant, 1981), including the well-studied Helianthus hybrid species (Box 2).
Figure 3.
Recombinational speciation contributes to homoploid hybrid speciation. Recombinational speciation involves the sorting of intrinsic incompatibilities between parents in a hybrid lineage. If some incompatibilities are resolved in favor of one parent and others are resolved in favor of the other, the hybrid lineage will have some degree of intrinsic incompatibility with each parent not exceeding the level of incompatibility between the parents. Incompatible alleles are connected by a dashed red line and an X. A, Chromosomal rearrangements such as inversions present in parent taxa may be differentially fixed in hybrid populations conferring partial incompatibility with both parent lineages. B, Similarly, recombinational speciation may involve the differential fixation of genic incompatibilities.
Hybrid speciation is more likely to be successful if hybrids can escape competition with initially much more numerous parental genotypes through ecological differentiation (Buerkle et al., 2000). As early as 1894, Anton Kerner emphasized the importance of hybrids colonizing open habitat, unoccupied by parent populations, for their persistence (Grant, 1981). Early support for the necessity of open habitat came from observing a high frequency of allopolyploid species in regions subject to glacial disturbance (Anderson, 1953). It has since been hypothesized that the rapid evolution of genome structure and gene expression following polyploidy can contribute to novel trait expression and ecological differentiation (Soltis and Soltis, 2009; Madlung, 2013). Synthetic allopolyploids have been created to verify the feasibility of this hypothesis (Gaeta et al., 2007). It has been demonstrated that, in some cases, allopolyploids can survive in a broader range of environments than their progenitors. This can be due to greater gene regulatory flexibility as a result of homolog-specific gene regulation (Dong and Adams, 2011; Combes et al., 2012) or alternative splicing (Zhou et al., 2011) in response to environmental perturbation. However, there remains a need for more empirical evidence demonstrating ecological differentiation facilitated by allopolyploidy (Abbott et al., 2013; Madlung, 2013; Soltis et al., 2014).
Homoploid hybrid species are thought to adapt to unique habitats primarily through the transgressive segregation of parental alleles. Stebbins (1950) highlighted the potential of transgressive segregation to produce new hybrid traits allowing the colonization of a novel, rather than intermediate, ecological niche. The Helianthus hybrid species provide the most detailed analysis of ecological differentiation contributing to homoploid hybrid speciation (Box 2).
The most recent advance in the study of hybridization is the use of genetic and genomic data to detect signatures of hybridization in wild populations. Genomic tools are particularly useful for identifying hybrid species because hybrid species are predicted to have transgressive phenotypes, thus making phenotypic intermediacy an inadequate criterion for identifying hybrid lineages, and homoploid hybrids are not necessarily expected to maintain equal proportions of both parental genomes. However, the application of genomic data to the identification of homoploid hybrid species is currently plagued by the difficulty of distinguishing between introgression that occurred after speciation and hybridization that was directly involved in the speciation process (Schumer et al., 2013). With so few thoroughly explored examples of homoploid hybrid speciation in plants, further investigations of putative cases that focus on demonstrating a role for hybridization in establishing reproductive isolation will elucidate which details of hybrid speciation are general patterns and which are exceptional. For instance, what is the relative importance of intrinsic incompatibilities versus ecological differentiation in establishing isolation from parent species? What is the level of incompatibility between parental species that create a hybrid species? And, is it common for a pair of species to harbor genetic variation that could produce transgressive hybrid phenotypes facilitating ecological divergence?
CONCLUSION
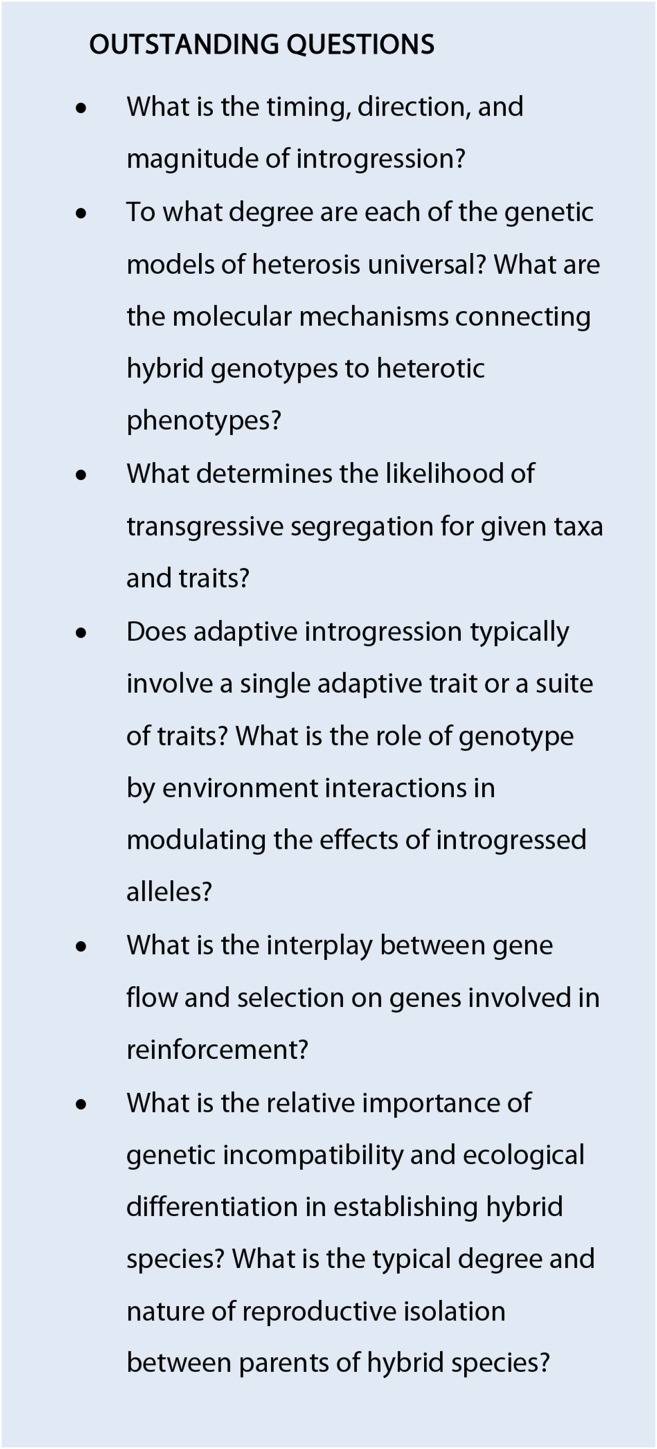
As plant evolutionary biologists, we should not be surprised by the growing realization that hybridization occurs across the tree of life. Botanists have been studying the existence and evolutionary consequences of hybridization since the birth of taxonomy, through the modern synthesis, and into the genomic era. We now have access to a torrent of genomic data unimaginable by earlier researchers in this field, which is revealing that many genomes are mosaics of fragments with different ancestry, some of which are more reticent to gene flow than others (Nosil et al., 2009; Mallet et al., 2016; but see Cruickshank and Hahn, 2014). As we begin to use these data to address outstanding questions in evolutionary biology, we must not lose sight of the historical foundations of the hypotheses we are testing. Our new challenge is to integrate advancements in genomic and genetic techniques with classical experimental protocols of genetic crosses, common garden field experiments, and controlled environment manipulations to better understand how and why hybridization has such important evolutionary repercussions. With these new techniques, we can gain insights into the causal mechanisms underlying phenomena such as heterosis and transgressive segregation; we can better understand how new combinations of alleles from parental taxa interact with novel environments to allow the persistence of hybrid lineages; and we can infer the history of gene flow and selection across specific genomic regions simply by looking at patterns of genetic variation. In addition to testing old hypotheses about how hybridization generates novel phenotypes and lineages, we are generating new hypotheses based on phenomena, such as small RNAs and epigenetics, that have been discovered only recently. Many of the outstanding questions about hybridization (see Outstanding Questions) are the same questions that plagued the founders of genetics, but our new tools give promise of new answers.
Acknowledgments
We thank James Mallet and Heather Briggs for helpful comments, Loren Rieseberg for assistance with Box 2, as well as Alice Cheung, Richard Amasino, Cris Kuhlemeier, and Thomas Dresselhaus for the invitation to participate in this special issue.
Glossary
- ILS
incomplete lineage sorting
- QTL
quantitative trait locus
Footnotes
Articles can be viewed without a subscription.
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, et al. (2013) Hybridization and speciation. J Evol Biol 26: 229–246 [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. (1948) Hybridization of the habitat. Evolution 2: 1–9 [Google Scholar]
- Anderson E. (1949) Introgressive Hybridization. John Wiley & Sons, New York [Google Scholar]
- Anderson E. (1953) Introgressive hybridization. Biol Rev Camb Philos Soc 28: 280–307 [Google Scholar]
- Anderson E, Hubricht L (1938) Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am J Bot 25: 396–402 [Google Scholar]
- Anderson E, Stebbins GL (1954) Hybridization as an evolutionary stimulus. Evolution 8: 378–388 [Google Scholar]
- Arnold ML. (2006) Evolution through Genetic Exchange. Oxford University Press, Oxford [Google Scholar]
- Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L, et al. (2013) Networks: expanding evolutionary thinking. Trends Genet 29: 439–441 [DOI] [PubMed] [Google Scholar]
- Barbash DA, Awadalla P, Tarone AM (2004) Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol 2: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA (2016) On the relative abundance of autopolyploids and allopolyploids. New Phytol 210: 391–398 [DOI] [PubMed] [Google Scholar]
- Blair WF. (1955) Mating call and stage of speciation in the Microhyla olivacea–M. carolinensis complex. Evolution 9: 469–480 [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH (2000) The likelihood of homoploid hybrid speciation. Heredity 84: 441–451 [DOI] [PubMed] [Google Scholar]
- Buerkle CA, Wolf DE, Rieseberg LH (2003) The origin and extinction of species through hybridization. In Population Viability in Plants. Springer, Berlin, pp 117–141 [Google Scholar]
- Butlin R. (1987) Speciation by reinforcement. Trends Ecol Evol 2: 8–13 [DOI] [PubMed] [Google Scholar]
- Chapman MA, Burke JM (2007) Genetic divergence and hybrid speciation. Evolution 61: 1773–1780 [DOI] [PubMed] [Google Scholar]
- Chen ZJ. (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14: 471–482 [DOI] [PubMed] [Google Scholar]
- Cockayne L. (1923) Hybridism in the New Zealand flora. New Phytol 22: 105–127 [Google Scholar]
- Combes MC, Cenci A, Baraille H, Bertrand B, Lashermes P (2012) Homeologous gene expression in response to growing temperature in a recent allopolyploid (Coffea arabica L.). J Hered 103: 36–46 [DOI] [PubMed] [Google Scholar]
- Crow JF. (1948) Alternative hypotheses of hybrid vigor. Genetics 33: 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW (2014) Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol 23: 3133–3157 [DOI] [PubMed] [Google Scholar]
- Dannemann M, Andrés AM, Kelso J (2016) Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human Toll-like receptors. Am J Hum Genet 98: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (1876) The Effects of Cross and Self Fertilisation in the Vegetable Kingdom. John Murray, London [Google Scholar]
- deVicente MC, Tanksley SD (1993) QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134: 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich-Reed DR, Fitzpatrick BM (2013) Transgressive hybrids as hopeful monsters. Evol Biol 40: 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. (1940) Speciation as a stage in evolutionary divergence. Am Nat 74: 312–321 [Google Scholar]
- Dobzhansky T, Ehrman L, Pavlovsky O, Spassky B (1964) The superspecies Drosophila paulistorum. Proc Natl Acad Sci USA 51: 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Koller PC (1938) An experimental study of sexual isolation in Drosophila. Biol Zentralblatt 58: 589–607 [Google Scholar]
- Dong S, Adams KL (2011) Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol 190: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M (2011) Testing for ancient admixture between closely related populations. Mol Biol Evol 28: 2239–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Rietz GE. (1930) The Fundamental Units of Biological Taxonomy. Svensk Botaniska Foreningen, Uppsala, Sweden [Google Scholar]
- East EM. (1936) Heterosis. Genetics 21: 375–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton DAR, Ree RH (2013) Inferring phylogeny and introgression using RADseq data: an example from flowering plants (Pedicularis: Orobanchaceae). Syst Biol 62: 689–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC, Meirmans P, Rong J, Bartsch D, Ghosh A, de Jong TJ, Haccou P, Lu BR, Snow AA, Stewart CN, et al. (2013) Introgression of crop alleles into wild or weedy populations. Annu Rev Ecol Evol Syst 44: 325–345 [Google Scholar]
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, van Dorp L, Lawson D (2016) A tutorial on how (not) to over-interpret STRUCTURE/ADMIXTURE bar plots. bioRxiv 66431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1981) Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution 35: 124–138 [DOI] [PubMed] [Google Scholar]
- Flint-Garcia SA, Buckler ES, Tiffin P, Ersoz E, Springer NM (2009) Heterosis is prevalent for multiple traits in diverse maize germplasm. PLoS ONE 4: e7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focke WO. (1881) Die Pflanzen-Mischling. Borntraeger, Berlin [Google Scholar]
- Gaeta RT, Pires JC, Iniguez-Luy F, Leon E, Osborn TC (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Farzaneh N, Grassa CJ, McKown AD, Guy RD, Mansfield SD, Douglas CJ, Cronk QCB (2014) Landscape genomics of Populus trichocarpa: the role of hybridization, limited gene flow, and natural selection in shaping patterns of population structure. Evolution 68: 3260–3280 [DOI] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA (2013) Analyses of genetic ancestry enable key insights for molecular ecology. Mol Ecol 22: 5278–5294 [DOI] [PubMed] [Google Scholar]
- Grant V. (1958) The regulation of recombination in plants. Cold Spring Harb Symp Quant Biol 23: 337–363 [DOI] [PubMed] [Google Scholar]
- Grant V. (1966) The selective origin of incompatibility barriers in the plant genus Gilia. Am Nat 100: 99–118 [Google Scholar]
- Grant V. (1975) Genetics of Flowering Plants. Columbia University Press, New York [Google Scholar]
- Grant V. (1981) Plant Speciation, Ed 2 Columbia University Press, New York [Google Scholar]
- Gravel S. (2012) Population genetics models of local ancestry. Genetics 191: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Gonzalez-Bayon R, Wang L, Zhu A, Liu PC, Groszmann M, Peacock WJ, Dennis ES (2015) Epigenetic changes in hybrids. Plant Physiol 168: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MHY, et al. (2010) A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Kane NC, Lexer C, Ludwig F, Rosenthal DM, Donovan LA, Rieseberg LH (2004) Reconstructing the origin of Helianthus deserticola: survival and selection on the desert floor. Am Nat 164: 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 29: 684–690 [DOI] [PubMed] [Google Scholar]
- Hagedoorn AL, Hagedoorn-Vorstheuvel La Brand AC (1921) The Relative Value of the Processes Causing Evolution. Martinus Nijhoff, The Hague, The Netherlands [Google Scholar]
- Hagiwara WE, Onishi K, Takamure I, Sano Y (2006) Transgressive segregation due to linked QTLs for grain characteristics of rice. Euphytica 150: 27–35 [Google Scholar]
- Hahn MW, Nakhleh L (2016) Irrational exuberance for resolved species trees. Evolution 70: 7–17 [DOI] [PubMed] [Google Scholar]
- Harris K, Nielsen R (2013) Inferring demographic history from a spectrum of shared haplotype lengths. PLoS Genet 9: e1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG. (1990) Hybrid zones: windows on evolutionary process. Oxford Surv Evol Biol 7: 69–128 [Google Scholar]
- Heiser CB. (1951) Hybridization in the annual sunflowers: Helianthus annuus × H. debilis var. cucumerifolius. Evolution 5: 42–51 [Google Scholar]
- Hey J, Nielsen R (2004) Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins R. (2013) Reinforcement in plants. New Phytol 197: 1095–1103 [DOI] [PubMed] [Google Scholar]
- Hopkins R, Guerrero RF, Rausher MD, Kirkpatrick M (2014) Strong reinforcing selection in a Texas wildflower. Curr Biol 24: 1995–1999 [DOI] [PubMed] [Google Scholar]
- Hopkins R, Rausher MD (2011) Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature 469: 411–414 [DOI] [PubMed] [Google Scholar]
- Hopkins R, Rausher MD (2012) Pollinator-mediated selection on flower color allele drives reinforcement. Science 335: 1090–1092 [DOI] [PubMed] [Google Scholar]
- Howard DJ. (1993) Reinforcement: origin, dynamics, and fate of an evolutionary hypothesis. In Harrison RG, ed, Hybrid Zones and the Evolutionary Process. Oxford University Press, New York, pp 46–69 [Google Scholar]
- Hufford MB, Lubinksy P, Pyhäjärvi T, Devengenzo MT, Ellstrand NC, Ross-Ibarra J (2013) The genomic signature of crop-wild introgression in maize. PLoS Genet 9: e1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DF. (1917) Dominance of linked factors as a means of accounting for heterosis. Proc Natl Acad Sci USA 3: 310–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler S. (2012) Heterosis: many genes, many mechanisms—end the search for an undiscovered unifying theory. ISRN Bot 2012: 1–12 [Google Scholar]
- Kiær LP, Felber F, Flavell A, Guadagnuolo R, Guiatti D, Hauser TP, Olivieri AM, Scotti I, Syed N, Vischi M, et al. (2009) Spontaneous gene flow and population structure in wild and cultivated chicory, Cichorium intybus L. Genet Resour Crop Evol 56: 405–419 [Google Scholar]
- Kirkpatrick M. (2000) Reinforcement and divergence under assortative mating. Proc Biol Sci 267: 1649–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Servedio MR (1999) The reinforcement of mating preferences on an island. Genetics 151: 865–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölreuter JG. (1766) Vorläufige nachricht von einigen das geschlecht der pflanzen betreffenden versuchen und beobachtungen, nebst fortsetzungen 1, 2 und 3 (1761-1766). Wilhelm Engelmann, Leipzig, Germany [Google Scholar]
- Krieger U, Lippman ZB, Zamir D (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet 42: 459–463 [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF (2009) The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet 5: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwit C, Moon HS, Warwick SI, Stewart CN Jr (2011) Transgene introgression in crop relatives: molecular evidence and mitigation strategies. Trends Biotechnol 29: 284–293 [DOI] [PubMed] [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH (2005) Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq JB, Nielly-Thibault L, Charron G, Eberlein C, Verta JP, Samani P, Sylvester K, Hittinger CT, Bell G, Landry CR (2016) Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat Microbiol 1: 15003. [DOI] [PubMed] [Google Scholar]
- Levin DA, editor (1979) Hybridization: An Evolutionary Perspective. Dowden, Hutchinson & Ross, Stroudsberg, PA [Google Scholar]
- Levin DA, Francisco-Ortega J, Jansen RK (1996) Hybridization and the extinction of rare plant species. Conserv Biol 10: 10–16 [Google Scholar]
- Levin DA, Kerster HW (1967) Natural selection for reproductive isolation in Phlox. Evolution 21: 679–687 [DOI] [PubMed] [Google Scholar]
- Lexer C, Welch ME, Durphy JL, Rieseberg LH (2003) Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species. Mol Ecol 12: 1225–1235 [DOI] [PubMed] [Google Scholar]
- Linnaeus C. (1760) Disquisitio de sexu plantarum Amoenitates Acad 10: 100–131 [Google Scholar]
- Liou LW, Price TD (1994) Speciation by reinforcement of premating isolation. Evolution 48: 1451–1459 [DOI] [PubMed] [Google Scholar]
- Littlejohn MJ, Loftus-Hills JJ (1968) An experimental evaluation of premating isolation in the Hyla ewingi complex (Anura: Hylidae). Evolution 22: 659–663 [DOI] [PubMed] [Google Scholar]
- Liu Y, Nyunoya T, Leng S, Belinsky SA, Tesfaigzi Y, Bruse S (2013) Softwares and methods for estimating genetic ancestry in human populations. Hum Genomics 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotsy JP. (1916) Evolution by Means of Hybridization. Martinus Nijhoff, The Hague, The Netherlands [Google Scholar]
- Lukhtanov VA, Shapoval NA, Anokhin BA, Saifitdinova AF, Kuznetsova VG (2015) Homoploid hybrid speciation and genome evolution via chromosome sorting. Proc Biol Sci 282: 20150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A. (2013) Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity (Edinb) 110: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailund T, Halager AE, Westergaard M, Dutheil JY, Munch K, Andersen LN, Lunter G, Prüfer K, Scally A, Hobolth A, et al. (2012) A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species. PLoS Genet 8: e1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20: 229–237 [DOI] [PubMed] [Google Scholar]
- Mallet J, Besansky N, Hahn MW (2016) How reticulated are species? BioEssays 38: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Liu T, Xu C, Li X, Xing Y (2011) Epistasis and complementary gene action adequately account for the genetic bases of transgressive segregation of kilo-grain weight in rice. Euphytica 180: 261–271 [Google Scholar]
- Marsden-Jones EM. (1930) The genetics of Geum intermedium Willd. Haud Ehrh., and its back-crosses. J Genet 23: 377–395 [Google Scholar]
- Mavárez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M (2006) Speciation by hybridization in Heliconius butterflies. Nature 441: 868–871 [DOI] [PubMed] [Google Scholar]
- Mayr E. (1942) Systematics and the origin of species from the viewpoint of a zoologist. Columbia University Press, New York [Google Scholar]
- Mayr E. (1986) Joseph Gottlieb Kölreuter’s contributions to biology. Osiris 2: 135–176 [Google Scholar]
- McNeilly T, Antonovics J (1968) Evolution in closely adjacent plant populations. IV. Barriers to gene flow. Heredity 23: 205–218 [Google Scholar]
- Müntzing A. (1930) Outlines to a genetic monograph for the genus Galeopsis: with special reference to the nature and inheritance of partial sterility. Hereditas 13: 185–341 [Google Scholar]
- Ng DWK, Lu J, Chen ZJ (2012) Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol 15: 154–161 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J (2001) Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson-Ehle H. (1911) Kreuzungsuntersuchungen an hafer und weizen. Lunds Univ Areskripft 7: 1–84 [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D (2009) Divergent selection and heterogeneous genomic divergence. Mol Ecol 18: 375–402 [DOI] [PubMed] [Google Scholar]
- Ortíz-Barrientos D, Noor MAF (2005) Evidence for a one-allele assortative mating locus. Science 310: 1467. [DOI] [PubMed] [Google Scholar]
- Pardo-Diaz C, Salazar C, Baxter SW, Merot C, Figueiredo-Ready W, Joron M, McMillan WO, Jiggins CD (2012) Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genet 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşaniuc B, Sankararaman S, Kimmel G, Halperin E (2009) Inference of Locus-Specific Ancestry in Closely Related Populations. Oxford University Press, Oxford, pp i213–i221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterniani E. (1969) Selection for reproductive isolation between two populations of maize, Zea mays L. Evolution 23: 534–547 [DOI] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH (2016) A genomic perspective on hybridization and speciation. Mol Ecol 25: 2337–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Haak DC, Hahn MW, Moyle LC (2016) Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol 14: e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Hahn MW (2015) Detection and polarization of introgression in a five-taxon phylogeny. Syst Biol 64: 651–662 [DOI] [PubMed] [Google Scholar]
- Pool JE, Nielsen R (2009) Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics 181: 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Hurtado L, Ruiz Y, Santos C, Phillips C, Carracedo A, Lareu MV (2013) An overview of STRUCTURE: applications, parameter settings, and supporting software. Front Genet 4: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Stephan W (2007) Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol Biol Evol 24: 306–314 [DOI] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S (2009) Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet 5: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E (2015) Evidence for archaic adaptive introgression in humans. Nat Rev Genet 16: 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29: 467–501 [Google Scholar]
- Rieseberg LH. (2006) Hybrid speciation in wild sunflowers. Ann Mo Bot Gard 93: 34–48 [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83: 363–372 [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Carney SE (1998) Plant hybridization. New Phytol 140: 599–624 [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Ellstrand NC, Arnold M (1993) What can molecular and morphological markers tell us about plant hybridization? CRC Crit Rev Plant Sci 12: 213–241 [Google Scholar]
- Rieseberg LH, Kim SC, Randell RA, Whitney KD, Gross BL, Lexer C, Clay K (2007) Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129: 149–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003a) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM (1996) Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745 [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Widmer A, Arntz AM, Burke JM (2003b) The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos Trans R Soc Lond B Biol Sci 358: 1141–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal DM, Rieseberg LH, Donovan LA (2005) Re-creating ancient hybrid species’ complex phenotypes from early-generation synthetic hybrids: three examples using wild sunflowers. Am Nat 166: 26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig BK, Pease JB, Besansky NJ, Hahn MW (2016) Powerful methods for detecting introgressed regions from population genomic data. Mol Ecol 25: 2387–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru D, Mao K, Zhang L, Wang X, Lu Z, Sun Y (2016) Genomic evidence for polyphyletic origins and interlineage gene flow within complex taxa: a case study of Picea brachytyla in the Qinghai-Tibet Plateau. Mol Ecol 25: 2373–2386 [DOI] [PubMed] [Google Scholar]
- Saether SA, Saetre GP, Borge T, Wiley C, Svedin N, Andersson G, Veen T, Haavie J, Servedio MR, Bures S, et al. (2007) Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318: 95–97 [DOI] [PubMed] [Google Scholar]
- Sankararaman S, Sridhar S, Kimmel G, Halperin E (2008) Estimating local ancestry in admixed populations. Am J Hum Genet 82: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scascitelli M, Whitney KD, Randell RA, King M, Buerkle CA, Rieseberg LH (2010) Genome scan of hybridizing sunflowers from Texas (Helianthus annuus and H. debilis) reveals asymmetric patterns of introgression and small islands of genomic differentiation. Mol Ecol 19: 521–541 [DOI] [PubMed] [Google Scholar]
- Schumer M, Cui R, Boussau B, Walter R, Rosenthal G, Andolfatto P (2013) An evaluation of the hybrid speciation hypothesis for Xiphophorus clemenciae based on whole genome sequences. Evolution 67: 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Cui R, Rosenthal GG, Andolfatto P (2015) Reproductive isolation of hybrid populations driven by genetic incompatibilities. PLoS Genet 11: e1005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Rosenthal GG, Andolfatto P (2014) How common is homoploid hybrid speciation? Evolution 68: 1553–1560 [DOI] [PubMed] [Google Scholar]
- Servedio MR. (2009) The role of linkage disequilibrium in the evolution of premating isolation. Heredity 102: 51–56 [DOI] [PubMed] [Google Scholar]
- Servedio MR, Kirkpatrick M (1997) The effects of gene flow on reinforcement. Evolution 51: 1764–1772 [DOI] [PubMed] [Google Scholar]
- Servedio MR, Noor M (2003) The role of reinforcement in speciation: theory and data. Annu Rev Ecol Evol Syst 34: 339–364 [Google Scholar]
- Shang L, Wang Y, Cai S, Wang X, Li Y, Abduweli A, Hua J (2015) Partial dominance, overdominance, epistasis and QTL by environment interactions contribute to heterosis in two upland cotton hybrids. G3 (Bethesda) 6: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G, Zhan W, Chen H, Xing Y (2014) Dominance and epistasis are the main contributors to heterosis for plant height in rice. Plant Sci 215-216: 11–18 [DOI] [PubMed] [Google Scholar]
- Shen H, He H, Li J, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, et al. (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24: 875–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull GH. (1908) The composition of a field of maize. J Hered Os 4: 296–301 [Google Scholar]
- Shull GH. (1911) The genotypes of maize. Am Nat 45: 234–252 [Google Scholar]
- Smadja CM, Loire E, Caminade P, Thoma M, Latour Y, Roux C, Thoss M, Penn DJ, Ganem G, Boursot P (2015) Seeking signatures of reinforcement at the genetic level: a hitchhiking mapping and candidate gene approach in the house mouse. Mol Ecol 24: 4222–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE. (1804) Flora Britannica, Vol. 3. J Taylor, London [Google Scholar]
- Snow AA, Pilson D, Rieseberg LH, Paulsen MJ, Pleskac N, Reagon MR, Wolf DE, Selbo SM (2003) A Bt transgene reduces herbivory and enhances fecundity in wild sunflowers. Ecol Appl 13: 279–286 [Google Scholar]
- Solís-Lemus C, Ané C (2016) Inferring phylogenetic networks with maximum pseudolikelihood under incomplete lineage sorting. PLoS Genet 12: 1509.06075v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS (2014) The polyploidy revolution then…and now: Stebbins revisited. Am J Bot 101: 1057–1078 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Sousa V, Hey J (2013) Understanding the origin of species with genome-scale data: modelling gene flow. Nat Rev Genet 14: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL., Jr (1947) Types of polyploids: their classification and significance. Adv Genet 1: 403–429 [DOI] [PubMed] [Google Scholar]
- Stebbins GL. (1950) Variation and Evolution in Plants. Columbia University Press, New York [Google Scholar]
- Stebbins GL. (1959) The role of hybridization in evolution. Proc Am Philos Soc 103: 231–251 [Google Scholar]
- Stelkens R, Seehausen O (2009) Genetic distance between species predicts novel trait expression in their hybrids. Evolution 63: 884–897 [DOI] [PubMed] [Google Scholar]
- Suarez-Gonzalez A, Hefer CA, Christe C, Corea O, Lexer C, Cronk QCB, Douglas CJ (2016) Genomic and functional approaches reveal a case of adaptive introgression from Populus balsamifera (balsam poplar) in P. trichocarpa (black cottonwood). Mol Ecol 25: 2427–2442 [DOI] [PubMed] [Google Scholar]
- Sweigart AL, Flagel LE (2015) Evidence of natural selection acting on a polymorphic hybrid incompatibility locus in Mimulus. Genetics 199: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Yan J, Ma X, Teng W, Wu W, Dai J, Dhillon BS, Melchinger AE, Li J (2010) Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor Appl Genet 120: 333–340 [DOI] [PubMed] [Google Scholar]
- Tang S, Presgraves DC (2009) Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than C, Ruths D, Nakhleh L (2008) PhyloNet: a software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinformatics 9: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hübner S, Heredia SM, Hahn MA, Caseys C, Bock DG, et al. (2016) Hybridization and extinction. Evol Appl 9: 892–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähä JP, Primmer CR (2006) Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15: 63–72 [DOI] [PubMed] [Google Scholar]
- Vallejo-Marín M, Hiscock SJ (2016) Hybridization and hybrid speciation under global change. New Phytol 211: 1170–1187 [DOI] [PubMed] [Google Scholar]
- Wallace AR. (1889) Darwinism: An Exposition of the Theory of Natural Selection with Some of Its Applications. Macmillan, London [Google Scholar]
- Wang L, Greaves IK, Groszmann M, Wu LM, Dennis ES, Peacock WJ (2015) Hybrid mimics and hybrid vigor in Arabidopsis. Proc Natl Acad Sci USA 112: E4959–E4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warschefsky E, Penmetsa RV, Cook DR, von Wettberg EJB (2014) Back to the wilds: tapping evolutionary adaptations for resilient crops through systematic hybridization with crop wild relatives. Am J Bot 101: 1791–1800 [DOI] [PubMed] [Google Scholar]
- Wen D, Yu Y, Hahn MW, Nakhleh L (2016) Reticulate evolutionary history and extensive introgression in mosquito species revealed by phylogenetic network analysis. Mol Ecol 25: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen M. (1978) Reproductive character displacement and floral diversity in Solanum section Androceras. Syst Bot 3: 77–86 [Google Scholar]
- Whitney KD, Broman KW, Kane NC, Hovick SM, Randell RA, Rieseberg LH (2015) Quantitative trait locus mapping identifies candidate alleles involved in adaptive introgression and range expansion in a wild sunflower. Mol Ecol 24: 2194–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH (2010) Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. New Phytol 187: 230–239 [DOI] [PubMed] [Google Scholar]
- Wiegand KM. (1935) A taxonomist’s experience with hybrids in the wild. Science 81: 161–166 [DOI] [PubMed] [Google Scholar]
- Winge O. (1917) The chromosomes: their numbers and general importance. Comptes Rendus des Trav du Lab Carlesb 13: 131–175 [Google Scholar]
- Yakimowski SB, Rieseberg LH (2014) The role of homoploid hybridization in evolution: a century of studies synthesizing genetics and ecology. Am J Bot 101: 1247–1258 [DOI] [PubMed] [Google Scholar]
- Yang X, Xia H, Wang W, Wang F, Su J, Snow AA, Lu BR (2011) Transgenes for insect resistance reduce herbivory and enhance fecundity in advanced generations of crop-weed hybrids of rice. Evol Appl 4: 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Chen Y, Yao W, Zhang C, Xie W, Hua J, Xing Y, Xiao J, Zhang Q (2012) Genetic composition of yield heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 109: 15847–15852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Moshgabadi N, Adams KL (2011) Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci USA 108: 16122–16127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirkle C. (1934) More records of plant hybridization before Koelreuter. J Hered 25: 3–18 [Google Scholar]