SUMOylation is required for the regulation of transcriptional gene silencing, which is dependent on Pol V pathway in Arabidopsis.
Abstract
The expression of genes with aberrant structure is prevented at both the transcriptional and posttranscriptional regulation levels. Aberrant gene silencing at the posttranscriptional level is well studied; however, it is not well understood how aberrant genes are silenced at the transcriptional level. In this study, through genetic screening a transgenic report line that harbors an aberrant gene (35S-LUC, lacking 3′-untranslated region [3′-UTR]) and lacks luciferase (LUC) activity, we identify that the small ubiquitin-like modifier (SUMO) protease OTS1 gene is required for maintaining the silence of the reporter 35S-LUC and an endogenous mutator-like element MULE-F19G14 at the transcriptional level, which requires DNA-dependent RNA polymerase (Pol) V and DDR complex, but not Pol IV. The increased transcripts in ots1 mutants are terminated by the 3′-UTRs of downstream genes. In addition to ots1 mutations, mutations in several known or putative SUMO proteases and two SUMO E3 ligases, SIZ1 and MMS21, have similar effects on this silencing regulation. Taken together, our results reveal that the enzymes involved in the SUMOylation process restrain aberrant gene transcription by using a downstream gene 3′-UTR, and this regulation requires a functional Pol V-dependent pathway in Arabidopsis (Arabidopsis thaliana).
Gene transcription is strictly controlled to partition genome and prevent transcriptional interference at neighboring genes in eukaryotes. Previous reports have shown that 3′-untranslated region (3′-UTR) participates in numerous regulatory processes, including premRNA cleavage, polyadenylation, stability, and localization of mRNA and translation efficiency, indicating that 3′-UTR plays a vital role in accurate regulation of gene expression (Proudfoot et al., 2002; Barrett et al., 2012). RDR6-mediated RNA silencing mechanism can be induced when some aberrant or missing 3′-UTR genes generate improperly terminated and unpolyadenylated readthrough mRNAs in Arabidopsis (Arabidopsis thaliana; Herr et al., 2006; Luo and Chen, 2007). In addition to RDR6-mediated RNA silencing pathway, organisms have developed several other mRNA quality control mechanisms during the long evolutionary period, such as nonsense-mediated mRNA decay, nonstop mRNA decay, and no-go mRNA decay (Isken and Maquat, 2007). These surveillance pathways contribute to the integrity of gene expression in a posttranscriptional regulation level; however, little is known how the expression of aberrant genes is controlled at a transcriptional regulation level.
SUMOylation is one class of dynamic and reversible posttranslational modifications (PTMs). The small ubiquitin-like modifier (SUMO) proteins are a member of highly conserved ubiquitin-like polypeptides that are covalently conjugated to targets, which affects the function of substrates in distinct ways, for example, by altering their cellular location, activity, stability, or interaction with other proteins (Melchior, 2000; Johnson, 2004; Hay, 2005; Geiss-Friedlander and Melchior, 2007; Wang and Dasso, 2009; van der Veen and Ploegh, 2012). In Arabidopsis, there are potentially eight genes encoding SUMO paralogs, of which only four (SUM1, SUM2, SUM3, and SUM5) have been shown to act as functional PTMs (Kurepa et al., 2003; Budhiraja et al., 2009; Park et al., 2011b). The SUMO conjugation process shows a very similar manner with ubiquitination pathway and requires its own suit of analogous E1, E2, and E3 enzymes that catalyze activation, conjugation, and ligation, respectively. Moreover, SUMO modification can be recycled by a class of Cys proteases that also involves generating mature SUMO (Mukhopadhyay and Dasso, 2007). In Arabidopsis, two SUMO E3 ligases, SIZ1 and MMS21, are identified, which have been reported to participate in many biological processes (Miura et al., 2005, 2007b, 2009, 2010; Yoo et al., 2006; Catala et al., 2007; Lee et al., 2007; Jin et al., 2008; Ishida et al., 2009; Chen et al., 2011; Park et al., 2011a; Ishida et al., 2012; Zheng et al., 2012; Zhang et al., 2013; Castro et al., 2015). On the basis of protein sequence comparison in the Arabidopsis database, there are at least seven genes encoding putative SUMO proteases; however, only AtULP1a/ELS1 (At3g06910), AtULP1c/OTS2 (At1g10570), AtULP1d/OTS1 (At1g60220), and AtESD4 (At4g15880) have been functionally characterized as SUMO proteases (Chosed et al., 2006; Colby et al., 2006; Park et al., 2011b; Murtas et al., 2003; Hermkes et al., 2011). Two SUMO proteases, OTS1 and OTS2, act redundantly to modulate salt stress response (Conti et al., 2008, 2014), mediate deconjugation of SUMO from phyB to positively regulate light-induced signaling (Sadanandom et al., 2015), and play a negative role in SA-mediated signaling (Bailey et al., 2016). However, little is known about how SUMOylation is involved in the regulation of transcriptional gene silencing (TGS; Shiio and Eisenman, 2003; Kang et al., 2010).
There are three essential DNA-dependent RNA polymerases, I, II, and III, in all eukaryotes. Plants have evolved two additional multisubunit RNA polymerases, Pol IV and Pol V, which are nonessential for viability but required for the RNA-directed DNA methylation (RdDM) pathway involved in de novo DNA methylation in the Arabidopsis genome within all sequence contexts (CG, CHG, and CHH; Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005; Pontier et al., 2005; Wierzbicki et al., 2008; Huang et al., 2009; Matzke et al., 2009). The RdDM pathway is a conserved mechanism for mediating TGS of many endogenous transposons and repeat elements. Pol IV collaborating with RDR2 and DCL3 is responsible for 24-nucleotide small-interfering RNA (siRNA) biogenesis, whereas with the help of chromatin-remodeling DDR complex (DRD1, DMS3, and RDM1), Pol V functions to generate scaffold RNAs for guiding sequence-specific DNA methylation by siRNAs (Onodera et al., 2005; Pontier et al., 2005; Wierzbicki et al., 2008; Zhou and Law, 2015). Loss of Pol V or DRD1 disrupts heterochromatin organization during interphase; on the contrary, loss of Pol IV, RDR2, or DCL3 does not (Pontes et al., 2009). Approximately 60% of Pol V-occupied regions overlap with the regions of 24-nucleotide siRNA complementarity and cytosine methylation; however, about one-quarter of Pol V peaks occur at loci (mostly genes) lacking significant cytosine methylation or siRNA signatures (Wierzbicki et al., 2012), indicating that Pol V may not function only in the RdDM pathway in Arabidopsis. Many transcription factors (including RNA pol I, II, and III) and premRNA processing factors are SUMOylated in yeast (Saccharomyces cerevisiae; however, whether Pol V is SUMO-modified has not been examined yet (Panse et al., 2004).
In this study, we isolated a mutant, ots1-3, which causes the production of longer transcripts of the transgene lacking 3′-UTR (35S-LUC) and endogenous mutator-like element MULE-F19G14 with aberrant gene structure. Both of the longer transcripts were terminated by the 3′-UTRs of downstream genes. Meanwhile, we found that the other SUMO proteases and two SUMO E3 ligases also involved in regulating this transcriptional process. Furthermore, Pol V and DDR complex require for the release of aberrant gene silencing in the ots1-3 mutant. Our results indicate that SUMOylation involved in the regulation of TGS of aberrant genes requires a functional Pol V-dependent pathway.
RESULTS
Genetic Screening Mutants Expressing 35S-LUC Transgene Lacking 3′-UTR
We previously developed a screening system in Arabidopsis to identify components involved in regulating TGS of aberrant genes (Zhou et al., 2010). This transgenic line, referred to as wild type, contains the 35S-LUC transgene, firefly LUCIFERASE (LUC) lacking 3′-UTR driven by the Cauliflower Mosaic Virus 35S promoter (35S), and the downstream 35S-hygromycin phosphotransferase (HPT)-3′UTR gene (Fig. 1A). No LUC luminance was detected in this transgenic line, which was mutagenized with ethyl methanesulfonate. From this mutagenesis population, we screened mutants that express LUC using a high-throughput luminescence imaging method. We previously identified a mutant, mom1-44, in which polyadenylated LUC was transcribed and terminated by the HPT 3′-UTR (Zhou et al., 2010).
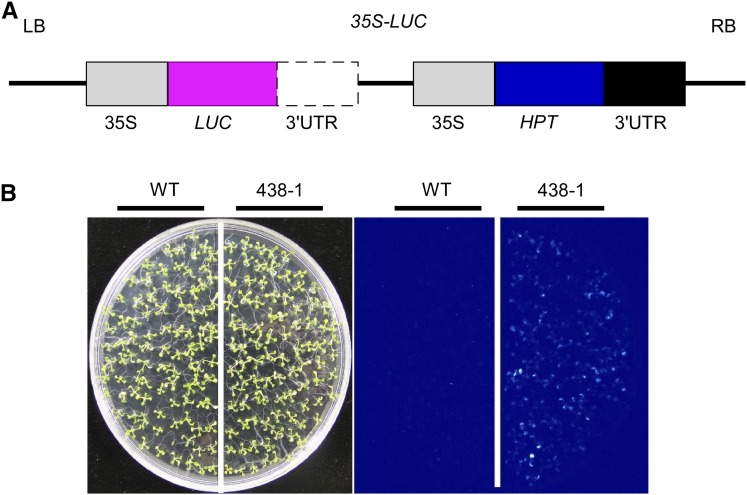
Figure 1.
Identification of the 438-1 mutant. A, Structure chart of 35S-LUC construct in the binary vector pCAMBIA1301. LUC CDS without 3′-UTR is driven by the CaMV 35S promoter (35S-LUC) following complete HPT gene. 35S, CaMV 35S promoter; LB, left border; RB, right border. B, Luminescence images showing LUC expression in wild-type and mutant 438-1 plants.
In this study, we obtained another mutant, 438-1, which displayed a weak LUC luminescence (Fig. 1B). We backcrossed the 438-1 mutant with wild-type plants. F1 plants showed a wild type-like luminescence phenotype and F2 progenies showed the ratio of 3:1 for wild type to mutant, suggesting that 438-1 is a recessive mutant controlled by a nuclear gene.
Polyadenylated Transcripts of the 35S-LUC and Endogenous Mutator-Like Element MULE-F19G14 Are Terminated by the 3′-UTRs of Downstream Genes in the 438-1 Mutant
The mom1-44 mutant rescues LUC activity because of transcription of 35S-LUC by using the 3′-UTR of downstream gene HPT (Zhou et al., 2010). We detected LUC transcript status in the 438-1 mutant. Through Reverse Transcription (RT)-PCR assays by means of total RNA reverse transcribed using oligo(dT) or random primers, the LUC transcript was detected in 438-1 using LUC specific primers (F0+R0), whereas LUC was at an undetectable level in wild type (Fig. 2A). The relative expression of LUC was also examined in wild type and 438-1 by real-time PCR using the oligo(dT) reverse-transcribed cDNA. The amount of LUC transcript in 438-1 was about 10 times more than that in wild type (Fig. 2B). These results suggest that LUC mRNA largely exists in the polyadenylated state in the 438-1 mutant. We next detected the LUC transcript by northern blot using total RNAs extracted from the LUC, wild-type, mom1-44, and 438-1 seedlings. The LUC plants harbor a regular 35S-LUC-3′-UTR transgene used as a positive control (Zhou et al., 2010). Compared to the mom1-44, a 4-kb LUC transcript also expressed in the 438-1 mutant that is longer than its regular size (2 kb) in LUC plants (Fig. 2C), suggesting that the 438-1 may be a new mutant using the 3′-UTR of downstream gene HPT. Using oligo(dT) reverse-transcribed cDNA as template, we conducted RT-PCR assays using a forward primer specific in the LUC CDS (F1) and reverse primers located between the LUC and HPT (R1), in the HPT promoter (R2), in the HPT CDS (R3), or in the HPT 3′-UTR (R4; Fig. 2D). The PCR products were acquired in four pairs of primer combinations from the 438-1 cDNA, but not from the LUC and wild-type cDNAs (Fig. 2E). As a control, Actin2 transcript was amplified from the LUC, wild-type, and 438-1 cDNAs, but not from the 438-1 genomic DNA template. The larger band amplified from the 438-1 genomic DNA using the Actin2 primers is due to an intron (Fig. 2E), suggesting that the cDNAs were not contaminated with genomic DNA. These results indicate that the LUC luminescence of 438-1 was due to the longer transcript of 35S-LUC by using the 3′-UTR of downstream HPT.
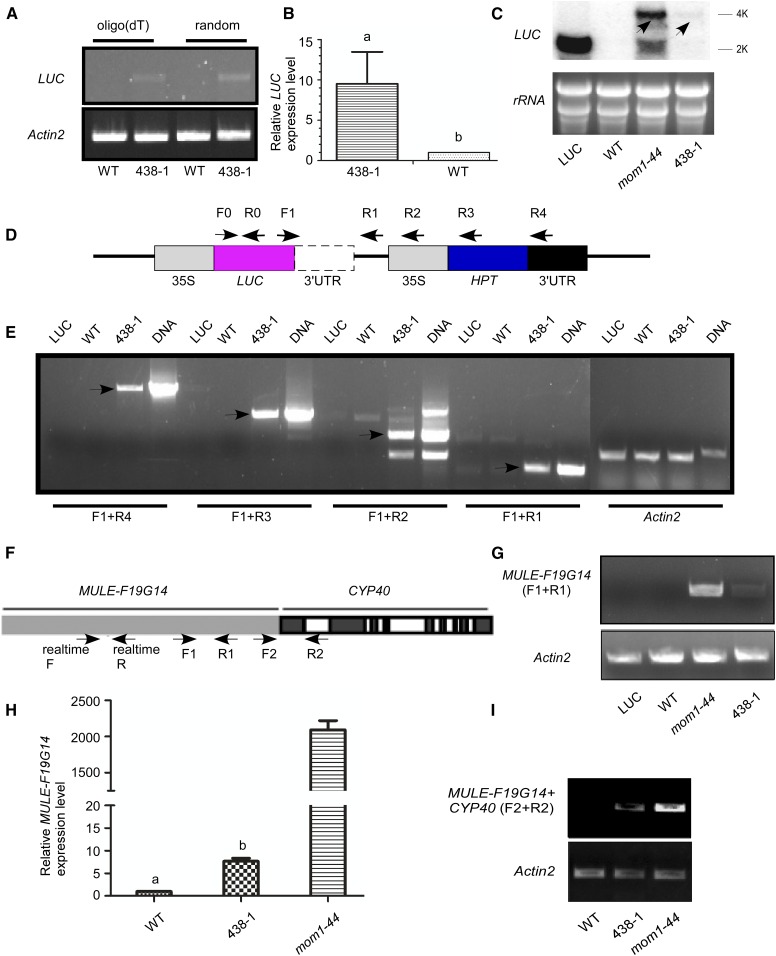
Figure 2.
Detection of 35S-LUC and MULE-F19G14 transcriptions by using the 3′-UTRs of downstream genes in the mutant 438-1. A, Semiquantitative RT-PCR was performed to detect the transcript levels of LUC in wild-type and 438-1 plants using total RNA reverse transcribed by oligo(dT) (left) or random (right) primers. Primers used were F0 and R0 shown in D. Actin2 was amplified as an internal control. B, Relative LUC expression in wild-type and 438-1 plants as shown by real-time PCR using total RNA reverse transcribed by oligo(dT). Primers used were F0 and R0. Actin2 was used as an internal control. Error bars represent SD (n = 3) for at least three replicate experiments. According to Student’s t test, significant differences (P ≤ 0.05) were indicated by different lowercase letters. C, In northern-blot analysis of mRNAs of LUC, the rRNA was used as an RNA loading control. The 4-kb LUC transcript was indicated with arrows. D, The positions of the primers used in A, B, and E are indicated by arrows in the 35S-LUC constructs, including F0, R0, F1, R1, R2, R3, and R4. E, Analysis of LUC expression by RT-PCR using primer pairs F1 and R4, F1 and R3, F1 and R2, or F1 and R1. Actin2 was amplified as an internal control. The cDNAs were reverse transcribed from LUC, wild-type, and 438-1 plants. DNA indicates genomic DNA extracted from 438-1. F, Schema of genetic structure of endogenous MULE-F19G14 and CYP40. The gray box represents MULE-F19G14; dark and open boxes represent exons and introns of CYP40, respectively. The primers used were indicated by arrows including realtime F, realtime R, F1, R1, F2 and R2. G, RT-PCR was performed to detect MULE-F19G14 transcript in LUC, wild-type, mom1-44, and 438-1 plants. Primers used were F1 and R1. Actin2 was used as an internal control. H, Relative MULE-F19G14 expression in wild-type, mom1-44, and 438-1 plants as shown by real-time PCR using total RNA reverse transcribed by oligo(dT). Primers used were realtime F and R. Error bars represent SD (n = 3) for at least three replicate experiments. According to Student’s t test, significant differences (P ≤ 0.05) were indicated by different lowercase letters. I, Expression analysis of MULE-F19G14 by RT-PCR using primers F2 and R2. Actin2 was used as an internal control.
In the mom1-44 mutant, an endogenous mutator-like element MULE-F19G14 with aberrant gene structure is up-regulated by using the 3′-UTR of downstream gene CYCLOPHILIN40 (CYP40; Zhou et al., 2010). Hence, we determined whether the MULE-F19G14 is also activated in the same manner as the 35S-LUC in the 438-1 mutant. First, we performed oligo(dT)-directed RT-PCR assays using a pair of primers (F1+R1, Fig. 2F) specific for MULE-F19G14. Indeed, MULE-F19G14 was up-regulated in the 438-1 mutant (Fig. 2, F and G). This result was confirmed by the following oligo(dT)-directed real-time PCR assays, and the mom1-44 was used as a positive control (Fig. 2H). We designed another pair of primers (F2+R2) respectively located in MULE-F19G14 and CYP40 to perform RT-PCR assays (Fig. 2F). The polyadenylated transcript of MULE-F19G14-CYP40 was detected in both mom1-44 and 438-1 mutants (Fig. 2I), showing that this endogenous MULE-F19G14 was also activated by using the 3′-UTR of downstream CYP40 gene in the 438-1 mutant.
Cloning of the OTS1 Gene
To clone the mutational gene causing release of aberrant gene silencing in the 438-1 mutant, the 438-1 (Col-0) was crossed with wild-type Landsberg erecta (Ler) plants. F1 selfed and F2 seedlings with LUC activity were selected to position the mutational gene. Initial mapping with simple sequence-length polymorphism markers located the gene to the BAC clone T13D8 of lower arm of chromosome I (Fig. 3A). We then sequenced all the candidate genes in this BAC. The sequencing results revealed that the last nucleotide G in the eighth intron of At1g60220 was replaced by a base A (G to A; Fig. 3B). Because the first base of the ninth exon is a G, this mutation causes a nucleotide G deletion in At1g60220 mRNA. To validate this result, we sequenced At1g60220 coding region (CDS) in the wild-type and 438-1 plants. Compared with the wild type, the cDNA of At1g60220 indeed lost a base G corresponding to the first base of the ninth exon in the 438-1 background (Fig. 3C), which in turn results in a truncated At1g60220 protein. At1g60220 encodes OTS1 protein that previously is published as OVERLY TOLERANT TO SALT 1 (OTS1) and UB-LIKE PROTEASE 1D (ULP1D; Chosed et al., 2006; Conti et al., 2008). We then renamed mutant 438-1 to ots1-3 (Fig. 3B).
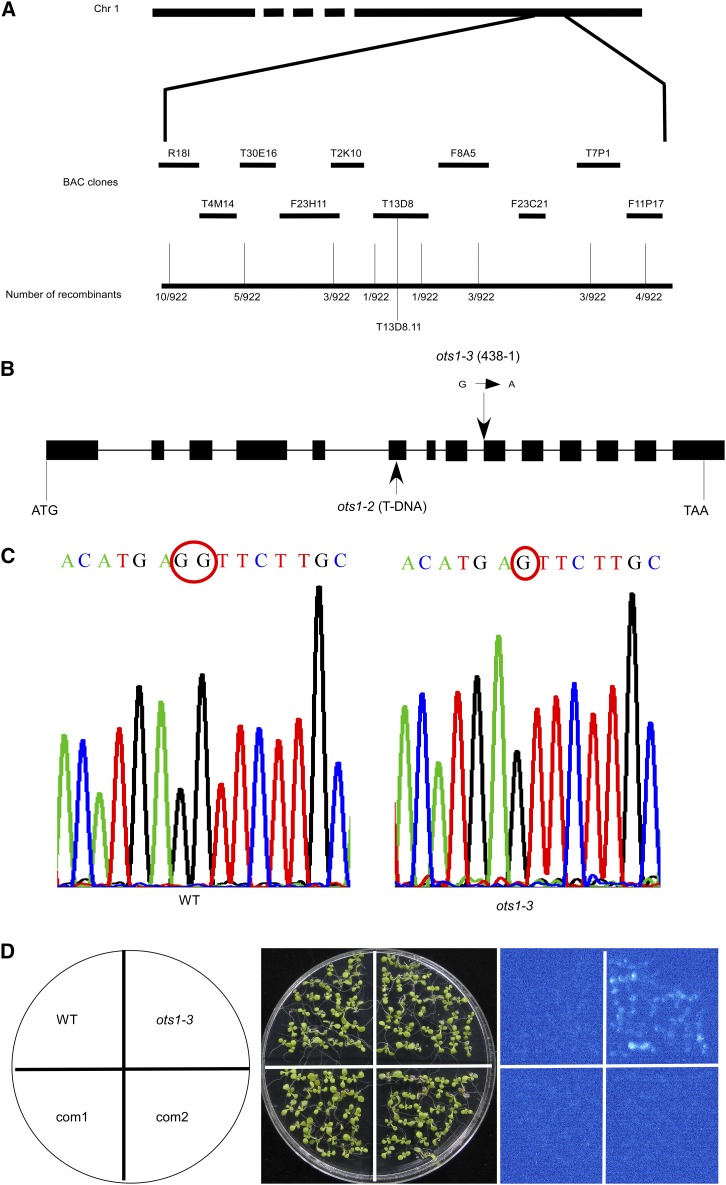
Figure 3.
Cloning of OTS1. A, Map-based cloning of the mutation in 438-1. The 438-1 mutation was mapped to BAC T13D8 of the low arm of chromosome 1. B, Structure of OTS1 gene with exons represented by boxes and introns represented by lines and mutation and insertion positions of the 438-1/ots1-3 and ots1-2 were indicated by arrows. C, The sequencing peaks of a fragment cDNA of OTS1 containing mutation site in wild type and ots1-3. The red circles indicate the loss of a base G in ots1-3 mutant compared with wild type. D, Complementation of the ots1-3 mutant with the full-length OTS1 genomic sequence. Luminescence images showing LUC expression in wild-type and 438-1 plants, and two independent complemented lines (com1 and com2).
To further determine whether the LUC phenotype of ots1-3 is by the reason of mutation in OTS1 gene, we transformed a full-length OTS1 genomic sequence (corresponding to the sequence from 2,779 bp upstream of the OTS1 translation start codon to 312 bp downstream of the stop codon) into mutant ots1-3. The LUC activity of ots1-3 was rescued to wild-type level in two transgenic T3 homozygous lines, com1 and com2 (Fig. 3D). Sequencing results showed that these two complementation lines expressed both wild-type OTS1 and ots1-3 mRNAs (Supplemental Fig. S1). We ordered a T-DNA insertion mutant line GK-207D11 (in the sixth exon of OTS1) from the Arabidopsis Biological Resource Center (ABRC), named as ots1-2 (Fig. 3B). The full-length CDS of OTS1 was not detected in ots1-2 by RT-PCR assays (Supplemental Fig. S2A). We crossed ots1-2 with wild-type plants and allowed the F1 progeny to self. The homozygous ots1-2 line harboring the 35S-LUC was identified. As with ots1-3, the LUC signal and polyadenylated LUC transcript were detected in ots1-2 (Supplemental Fig. S2, B and C). Our results demonstrate that the mutations in OTS1 cause the 35S-LUC expression.
OTS1 Does Not Function Redundantly with OTS2 and Functions in a Different Regulatory Pathway with MOM1 in Regulating TGS
SUMO protease OTS2 shares high similarity with the OTS1 (full-length protein 56% and C-terminal protease domain 73%; Conti et al., 2008). OTS1 and OTS2 act redundantly to regulate salt stress response and light-induced and SA-mediated signaling in Arabidopsis (Conti et al., 2008, 2014; Bailey et al., 2016; Sadanandom et al., 2015). We assumed that the weak LUC signal in the ots1 mutants is because OTS1 and OTS2 have a redundant role regarding to this function. To confirm our speculation, we obtained a knock-out OTS2 T-DNA insertion mutant (SALK_001579; ots2-1; Conti et al., 2008; Supplemental Fig. S2A). We crossed the ots2-1 with wild-type and ots1-3 plants, respectively, and obtained the homozygous ots2-1 and double mutant ots1-3 ots2-1 containing the 35S-LUC transgene. Similar as reported previously, salt-sensitive and early-flowering phenotypes were detected in the double mutant (Supplemental Fig. S2, D and E). However, the LUC signal was not detected in the ots2-1/35S-LUC and was not enhanced in the ots1-3 ots2-1/35S-LUC double mutant seedlings (Supplemental Fig. S2B). Meanwhile, the polyadenylated transcripts of transgene LUC-HPT and MULE-F19G14-CYP40 were detected only in the ots1 mutants and that was not affected by the deletion of OTS2 (Supplemental Fig. S2C). These results suggest that OTS1 and OTS2 have no redundancy on maintaining aberrant gene silencing.
To determine if the OTS1 functions in the same regulatory pathway with the MOM1 in regulation of aberrant gene silence, we crossed the ots1-3 with mom1-44 to generate their double mutant. As shown in Supplemental Figure S3, the double mutant displayed a stronger LUC activity and more polyadenylated transcripts of the 35S-LUC and endogenous MULE-F19G14 than either single mutant. Our results suggest that the mutations in OTS1 and MOM1 have an additive effect on silencing of the 35S-LUC and endogenous MULE-F19G14.
OTS1 SUMO Protease Activity Is Required for Regulating Aberrant Gene Silence
OTS1 encodes a SUMO protease, which localizes in the nucleus (Conti et al., 2008). Previous reports have demonstrated that the residue Cys-525 in the conserved C-terminal catalytic domain is critical for its SUMO protease activity (Li and Hochstrasser, 1999; Chosed et al., 2006; Colby et al., 2006; Conti et al., 2008, 2014). To determine if the OTS1 SUMO protease activity is necessary for the regulation of gene silence, we generated the transgenic plants in the ots1-3 background, which harbored the full-length OTS1 genomic sequence used in the complementation assay but with a mutation causing the C525S substitution in the C-terminal catalytic domain. We used two independent, T3 homozygous transgenic lines for further analysis (Supplemental Fig. S4). Neither the LUC activity nor polyadenylated transcripts of 35S-LUC and endogenous MULE-F19G14 were rescued by the SUMO protease-inactive form (Fig. 4, A and B). These results suggest that the deSUMOylation activity of OTS1 is required for regulating the aberrant gene silence of 35S-LUC and MULE-F19G14.
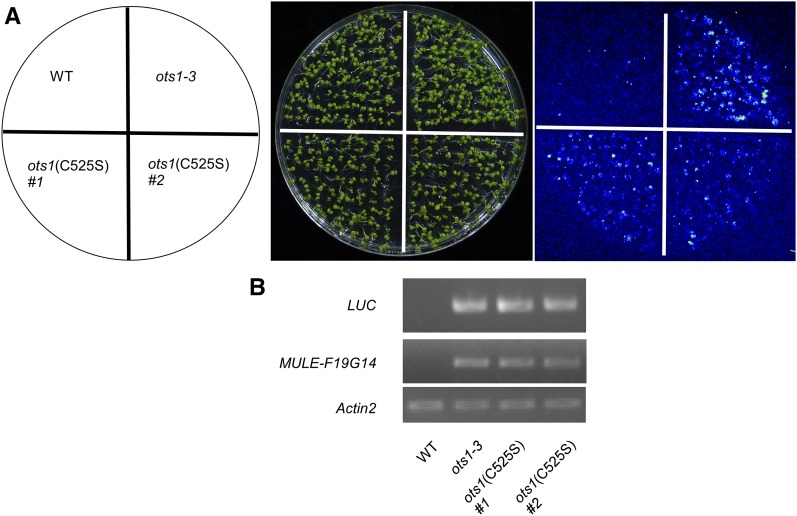
Figure 4.
The deSUMOylation activity of OTS1 is required for maintaining gene silencing. A, Luminescence images showing LUC expression in wild type, ots1-3, and two independent transgenetic lines OTS1(C525S)#1 and #2 in ots1-3 background. B, RT-PCR analysis of expression of LUC and MULE-F19G14 from total RNA extracted from wild-type, ots1-3, OTS1(C525S)#1, and #2 plants. Primers F2 and R2 of MULE-F19G14 was used and Actin2 was used as an internal control.
Expression of Aberrant Genes in ots1-3 Requires Pol V and DDR Complex
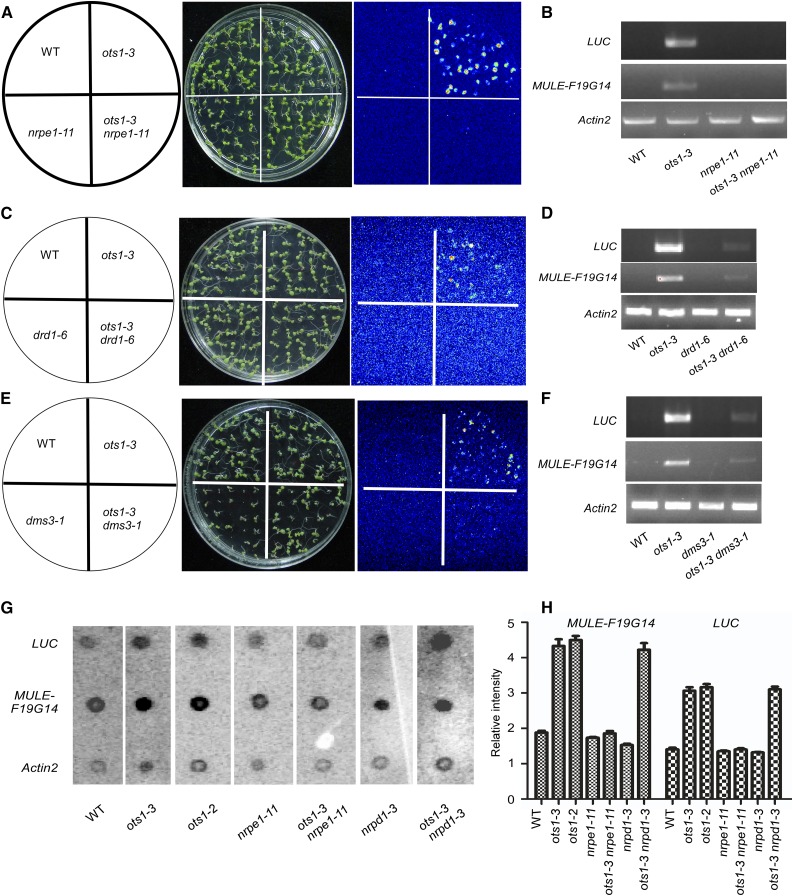
RdDM mediates TGS of transposons, repeated sequences, and some neighboring genes, which has been well-characterized in Arabidopsis (Matzke and Mosher, 2014). To explore if the RdDM functions in the OTS1-mediated regulation of aberrant gene expression, we crossed some mutants of the RdDM components with the wild-type and ots1-3 plants. We first obtained the nrpe1-11 (NRPE1, the largest subunit of plant-specific RNA Pol V) and double mutant ots1-3 nrpe1-11 harboring the 35S-LUC. The LUC signal was not detected in the nrpe1-11/35S-LUC and was suppressed by the nrpe1-11 mutant in the ots1-3 background (Fig. 5A), suggesting that aberrant gene expression in ots1-3 requires Pol V. To confirm this result, we performed oligo(dT)-directed RT-PCR analyses to detect the polyadenylated transcripts of 35S-LUC and endogenous MULE-F19G14 in the ots1-3 nrpe1-11 plants, and both of the transcripts were at an undetectable level (Fig. 5B).
Figure 5.
Pol V and DDR complex are required for aberrant gene expression in ots1-3. A, Luminescence images showing LUC expression in wild-type plants, ots1-3, nrpe1-11, and ots1-3 nrpe1-11 mutants. B, RT-PCR analysis of expression of LUC and MULE-F19G14 from total RNA extracted from wild type, ots1-3, nrpe1-11, and ots1-3 nrpe1-11. Primers F2 and R2 of MULE-F19G14 were used, and Actin2 was used as an internal control. C, Luminescence images showing LUC expression in wild-type plants, ots1-3, drd1-6, and ots1-3 drd1-6 mutants. D, RT-PCR analysis of expression of LUC and MULE-F19G14 from total RNA extracted from wild type, ots1-3, drd1-6, and ots1-3 drd1-6. Primers F2 and R2 of MULE-F19G14 was used and Actin2 was used as an internal control. E, Luminescence images showing LUC expression in wild-type plants, ots1-3, dms3-1, and ots1-3 dms3-1 mutants. F, RT-PCR analysis of expression of LUC and MULE-F19G14 from total RNA extracted from wild type, ots1-3, dms3-1, and ots1-3 dms3-1. Primers F2 and R2 of MULE-F19G14 were used and Actin2 was used as an internal control. G, Analysis of nascent transcription of LUC and MULE-F19G14 by nuclear run-on assays in wild-type plants, ots1-3, ots1-2, nrpe1-11, ots1-3 nrpe1-11, nrpd1-3, and ots1-3 nrpd1-3 mutants. Actin2 was used as a control. H, Quantification of nascent transcription of LUC and MULE-F19G14 in G, was measured with ImageQuant 5.0 software and normalized to the intensity of the corresponding Actin2 band (n = 3; data are presented as mean ± sd).
Pol V requires DDR complex (DRD1, DMS3, and RDM1) for function in producing noncoding scaffold RNAs and guiding sequence-specific DNA methylation (Kanno et al., 2004, 2005, 2008; Herr et al., 2005; Onodera et al., 2005; Pontier et al., 2005; Gao et al., 2010; Law et al., 2010; Zhong et al., 2012; Zhou and Law, 2015). To test whether the DDR complex is required for the OTS1-mediated regulation of aberrant gene expression, we introduced drd1-6 or dms3-1 into ots1-3 background and obtained the corresponding double mutants ots1-3 drd1-6 and ots1-3 dms3-1. The mutation in DRD1 or DMS3 also suppressed the expression of 35S-LUC and MULE-F19G14 in the ots1-3 mutant (Fig. 5, C–F), indicating that the DDR complex is also involved in the OTS1-mediated aberrant gene expression regulation. Pol IV is another plant-specific RNA polymerase, whose function is to generate siRNAs in the RdDM pathway (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005; Pontier et al., 2005; Matzke et al., 2009). We tested if Pol IV is involved in the OTS1-mediated aberrant gene expression regulation. We obtained double mutant ots1-3 nrpd1-3 (Supplemental Fig. S5A; nrpd1-3, NRPD1, the largest subunit of Pol IV; Wierzbicki et al., 2008) harboring the 35S-LUC by crossing nrpd1-3 into the ots1-3 background. The expression of 35S-LUC and MULE-F19G14 in nrpd1-3 ots1-3 was similar as in the ots1-3 mutant (Supplemental Fig. S5, B and C), suggesting that Pol IV is not required for the generation of polyadenylated transcripts of aberrant genes. These results suggest that Pol V and DDR complex are key components in OTS1-mediated pathway to prevent aberrant gene expression by using the 3′-UTR of downstream gene, which may be distinct from their functions in RdDM pathway.
To determine that OTS1-mediated aberrant gene silence is at the transcriptional level or posttranscriptional level, we further detected the nascent transcription levels of the 35S-LUC and MULE-F19G14 in the wild-type, ots1, nrpel-11, nrpd1-3, ots1 nrpel-11, and ots1 nrpd1-3 plants by nuclear run-on assays. The nascent 35S-LUC and MULE-F19G14 transcripts were detected in the wild-type, nrpel-11 and nrpd1-3 plants and enhanced in the ots1 mutants (Fig. 5, G and H). The nrpel-11 but not nrpd1-3 mutation repressed this enhancement (Fig. 5, G and H). These results suggest that the OTS1 maintains aberrant gene silencing and inhibits the formation of polyadenylated mRNAs at the transcriptional level that requires a functional Pol V-dependent pathway.
SUMO Modification Plays an Important Role in Regulating Aberrant Gene TGS
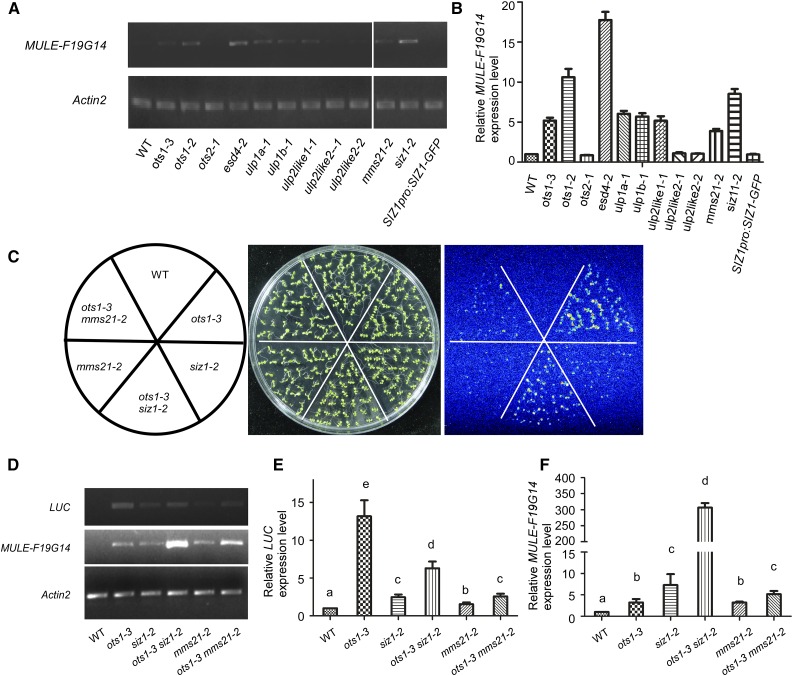
Comparing to mom1-44 mutant, the ots1 mutants displayed weak phenotype in mediating the expression of 35S-LUC. One possibility is due to its redundant function with other SUMO proteases, but not OTS2. To test if other SUMOylation-related proteins are involved in the regulation of aberrant gene expression, we obtained the T-DNA insertion mutants in other known SUMO proteases, ESD4 (At4G15880) and AtULP1a/ELS1 (At3g06910), and putative SUMO proteases, AtULP1b (At4g00690), ULP2like1 (At4g33620), and ULP2like2 (At1g09730; Chosed et al., 2006; Colby et al., 2006; Park et al., 2011b; Murtas et al., 2003; Hermkes et al., 2011; Novatchkova et al., 2004), and identified their knock-out or knock-down features by RT-PCR analysis (Supplemental Fig. S6). The resulting mutants were used to perform RT-PCR and real-time PCR to determine the polyadenylated mRNAs using the 3′-UTR of downstream gene of MULE-F19G14 (Fig. 6, A and B). This transcript was detected in the esd4, ulp1a, ulp1b, and ulp2like1 mutants, but not in the ulp2like2 mutant. These results suggest that SUMO modification plays an important role in regulating TGS. To test if SUMO E3 ligase also is involved in this regulation, we obtained two mutants of SIZ1 and MMS21 (HYP2), siz1-2 and mms21-2 (hpy2-2), and siz1-2 harboring SIZ1pro:SIZ1-GFP plants (Ishida et al., 2012). We also generated ots1-3 siz1-2 and ots1-3 mms21-2 double mutants containing the 35S-LUC. These materials were then used to test the transcript status of 35S-LUC and MULE-F19G14. To our surprise, two E3 ligases also inhibited the expression of 35S-LUC and MULE-F19G14, and SIZ1pro:SIZ1-GFP rescued the phenotype of siz1-2 mutant (Fig. 6, A–C). We predicted that the LUC-HPT and MULE-F19G14-CYP40 transcripts in the ots1-3 mutant may be counteracted by the mutations of SUMO E3 ligases. Indeed, in the 35S-LUC locus, the LUC-HPT transcript was reduced in the double mutant compared with that in the ots1-3, especially in the ots1-3 mms21-2 double mutant (Fig. 6, C–E). However, many more transcripts of MULE-F19G14 than that in their single mutants were detected in the ots1-3 siz1-2 and ots1-3 mms21-2 double mutants, especially in ots1-3 siz1-2 (Fig. 6, D and F). These results suggest that TGS of aberrant genes regulated by SUMOylation in different loci involves various SUMOylation targets that may function in distinct pathways.
Figure 6.
SUMO modification plays a role in the regulation of aberrant gene TGS. A, RT-PCR analysis of MULE-F19G14 expression using primers F2 and R2 (Fig. 2F) in wild-type, ots1-3, ots1-2, ots2-1, esd4-2, ulp1a-1, ulp1b-1, ulp2like1-1, ulp2like2-1, ulp2like2-2, mms21-2, siz1-2, and SIZ1pro:SIZ1-GFP in siz1-2 plants. Actin2 was used as an internal control. B, Analysis of the polyadenylated MULE-F19G14 expression in wild-type, ots1-3, ots1-2, ots2-1, esd4-2, ulp1a-1, ulp1b-1, ulp2like1-1, ulp2like2-1, ulp2like2-2, mms21-2, siz1-2, and SIZ1pro:SIZ1-GFP in siz1-2 plants by real-time PCR using total RNA reverse transcribed by oligo(dT). Actin2 was used as an internal control. Error bars represent SD (n = 3) for at least three replicate experiments. C, Luminescence images showing LUC expression in wild-type, ots1-3, siz1-2, ots1-3 siz1-2, mms21-2 and ots1-3 mms21-2 plants, which all carry the transgene 35S-LUC. D, RT-PCR analysis of expression of LUC and MULE-F19G14 in wild-type, ots1-3, siz1-2, ots1-3 siz1-2, mms21-2, and ots1-3 mms21-2 plants. Primers F2 and R2 of MULE-F19G14 was used and Actin2 was used as an internal control. E and F, Expression analysis of the polyadenylated LUC and MULE-F19G14 RNA in wild-type, ots1-3, siz1-2, ots1-3 siz1-2, mms21-2, and ots1-3 mms21-2 plants by real-time PCR using total RNA reverse transcribed by oligo(dT). Actin2 was used as an internal control. Error bars represent SD (n = 3) for at least three replicate experiments. According to Student’s t test, significant differences (P ≤ 0.05) were indicated by different lowercase letters.
DISCUSSION
SUMO is an evolutionarily conserved, reversible posttranslational modifier that covalently conjugates to cellular target proteins and modulates their activity, resulting in important consequences to the cellular machinery (Melchior, 2000; Johnson, 2004; Hay, 2005; Geiss-Friedlander and Melchior, 2007; Wang and Dasso, 2009; van der Veen and Ploegh, 2012). Many previous studies have established the roles of SUMOylation in abiotic stress responses, defense reactions, hormone signaling, and in the regulation of flowering time, cell growth, and development in plants (Miura et al., 2007a; Park et al., 2011b). In this study, we identify OTS1, a SUMO protease, whose activity is required for maintaining transcriptional aberrant gene silencing. Our study provides a genetic link that SUMOylation is involved in regulating aberrant gene TGS and preventing aberrant gene transcription by using the 3′-UTR of downstream gene and reveals a unique role of Pol V that positively regulates aberrant gene expression.
In Arabidopsis, there are about seven genes being considered encoding SUMO proteases. Our results showed that five of them function in the aberrant gene TGS regulatory process except for OTS2 and ULP2like2, suggesting that SUMO proteases may act redundantly to maintain aberrant gene silencing. This is also supported by the ots1 mutants that displayed a weak phenotype of the LUC activity. The SUMO E3 ligases, SIZ1 and MMS21, have a similar effect as these SUMO proteases on the regulation of aberrant gene expression. Recently, it has been shown that mutations in SUMO proteases and SUMO E3 ligase cause elevated SA levels and enhanced resistance to pathogen (Villajuana-Bonequi et al., 2014; Bailey et al., 2016). To date, a detailed mechanism explaining these contradictory results has not been characterized. The ots1 siz1 and ots1 mms21 double mutants display an enhanced transcriptional activity for the MULE-F19G14, but a neutral activity for the 35S-LUC. These results reveal that, first, SUMOylation is an important PTM for maintaining aberrant gene silencing; second, precisely regulated SUMO homeostasis in different components is indispensable for this process; last, different regulatory mechanisms may require for this process from different loci. It is not known whether and how other protein modification systems are involved in the TGS regulation. To precisely function in a regulatory process, most proteins have to be posttranslationally modified. For instance, a few key players of epigenetic regulation, such as NRPE1, DDM1, and Methyltransferase 1, are identified experimentally to be phosphorylated in plant (The Arabidopsis Information Resource, PhosPhAt), which implies that the phosphorylation modification is involved in the TGS regulation. However, further studies are required to determine if these phosphorylation modifications do function in this biological function.
MOM1 plays a role in regulating aberrant gene TGS and is a component of a TGS mechanism independent of DNA methylation marks (Amedeo et al., 2000; Habu et al., 2006; Vaillant et al., 2006). Although both OTS1 and MOM1 maintain aberrant gene silencing at transcription level and prevent aberrant gene transcription by using the 3′-UTR of a downstream gene, the double mutant ots1-3 mom1-44 displays a stronger LUC activity than any single mutant. Although it is likely that the endogenous gene MULE-F19G14 is not the target of Pol V (Wierzbicki et al., 2012), the loss of function of Pol V rescues the gene silencing and inhibits aberrant gene expression in ots1-3 mutant but does not affect MULE-F19G14 expression in mom1 mutants (Yokthongwattana et al., 2010). These results suggest that OTS1 and MOM1 function in different pathways in regulating aberrant gene TGS.
RdDM pathway usually positively regulates TGS (Matzke and Mosher, 2014). However, the Pol V and DDR complex, rather than Pol IV, is required for TGS release of the aberrant genes in mutant ots1-3 and plays a negative role for TGS. We currently do not know how the SUMOylation affects the Pol V functions of protein activity, stability, localization, partner interaction, which in turn mediate the TGS of aberrant genes. Previous reports have demonstrated that Pol V affects heterochromatin organization, which is independent of Pol IV (Douet et al., 2009; Pontes et al., 2009). Our study provides another case that Pol V functions in the regulation of gene expression, which does not rely on Pol IV. Meanwhile, our data give a new clue to recognize the biological functions of Pol V in plants and may partially explain why one-quarter of Pol V peaks occur at loci (mostly genes) lacking significant cytosine methylation or siRNA signatures (Wierzbicki et al., 2012).
MATERIALS AND METHODS
Plant Materials, Mutant Isolation, and Growth Conditions
In all experiments, Arabidopsis (Arabidopsis thaliana; Col-0) with the transgene 35S-LUC (3′-UTR deletion) and 35S-LUC-3′UTR reporter genes are referred to as wild-type and LUC plants, respectively. Wild type was mutagenized with ethyl methanesulfonate (Redei and Koncz, 1992). The construction of 35S-LUC and 35S-LUC-3′UTR plasmids, mutant ots1-3 (438-1) screening, and plant growth was described (Zhou et al., 2010). The ots1-2 (GK-207D11-014550) was obtained from the ABRC. The mom1-44 has been described (Zhou et al., 2010). The ots2-1 (SALK_001579) was previously described (Conti et al., 2008). The mutants nrpe1-11 (SALK_029919C) and nrpd1-3 (SALK_128428) have been described (Wierzbicki et al., 2008), and esd4-2 was described previously (Murtas et al., 2003). The siz1-2 (SALK_065397), mms21-2 (hpy2-2, SAIL_77_G06), and SIZ1pro:SIZ1-GFP were described (Ishida et al., 2012). The ulp1a-1 (SAIL_239_B08), ulp1b-1 (SALK_110379), ulp2like1-1 (SAIL_140_B10), ulp2like2-1 (SALK_049255), and ulp2like2-2 (SALK_022079) mutants from the Col-0 background were obtained from the ABRC. The dms3-1 was described (Kanno et al., 2008), as was drd1-6 (Kanno et al., 2004).
For salt treatment and flowering time measurement, we controlled the conditions as described previously (Conti et al., 2008).
Positional Cloning and Complementation
For genetic mapping, homozygous ots1-3 plants (Col-0 background) were crossed to Ler plants, and the F1 progenies were allowed to self. We selected homozygous ots1-3 plants by picking out LUC-positive seedlings in the F2 plants determined through luminescence imaging. A total of 922 homozygous ots1-3 mutant seedlings were selected for mapping with molecular markers that are polymorphic between Ler and Col-0. Genetic mapping was performed as described previously (Liu and Zhu, 1998). By using simple sequence-length polymorphism markers, the mutational gene was mapped to BAC clone T13D8 of chromosome 1. To identify the mutation in ots1-3, candidate genes in T13D8 from ots1-3 mutant plants were sequenced.
As complementation of ots1-3, the full-length OTS1 genomic DNA fragment 6,669 bp (corresponding to the sequence from 2,779 bp upstream of the OTS1 translation start codon to 312 bp downstream of the stop codon) or mutational OTS1 encoding SUMO protease-inactive (C525S) form were cloned into the BamHI/HindIII sites in pCAMBIA3300 vector. The resulting plasmids were transformed into the ots1-3 mutant using Agrobacterium tumefaciens-mediated floral transformation, respectively (Clough and Bent, 1998), and homozygous T3 transgenic plants were used for analysis. Primers are listed in Supplemental Table S1.
RNA Analysis
Total RNA was extracted with TRIzol reagent (Invitrogen) from 2-week-old seedlings grown on Murashige and Skoog medium. Total RNA was treated with RNase-free DNase I (Invitrogen) to remove genomic DNA. RNA was used for RNA gel-blot, RT-PCR, or quantitative real-time PCR analysis.
For northern-blot analysis, 15 μg of total RNA of each sample was separated on 1.2% denaturing agarose gels containing 2% formaldehyde and was transferred to Hybond-N+ membranes (GE Healthcare) for northern hybridization. Membranes were cross-linked by UV and hybridized overnight at 65°C with [α-32P]CTP-labeled DNA oligonucleotides in hybridization buffer. All labeling involved use of the Random Primer-labeled kit (TaKaRa). Northern blots were washed twice with 2× SSC, 0.1% SDS and then once with 1× SSC, 0.1% SDS. Washed blots were exposed to x-ray films.
For semiquantitative RT-PCR, cDNA was synthesized with oligo(dT) or random primers from 5 μg of total RNA using Moloney murine leukemia virus reverse transcriptase according to the manufacturer’s instructions (Promega). PCR amplification included a preincubation at 94°C for 2 min followed by 32 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 30 s, and extension at 68°C for 30 s with KOD-Plus-Neo polymerase (Takara). Quantitative real-time PCR analysis was performed using SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara). Analysis was performed using the Applied Biosystems 7500 Fast Real-Time PCR System. The reaction program started from predenaturation at 95°C for 30 s followed by 40 cycles of denaturation at 95°C for 5 s and extension at 60°C for 34 s. All primers are listed in Supplemental Table S1.
Nuclear Run-On Assay
We performed nuclear run-on assays as described previously (Folta and Kaufman, 2006). We obtained 2 μg CDS sequences of LUC, MULE-F19G14, and Actin2, denatured them at 95°C for 5 min, and dotted them on the Hybond-N+ membrane (GE Healthcare). After nuclei isolation from 2-week-old seedlings, 1 × 107 freshly prepared nuclei were incubated with ribonucleotides including [α-32P]UTP at 30°C for 30 min. After hybridization, radioactive signals were quantified by phosphorimaging. Primers are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OTS1, AT1G60220; NRPE1, AT2G40030; SIZ1, AT5G60410; OTS2, AT1G10570; DRD1, AT2G16390; ESD4, AT4G15880; MMS21, AT3G15150.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The sequencing chromatogram of OTS1 cDNA of two independent complemented lines of mutant 438-1, com1, and com2.
Supplemental Figure S2. OTS2 has no functional redundancy with OTS1 for the regulation of gene silence.
Supplemental Figure S3. OTS1-mediated gene silence is independent of MOM1.
Supplemental Figure S4. The sequencing chromatogram of OTS1 cDNA in two independent transgenic lines, OTS1(C525S)#1 and #2.
Supplemental Figure S5. Mutation of Pol IV does not affect the gene silence in the ots1-3 mutant.
Supplemental Figure S6. RT-PCR analysis of transcript levels of SUMO proteases, ULP1A, ULP1B, ULP2like1, and ULP2like2 in their corresponding T-DNA lines.
Supplemental Table S1. Primer list for nuclear run-on, northern blot, PCR, and plasmid construction.
Supplementary Material
Acknowledgments
We thank Yongfu Fu for providing the ots2-1 (SALK_001579) mutant.
Glossary
- PTM
posttranslational modification
- RdDM
RNA-directed DNA methylation
- siRNA
small-interfering RNA
- TGS
transcriptional gene silencing
Footnotes
This work was supported by National Natural Science Foundation of China (grant no. 31430012) and Foundation for Innovative Research Group of the National Natural Science Foundation of China (grant no. 31421062).
Articles can be viewed without a subscription.
References
- Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J (2000) Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405: 203–206 [DOI] [PubMed] [Google Scholar]
- Bailey M, Srivastava A, Conti L, Nelis S, Zhang C, Florance H, Love A, Milner J, Napier R, Grant M, et al. (2016) Stability of small ubiquitin-like modifier (SUMO) proteases OVERLY TOLERANT TO SALT1 and -2 modulates salicylic acid signalling and SUMO1/2 conjugation in Arabidopsis thaliana. J Exp Bot 67: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD (2012) Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 69: 3613–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhiraja R, Hermkes R, Müller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A (2009) Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol 149: 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Verde N, Lourenço T, Magalhães AP, Tavares RM, Bejarano ER, Azevedo H (2015) SIZ1-dependent post-translational modification by SUMO modulates sugar signalling and metabolism in Arabidopsis thaliana. Plant Cell Physiol 56: 2297–2311 [DOI] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Chen YY, Tang IC, Liang HM, Lai CC, Chiou JM, Yeh KC (2011) Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol 156: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R, Mukherjee S, Lois LM, Orth K (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J 398: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colby T, Matthäi A, Boeckelmann A, Stuible HP (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142: 318–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, et al. (2014) Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 28: 102–110 [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet J, Tutois S, Tourmente S (2009) A Pol V-mediated silencing, independent of RNA-directed DNA methylation, applies to 5S rDNA. PLoS Genet 5: e1000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta KM, Kaufman LS (2006) Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat Protoc 1: 3094–3100 [DOI] [PubMed] [Google Scholar]
- Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, et al. (2010) An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465: 106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Habu Y, Mathieu O, Tariq M, Probst AV, Smathajitt C, Zhu T, Paszkowski J (2006) Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep 7: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Hermkes R, Fu YF, Nürrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G (2011) Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Molnàr A, Jones A, Baulcombe DC (2006) Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA 103: 14994–15001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC (2009) An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol 16: 91–93 [DOI] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K (2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Yoshimura M, Miura K, Sugimoto K (2012) MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS One 7: e46897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE (2007) Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES. (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ETH (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell 38: 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJM (2008) A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet 40: 670–675 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJM (2004) Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14: 801–805 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE (2010) A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol 20: 951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251 [DOI] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Luo Z, Chen Z (2007) Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19: 943–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Melchior F. (2000) SUMO--nonclassical ubiquitin. Annu Rev Cell Dev Biol 16: 591–626 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM (2007a) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10: 495–502 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007b) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Miura T, Hasegawa PM (2010) SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol 51: 103–113 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32: 286–295 [DOI] [PubMed] [Google Scholar]
- Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15: 2308–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A (2004) SUMO conjugation in plants. Planta 220: 1–8 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E (2004) A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem 279: 41346–41351 [DOI] [PubMed] [Google Scholar]
- Park BS, Song JT, Seo HS (2011a) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ (2011b) SUMO and SUMOylation in plants. Mol Cells 32: 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Costa-Nunes P, Vithayathil P, Pikaard CS (2009) RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway. Mol Plant 2: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Redei GP, Koncz C (1992) Classical mutagenesis. In Koncz C, Chua N-H, Schell J, eds, Methods in Arabidopsis Research. World Scientific Publishing Co., Singapore, pp 16–82 [Google Scholar]
- Sadanandom A, Ádám É, Orosa B, Viczián A, Klose C, Zhang C, Josse EM, Kozma-Bognár L, Nagy F (2015) SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I, Schubert I, Tourmente S, Mathieu O (2006) MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep 7: 1273–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Ploegh HL (2012) Ubiquitin-like proteins. Annu Rev Biochem 81: 323–357 [DOI] [PubMed] [Google Scholar]
- Villajuana-Bonequi M, Elrouby N, Nordström K, Griebel T, Bachmair A, Coupland G (2014) Elevated salicylic acid levels conferred by increased expression of ISOCHORISMATE SYNTHASE 1 contribute to hyperaccumulation of SUMO1 conjugates in the Arabidopsis mutant early in short days 4. Plant J 79: 206–219 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dasso M (2009) SUMOylation and deSUMOylation at a glance. J Cell Sci 122: 4249–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS (2012) Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev 26: 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokthongwattana C, Bucher E, Caikovski M, Vaillant I, Nicolet J, Mittelsten Scheid O, Paszkowski J (2010) MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J 29: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM (2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142: 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Qi Y, Liu M, Yang C (2013) SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F). J Integr Plant Biol 55: 83–95 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE (2012) DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Law JA (2015) RNA Pol IV and V in gene silencing: rebel polymerases evolving away from Pol II’s rules. Curr Opin Plant Biol 27: 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang J, Lin H, Guo G, Guo Y (2010) MORPHEUS’ MOLECULE1 is required to prevent aberrant RNA transcriptional read-through in Arabidopsis. Plant Physiol 154: 1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.