KNS4/UPEX1 is involved in the synthesis of type II AG of AGPs/pectins that are essential components of pollen grain walls providing the scaffold for exine construction/architecture.
Abstract
Pollen exine is essential for protection from the environment of the male gametes of seed-producing plants, but its assembly and composition remain poorly understood. We previously characterized Arabidopsis (Arabidopsis thaliana) mutants with abnormal pollen exine structure and morphology that we named kaonashi (kns). Here we describe the identification of the causal gene of kns4 that was found to be a member of the CAZy glycosyltransferase 31 gene family, identical to UNEVEN PATTERN OF EXINE1, and the biochemical characterization of the encoded protein. The characteristic exine phenotype in the kns4 mutant is related to an abnormality of the primexine matrix laid on the surface of developing microspores. Using light microscopy with a combination of type II arabinogalactan (AG) antibodies and staining with the arabinogalactan-protein (AGP)-specific β-Glc Yariv reagent, we show that the levels of AGPs in the kns4 microspore primexine are considerably diminished, and their location differs from that of wild type, as does the distribution of pectin labeling. Furthermore, kns4 mutants exhibit reduced fertility as indicated by shorter fruit lengths and lower seed set compared to the wild type, confirming that KNS4 is critical for pollen viability and development. KNS4 was heterologously expressed in Nicotiana benthamiana, and was shown to possess β-(1,3)-galactosyltransferase activity responsible for the synthesis of AG glycans that are present on both AGPs and/or the pectic polysaccharide rhamnogalacturonan I. These data demonstrate that defects in AGP/pectic glycans, caused by disruption of KNS4 function, impact pollen development and viability in Arabidopsis.
Pollen, the male gametophyte of all seed plants, is crucial for reproductive success. Due to the harsh environmental conditions that pollen must survive in, it has developed an elaborate and specialized cell wall. However, despite its importance our knowledge of the fine structure and assembly of the pollen grain wall is poorly understood (Newbigin et al., 2009; Ariizumi and Toriyama, 2011; Quilichini et al., 2015; Shi et al., 2015). Arabidopsis (Arabidopsis thaliana) provides an ideal genetic and cell biological model to dissect the assembly of the components of the pollen grain wall. Arabidopsis pollen grains have a typical surface structure that consists of an inner intine layer, which is usually made up of cellulose and pectin, and the outer exine, which is largely composed of sporopollenin (Li et al., 1995). The exine is further subdivided into inner nexine and outer sexine. The sexine has a three-dimensional (3D) structure composed of many columns called baculae and a roof called tectum. These two structures give the Arabidopsis pollen surface a reticulate appearance with a uniform mesh size. The nexine is composed of outer nexine I (foot layer), which contains sporopollenin, and inner nexine II (endexine; Ariizumi and Toriyama, 2011; Jiang et al., 2013). A recent study suggests that arabinogalactan proteins (AGPs) are key constituents of nexine with expression of AtAGP6 in the nexine-deficient mutant transposable element silencing via at-hook (tek) partially restoring nexine formation (Jia et al., 2015). The materials comprising the exine are derived from the cells of the tapetum and the deposition pattern of the exine is determined during the tetrad stage (Zhou et al., 2015). Surrounding the outer exine layer is the pollen coat, containing proteins and lipids that accumulate in the exine cavities (Quilichini et al., 2015).
Exine is composed of sporopollenin, a structurally robust biopolymer made from aliphatic and aromatic precursors (Ariizumi and Toriyama, 2011; Quilichini et al., 2015). Sporopollenin precursors are synthesized mainly in tapetal cells and transported to the microspore surface by the involvement of many genes in which mutations often cause strong defects in exine development. These genes include: MALE STERILITY2 (MS2) (Aarts et al., 1997), FACELESS POLLEN1 (FLP1) (Ariizumi et al., 2003), NO EXINE FORMATION1 (NEF1) (Ariizumi et al., 2004), CYP703A2 (Morant et al., 2007), ACYL-CoA SYNTHETASE5 (ACOS5) (de Azevedo Souza et al., 2009), LESS ADHERENT POLLEN3 (LAP3), LAP5/POLYKETIDE SYNTHASE B (PKSB), LAP6/POLYKETIDE SYNTHASE A (PKSA) (Dobritsa et al., 2009a; Dobritsa et al., 2010; Kim et al., 2010), CYP704B1 (Dobritsa et al., 2009b), DIHYDROFLAVONOL 4-REDUCTASE-LIKE1 (DRL1)/TETRAKETIDEα-PYRONE REDUCTASE2 (TKPR2) (Tang et al., 2009; Grienenberger et al., 2010), TKPR1 (Grienenberger et al., 2010), and a member of ATP-binding cassette transporter gene ABCG26 (Quilichini et al., 2010; Choi et al., 2011; Dou et al., 2011; Kuromori et al., 2011). Although many of these genes are specifically expressed in tapetal cells, it has been proposed that microspores also synthesize and secrete sporopollenin at the initial stage of exine development in tetrads (Wallace et al., 2011).
Exine development is initiated with the formation of primexine, a layer of matrix between the microspore plasma membrane and callosic wall that temporarily separates the microspores in the pollen tetrad (Heslop-Harrison, 1968; Owen and Makaroff, 1995; Paxson-Sowders et al., 1997). Primexine is believed to act as a scaffold for the attachment of sporopollenin monomers (Ariizumi and Toriyama, 2011). After primexine formation, undulation of the microspore plasma membrane occurs, which contributes to determining the position of bacula formation (Paxson-Sowders et al., 1997; Zhou et al. 2015). An Arabidopsis gene, DEFECTIVE IN EXINE FORMATION1 (DEX1), encodes a membrane-associated protein required for primexine development and plasma membrane undulation, and a mutation of the gene results in random deposition of sporopollenin at the plasma membrane surface (Paxson-Sowders et al., 1997; Paxson-Sowders et al., 2001). Mutants of TRANSIENT DEFECTIVE EXINE1 (TDE1)/DE-ETIOLATED2 (DET2) revealed the involvement of brassinosteroids in primexine formation (Ariizumi et al., 2008). The callosic wall in tetrads also plays a crucial role in exine development. Mutations in CALLOSE SYNTHASE5 (CALS5)/GLUCAN SYNTHASE LIKE2 (GSL2) cause a severe defect in exine development despite normal deposition of primexine (Dong et al., 2005; Nishikawa et al., 2005). Furthermore, RUPTURED POLLEN GRAIN1 (RPG1), NO PRIMEXINE AND PLASMA MEMBRANE UNDULATION (NPU), and EXINE FORMATION DEFECT (EFD) are required for CALS5/GSL2 expression, primexine deposition, and plasma membrane undulation (Guan et al., 2008; Chang et al., 2012; Sun et al., 2013; Hu et al., 2014). Mutations in these genes almost completely abolish the structure of exine and often affect the viability of pollen grains.
Despite the accumulating knowledge about sporopollenin synthesis, little is known about how the 3D structure/organization of exine is formed (Quilichini et al., 2015; Shi et al., 2015). To identify the genes determining exine structure, we previously screened an EMS-mutagenized population of Arabidopsis by scanning electron microscopy (SEM) and identified 12 mutants, named kaonashi (kns1 to kns12), showing abnormal exine morphology and classified them into four types (Suzuki et al., 2008). Type-1 mutants showed collapsed exine structures whereas type-2 mutants make a remarkably thin exine. The type-3 mutants showed defective tectum formation and the type 4 mutants showed densely distributed baculae. All identified kns mutants are recessive, suggesting that these KNS genes are expressed in diploid cells (Suzuki et al., 2008). In this article, we focus on a unique type-2 mutant, kns4, in which the exine is less than half the thickness of wild type but still has a reticulate structure. Another characteristic of kns4 is that mature pollen grains of the mutant often aggregate (Suzuki et al., 2008).
A similar genetic screen was carried out by Dobritsa et al., (2011) with an Arabidopsis T-DNA insertion population and many mutants with a variety of defects in exine structure were found. For example, the exine of spongy2 (spg2) has irregularly distributed and elongated tectum elements and lacunae (holes within the tectum) of variable size and shape. Another mutant, uneven pattern of exine1 (upex1), shows a grossly disorganized exine (Dobritsa et al., 2011). Only part of the upex1 pollen surface maintains a reticulate structure; other areas either lack exine or have essentially smooth exine with small lacunae (Dobritsa et al., 2011). Observations at high magnification revealed that the upex1 exine consisted of very short baculae and an overdeveloped tectum that had fewer and smaller lacunae than in the wild type (Dobritsa et al., 2011). The gene underlying the upex1 mutation was identified as At1g33430, a gene that is expressed specifically in anthers at bud stages 9 to 10 when exine development occurs in microspores (Dobritsa et al., 2011).
During microspore development, many kinds of cell walls are successively synthesized and degraded in the following sequence. In Arabidopsis, the primary cell wall of the pollen mother cell (PMC) characteristically contains methylesterified pectin (homogalacturonan, HG) and AGPs (Rhee and Somerville, 1998; Coimbra et al., 2007). Beneath the PMC primary wall, a callosic wall forms that becomes thicker in tetrads and separates the microspores generated by meiosis. In the tetrad, microspores form a layer of primexine matrix consisting of at least methylesterified HG and rhamnogalacturonan II (RG-II; Rhee and Somerville, 1998), and exine construction is initiated in the primexine. Enzymatic degradation of the PMC primary cell wall and callosic wall occurs and the microspores are released into the locular space. Free microspores are still coated with a layer of pectic polysaccharides (also referred to as “primexine” in the literature), although the pectin undergoes demethylesterification (Rhee and Somerville, 1998). Intine forms beneath the exine, and two rounds of mitotic cell division produce two sperm cells surrounded by an AGP-containing wall (Coimbra et al., 2007). The intine in mature pollen grains consists mainly of cellulose, noncellulosic polysaccharides, and pectin (van Aelst and van Went, 1992; Ariizumi and Toriyama, 2011), whereas an intine-like structure (“germination plaque”) produced upon pollen germination consists of cellulose, callose, and partly demethylesterified pectin (Hoedemaekers et al., 2015), with the pollen tube wall also including AGPs (Nguema-Ona et al., 2012).
AGPs are a class of Hyp-rich glycoproteins found ubiquitously in plant cell walls, secretions, and at the plasma membrane (Nguema-Ona et al., 2012). Most members of this family of proteins are defined by a Hyp-rich protein backbone decorated by type II AGs, that is, a galactan backbone of β-(1,3)-linked-d-galactopyranose residues, with branches (between one and three residues long) of β-(1,6)-linked-d-galactopyranose, as well as other terminal monosaccharides such as Fuc, GlcA, and rhamnose, among others (Ellis et al., 2010). AGPs (mostly through their β-(1,3)-galactan backbone) have the ability to bind selectively to and be precipitated by (β-d-glucosyl)3 and (β-d-galactosyl)3 Yariv reagents (Fincher et al., 1983). These reagents are widely used to perturb the molecular functions of AGPs as well as for their detection, quantification, purification, and staining (Fincher et al., 1983).
AGPs are proposed to have a wide variety of functions and play a pivotal role in plant growth and development, particularly in sexual reproduction (for review, see Ellis et al., 2010; Tan et al., 2012). In light of the observation that AGP epitope-directed antibodies show a differential spatio-temporal distribution pattern during both anther and ovule development, it has been suggested that AGPs could be used as gametophytic cell differentiation markers (Coimbra et al., 2007). A few AGPs, for example, AtAGP6 and AtAGP11, have been implicated in pollen development (Coimbra et al., 2009). A double null agp6 agp11 mutant has collapsed pollen grains, reduced (and precocious) pollen grain germination and tube elongation, and displays shorter fruits with less seeds; these phenotypes are due to pollen abortion (Coimbra et al., 2009, 2010).
BLAST searches of the Arabidopsis genome using mammalian β-(1,3)-GalT sequences as queries identified 20 genes of the CAZy Glycosyltransferase (GT) 31 family (www.cazy.org/) as potentially being involved in the synthesis of the type II AG backbone of AGPs (Qu et al., 2008). Based on phylogenetic analysis, the Arabidopsis GT31 family GalTs were grouped into four clades [clades 1 (VIII and IX), 7 (V and VI), 10 (I to IV), and 11 (VII); Qu et al., 2008; Egelund et al., 2011]. Subsequently, several Arabidopsis GT31 clades 7 and 10 members have been shown to have AGP-glycan synthase activity: AtGALT2, 3, 4, 5, and 6 (clade 7; Basu et al., 2013; Basu et al., 2015), AtGALT31A (Geshi et al., 2013), as well as three Hyp O-GALTs, HPGT1, 2, and 3 (clade 10; Ogawa-Ohnishi and Matsubayashi, 2015). In addition to biochemical activity assay data, aberrant growth phenotypes, in varying degrees of severity, have been observed in the aforementioned loss-of-function galt mutants. Hence, both biochemical and genetic evidence exists supporting the view that the AG glycan moieties of AGPs are critical for their biological functions.
In this study, we used genetic and cell biology approaches to identify and characterize KNS4. The gene was found to be At1g33430, a clade 10 member of the GT31 family (Egelund et al., 2011), and identical to UPEX1 (Dobritsa et al., 2011). Heterologous expression and in vitro enzymatic activity assays provided biochemical evidence that KNS4 is a β-(1,3)-GalT likely synthesizing the β-(1,3)-galactan backbone of AGPs and/or the side-chains of RG-I. Immunolabeling and β-Glc Yariv staining show that kns4 mutants have reduced AGP content in the primexine of developing microspores, suggesting AGPs play an important role in this structure. Furthermore, kns4 mutants exhibit pollen aggregation and reduced fertility, as indicated by shorter fruit lengths and lower seed set compared to the wild type, confirming that KNS4 is critical for pollen development.
In the accompanying article by Li et al. (2017), the characterization of two exine mutants, upex1 and spg2, is reported. They showed that SPG2 (At1g27600) encodes the previously characterized IRREGULAR XYLEM9-LIKE (IRX9L) GT 43 protein involved in xylan backbone synthesis and that xylan, as well as AGP, is a component of the primexine wall (Li et al., 2017). Thus, our complementary articles provide evidence that AGPs, pectins, and xylans in primexine contribute to the deposition of sporopollenin onto the microspore surface, and to the construction of a regular reticulate exine pattern.
RESULTS
Identification of the KNS4 Gene
The causative mutation of the EMS-induced kns4 mutant (hereafter kns4-1) was previously reported to be mapped to chromosome 1 (Suzuki et al., 2008). Using a PCR marker approach, the kns4-1 mutation was mapped further to a 300 kb region between single nucleotide polymorphisms PERL0098471 and PERL0101839. A survey of the expression pattern of the 60 genes mapped in the marker region using AtGenExpress microarray data (http://www.arabidopsis.org/) found that At1g33430 was expressed specifically in young flower buds at floral developmental stages 9 to 11 (as defined by Smyth et al., 1990). These stages coincide with the development of the exine in microspores. Mutant alleles of this gene (upex1, upex2, SALK_091466, SAIL_544_C02) have previously been shown to have an uneven pattern of exine, which is also thin and composed of a very short baculae and overdeveloped tectum (Dobritsa et al., 2011). In addition, it was also reported that some regions of the exine of upex mutants partly lacked reticulate structure, being smooth with small lacunae; that their exine dissociated easily from the pollen surface; and that pollen grains were sticky (Dobritsa et al., 2011). These observations were similar and mostly consistent with our observations of kns4-1 (Suzuki et al., 2008; Fig. 1, A–F), although we did detect subtle differences, such as the entire surface of kns4 pollen grains being covered with reticulated exine to which pieces of debris clung (Fig. 1D), rather than a complete lack of exine. In this article, we use the term “lacunae” as the holes within the tectum (including the space directly beneath the tectum hole). When referring to the whole space under the tectum, including lacunae and gaps between neighboring baculae, we use the term “exine cavities”. The lacunae of kns4-1 were smaller and shallower, while their density was almost similar to that of wild type (compare Fig. 1, C and F). Taken together, these data suggested that At1g33430 was a likely candidate to contain the causative mutation of kns4-1. The kns4-1 mutant was shown to possess a G to A substitution in the fifth coding exon of At1g33430. This caused a change in the 276th codon from a Trp residue to a stop codon in the corresponding protein (Fig. 1G).
Figure 1.
Pollen phenotypes of kns4 mutants. A to F, Surface view of wild-type (Ler; A–C) and kns4-1 (D–F) pollen grains by SEM. Arrow in D indicates cell wall debris adhered to pollen grains. G, Schematic structure of At1g33430/KNS4. The shorter splice variant, At1g33430.1, is depicted, as only this form is detected in flower buds. Rectangles represent exons [noncoding exon 1 (nc1) and coding exons 1 to 6], and coding region and 5′ and 3′ UTRs are indicated with white, black, and gray, respectively. The thick black line represents noncoding sequence/introns. The positions of point mutation (kns4-1) and T-DNA insertions (kns4-2 and kns4-3) as well as the SALK and SAIL line numbers are indicated. Gray arrows represent the position of primers used in the RT-PCR experiment shown in Fig. 4A. H to J, Surface view of kns4-2 (H and I) and kns4-3 (J) pollen grains by SEM. K, Rescued pollen phenotype of the kns4-1 mutant after transformation with the ProKNS4:KNS4 construct. Scale bars = 20 μm in (A, D, H, J, and K), 10 μm in (B, E, and I), and 2 μm in (C and F).
To validate that the genetic lesion in At1g33430 leads to the pollen phenotype, the two aforementioned T-DNA lines (SALK_091466 and SAIL_544_C02, here designated kns4-2 and kns4-3, respectively) were obtained that had insertions present either in the noncoding exon or the fourth exon, respectively (Fig. 1G). Both lines showed pollen phenotypes indistinguishable from those observed in kns4-1, namely a thin exine with a poorly reticulate tectum and considerable debris adhered to the pollen grain surface (Fig. 1, H–J).
To further confirm that the phenotypes observed in kns4-1 mutants were due to a loss of At1g33430 function, a complementation experiment was performed by generating a construct containing the wild-type coding sequence of At1g33430 downstream of its own promoter (defined as 2006 bp upstream of the first Met, designated ProKNS4:KNS4) that was transformed into kns4-1 mutant plants. The aberrant pollen phenotype was completely restored in all 17 individual T1 transformants (Fig. 1K). Taken together, we conclude that the phenotypes observed in kns4-1 mutants are due to the loss of At1g33430 function and that kns4 and upex mutants (Dobritsa et al., 2011; Li et al., 2017) are synonymous.
KNS4 Is Required for Separation of PMCs and Microspores in Anthers
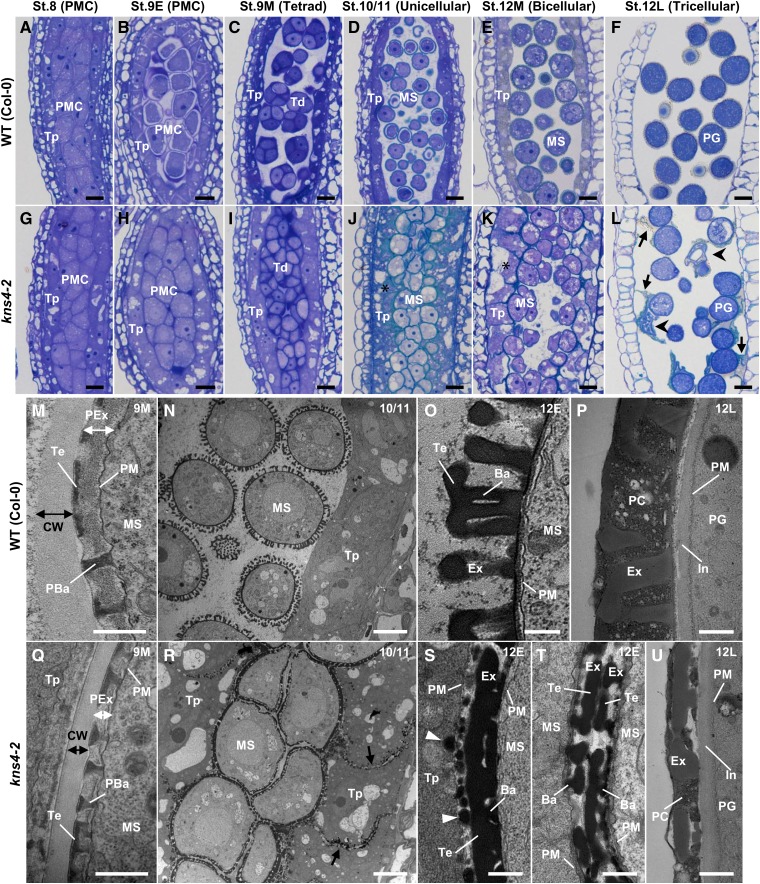
To identify the initial defect in pollen development of kns4 mutants, sections of resin-embedded kns4-2 anthers were prepared at several floral bud developmental stages (stages 8 to 12; as defined by Smyth et al., 1990) and counterstained with toluidine blue for visualization. Because stage 12 is rather long (approximately 48 h), we divided this stage into a further three substages: 12E (until the petals reach the style), 12M (until anthers begin to turn yellow), and 12L (thereafter). Furthermore, because microspores drastically change their morphology during stage 9, we also divided this stage into three substages: 9E, 9M, and 9L. For clarity, the morphological microspore/pollen development phenotype at each of these floral developmental stages is defined as follows: stage 8 (pollen mother cell [PMC] before initiation of callosic wall formation), stage 9E (PMC surrounded by the callosic wall), stage 9M (tetrad), stage 9L (newborn unicellular microspore), stage 10/11 (unicellular), stage 12E (unicellular to bicellular), stage 12M (bicellular to tricellular), and stage 12L (tricellular).
At stage 8 (PMC), all the PMCs were tightly adhered, either to each other or to tapetal cells and no obvious morphological differences were observed between wild type and kns4 mutants (compare Fig. 2, A and G, respectively). The first sign of developmental abnormality in kns4 mutants appeared in stage 9E (PMC), when callosic wall formation had been initiated. At this stage, the wild-type PMCs began to separate from each other and from tapetal cells, whereas in the kns4 mutant PMCs remained adherent and tapetal cells were slightly vacuolated and swollen (compare Fig. 2, B and H, respectively). Moreover, the protoplast of the wild-type PMCs was often detached from the primary cell wall, although this is likely to be a sample preparation artifact frequently observed during this developmental stage in wild type (Quilichini et al., 2014). Interestingly, this was never observed in mutant PMCs, perhaps indicating some structural differences in connectivity between the plasma membrane-cell wall interface between wild type and mutants. At stage 9M (tetrad), when the callosic wall of tetrads was clearly observed to stain strongly with toluidine blue, wild-type tetrads were separated and dispersed in anther locules, whereas the kns4 tetrads remained attached to tapetal cells and appeared to adhere to each other (compare Fig. 2, C and I, respectively). In the kns4 mutant the tapetal layer is thicker relative to wild type, resulting in a compression of the tetrads.
Figure 2.
Light and TEM micrographs reveal defects in pollen development in kns4-2 mutants. A to L, Light micrographs of longitudinal sections of resin-embedded anthers of wild-type (Col-0; A–F) and kns4-2 (G–L) plants stained with toluidine blue. Developmental stages (stages 8L to 12L) of anthers are indicated along with the developmental stage of pollen development in parentheses. Asterisks in (J) and (K) indicate swollen and vacuolated tapetal cells in the mutant. Arrows and arrowheads in (L) indicate cell wall debris and crushed pollen grains, respectively. M to U, TEM images of wild-type (Col-0; M to P) and kns4-2 (Q to U) anthers. M and Q, Primexine layers and developing exine in tetrads at stage 9M. (N, O, R, S, and T), Longitudinal section of stage 12E anthers containing unicellular microspores. (P) and (U), Exine and pollen coat of pollen grains (tricellular stage) at stage 12L. Arrows in (R) show walls between tapetal cells. Arrowheads in (S) show granules containing sporopollenin deposited on the surface of tapetal cell wall. Ba, Bacula; CW, callosic wall; Ex, exine; In, intine; MS, microspore; PBa, probacula; PC, pollen coat; PG, pollen grain; PM, plasma membrane; PMC, pollen mother cell; Td, tetrad; Te, tectum; Tp, tapetal cell. Scale bars = 10 μm in (A–L), 5 μm in (N and R), and 0.5 μm in (M, O, P, Q, S, T, and U).
After the dissolution of the callosic wall at stage 9L, spherical microspores were scattered in anther locules at stage 10/11 (unicellular) in wild type, but deformed microspores of the mutant did not separate from each other and filled the locules (compare Fig. 2, D and J, respectively). Interestingly, the tapetal cells in kns4 mutants appeared to be vacuolated compared with the wild type. Microspores of both wild type and kns4 mutants gradually grew larger during stage 12E, although their morphology was similar to the previous stage. At stage 12M (bicellular), most of the mutant microspores were still in contact with each other and many seemed tightly adhered to tapetal cells, even though a small space appeared in the center of each locule (compare Fig. 2, E and K, respectively). Strikingly, the vacuolated tapetal cell phenotype observed in the kns4 mutants at stage 10/11 appeared to be more obvious, particularly when compared with the wild type (compare Fig. 2, D and E with Fig. 2, J and K, respectively). At stage 12L (tricellular) in wild type, when tricellular mature pollen grains were developed, the tapetal cells disappear and only pollen grains can be observed (Fig. 2F). However, in kns4 locules, debris that stained weakly with toluidine blue remained attached to the surface of pollen grains (Fig. 2L). In these kns4 locules, many pollen grains were found to adhere to each other and to the surface of the anther cell wall (endothecium), presumably due to the presence of the debris. Some collapsed pollen grains were also observed (Fig. 2L). These observations suggest that pollen adhesion and debris formation in the kns4 mutant is a consequence of the defect in separation of the PMC at stage 9E.
KNS4 Is Required for Sporopollenin Deposition and Exine Development
Exine development in kns4 mutants was further investigated by transmission electron microscopy (TEM). In the tetrads of wild-type stage 9M anthers, a matrix of primexine appeared as a layer between the plasma membrane of the microspore and the callosic wall, in which the development of the exine framework consisting of the probaculae and tectum is initiated (Fig. 2M). The primexine is thinner and consequently the probaculae are shorter in the kns4 mutant than those in wild type, whereas tectum development was essentially normal in the mutant (Fig. 2Q). These results indicate that the characteristic exine phenotype in the kns4 mutant is related to an abnormality of thin primexine in the tetrads.
At stage 10/11, mutant microspores clumped together, whereas wild-type microspores were already separated, with distinct exine walls (compare Fig. 2, R and N, respectively). By the end of stage 12E, sporopollenin supplied by tapetal cells settles onto the surface of each microspore and exine development is nearly complete in wild type (Fig. 2O). In contrast, exine walls in the kns4 mutant microspores appeared compacted and tectum and baculae structures were not well defined (Fig. 2S). A considerable amount of sporopollenin secreted by kns4 mutant tapetal cells was deposited both on the surface of microspores and tapetal cells adjacent to the microspores, which appeared to result in the adhesion of microspores to tapetal cell walls (Fig. 2, R and S). Electron-dense particles, presumably containing sporopollenin, were also deposited on the walls separating adjacent tapetal cells (Fig. 2R). Interestingly, exine structures of two adjacent microspores were always separated and never fused (Fig. 2, R and T). At stage 12L in wild type, the tapetal cells have already collapsed and osmiophilic materials have filled the exine cavities of tricellular pollen grains to form the pollen coat (Fig. 2P). Pollen coat formation was similarly observed on the surface of kns4 pollen grains, although the amount of material was very low compared to wild type due to the short baculae length (Fig. 2U).
Exine development in wild type and kns4 was observed and exine thickness measured by use of optical sections of isolated microspores and pollen grains (Supplemental Fig. S1). The thickness of wild-type exine gradually increased from stage 9L to stage 10/11 and reached a peak at stage 12E. Thereafter, it decreased slightly until the mature pollen grain stage in open flowers. The exine of kns4 microspores was 24% thinner than wild-type exine at stage 9L, and decreased to less than 50% at stage 12E and later stages. These results suggested that KNS4 function is also required for exine thickening from stages 9L through to 12E, which is consistent with the expression pattern of KNS4 (described below).
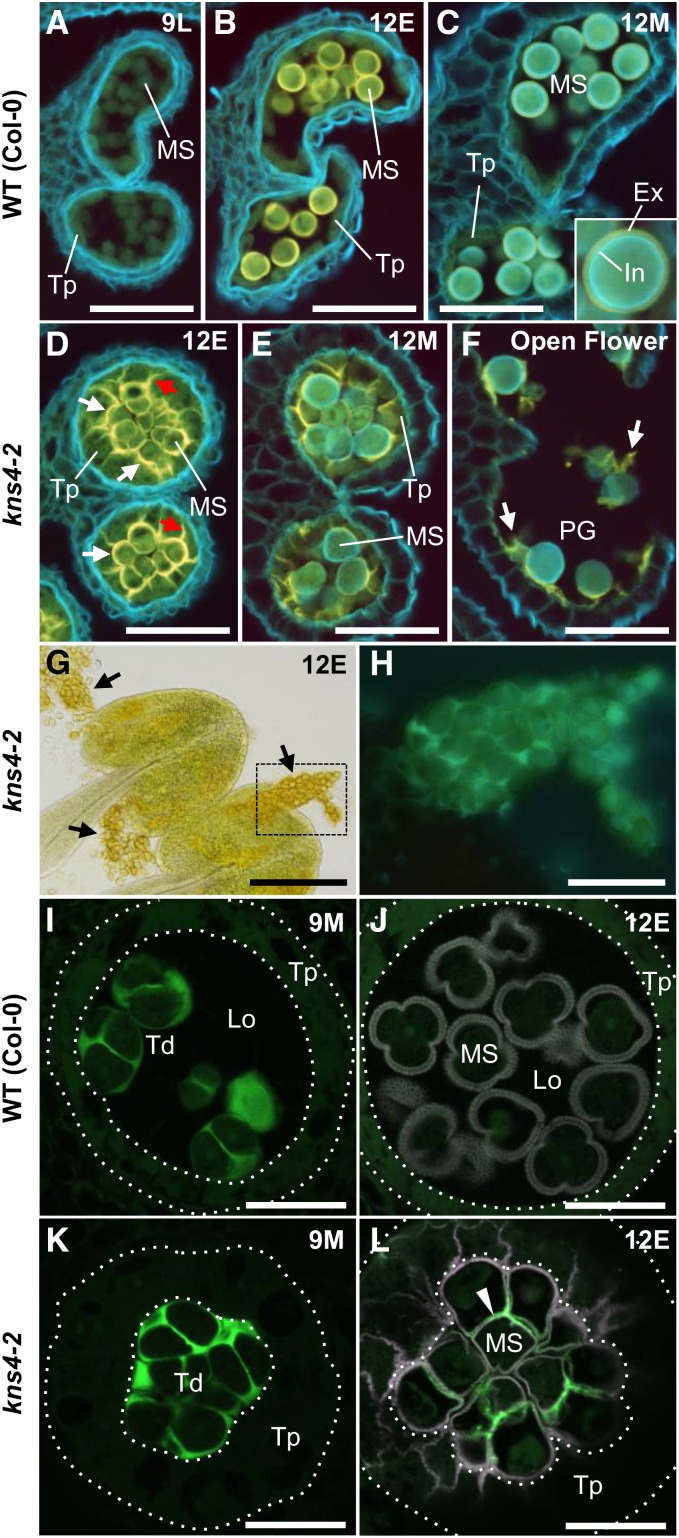
To examine the origin of the debris adhering to the kns4 pollen grains, cross sections of paraffin-embedded anthers were prepared, stained with calcofluor white, and viewed under UV excitation (Fig. 3, A–F). In wild-type anthers at stage 9L, most cell walls fluoresced blueish-white, whereas tapetal cells and microspores showed weak yellow fluorescence (Fig. 3A), suggesting that the walls of tapetal cells and early unicellular microspores did not contain much cellulose or callose. The yellow autofluorescence of polymerizing sporopollenin appeared on the developing exine and reached maximal brightness at stage 12E (Fig. 3B). When the anthers reached stage 12M, intine appeared on pollen grains and fluoresced blueish-white, while yellow exine autofluorescence became weaker (Fig. 3C). In the mutant stage 12E anthers, the yellow autofluorescence was observed not only on the developing exine but also, and more strongly, on the boundaries between tapetal cells and microspores (white arrows) and the walls separating adjacent tapetal cells (red arrows; Fig. 3D). This is consistent with the abnormal sporopollenin deposition in these walls observed by TEM (Fig. 2, R and S). The yellow fluorescent walls in kns4-2 were still observed at stage 12M (Fig. 3E), and even after anther dehiscence (Fig. 3F). It therefore appears that tapetal cell walls were not completely degraded when these cells collapsed, and remained adhered to the kns4 microspore surface as pollen grain development was completed. Thus, we suggest that the material adhering to kns4 pollen grains is not dissociated exine, but rather remnants of tapetal cell walls onto which sporopollenin was deposited (Fig. 3F).
Figure 3.
Abnormal sporopollenin deposition and delayed callose degradation in kns4-2. A to F, Calcofluor white staining of anther cross sections in wild type (Col-0; A to C), and kns4-2 (D–F) observed by fluorescence microscopy. Sections of stage 9L (newborn unicellular; A), stage 12E (unicellular; B and D), stage 12M (bicellular; C and E), and open flower (stage 14, tricellular; F) are shown. Cell walls containing cellulose and callose are stained whitish blue, and autofluorescence of sporopollenin is yellow. White and red arrows in (D) indicate boundary between tapetal cells and microspores and the walls separating adjacent tapetal cells, respectively. White arrows in (F) indicate tapetal wall remnant onto which sporopollenin is deposited. G and H, Aggregates of microspores (black arrows) squeezed out from kns4-2 anthers at stage 12E were observed with transmitted light (G) and fluorescence microscopy (H) after aniline blue staining; (H) shows the framed area in (G). I to L, Aniline blue staining of anther cross sections in wild-type (Col-0; I and J) and kns4-2 (K and L) observed by CLSM. Stages are 9M (tetrad; I and K) and 12E (unicellular; J and L). Green and magenta channels show fluorescence of aniline blue and sporopollenin autofluorescence, respectively. Dotted lines indicate the position of the tapetum. Arrowhead in (L) indicates callose in walls that remained in late unicellular stage. Ex, Exine; In, intine; Lo, locular space; MS, microspore; PG, pollen grain; Td, tetrad; Tp, tapetal cell. Scale bars = 50 μm in (A– F) and H, 200 μm in (G), 20 μm in (I–L).
While neighboring kns4 mutant pollen grains were never observed to fuse (Fig. 2T), mutant pollen grains did form clumps that could be squeezed out from anthers (Fig. 3G). This observation implies the existence of adherent material(s) that connect the microspores. During tetrad stage (stage 9M), microspores are separated by walls rich in callose, which can be visualized by confocal laser scanning microscopy (CLSM) using the aniline blue fluorochrome. No staining differences were readily observed when comparing wild type to kns4 mutants (compare Fig. 3, I and K). However, at stage 12E, when the callosic walls are completely degraded in wild type and hence the aniline blue signal is absent, kns4 microspores continued to show aniline blue signal between the microspores within the tetrad (compare Fig. 3, J and L). Aniline blue fluorescence was also detected in the walls of kns4 microspore clumps squeezed from mature anthers (Fig. 3H). These results suggest that the remnant callose in the walls between microspores prevents the developing exine from fusing in the kns4 mutant.
Expression Analysis of KNS4
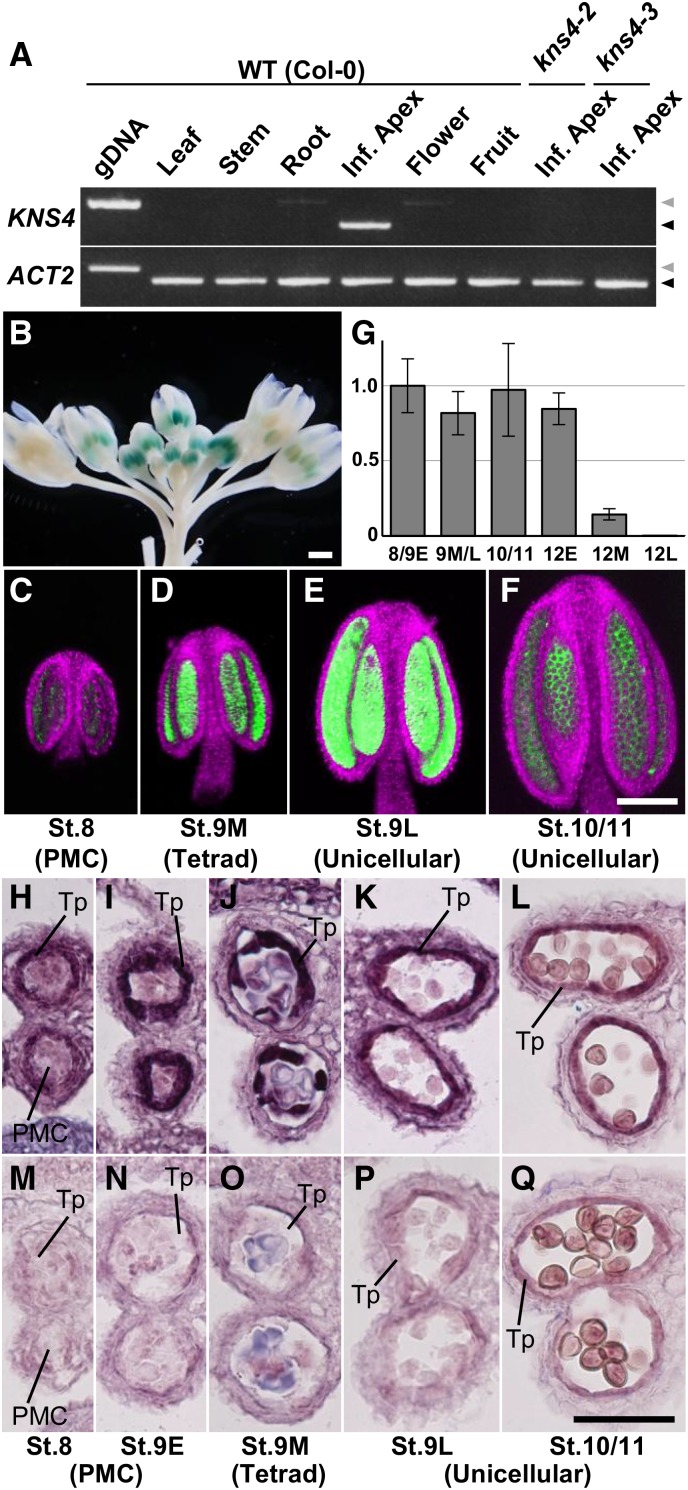
Reverse transcription PCR (RT-PCR) analysis of a variety of tissue types revealed that KNS4 expression (mRNA) is only detected in the inflorescence apices containing shoot apical meristems and flower buds up to stage 12L (Fig. 4A). This observation is consistent with transcript levels reported in AtGenExpress microarray datasets (http://www.arabidopsis.org/). No KNS4 transcripts were detected in either kns4-2 or kns4-3 inflorescence apices (Fig. 4A), indicating that both T-DNA insertions disrupted gene function.
Figure 4.
KNS4 is specifically expressed in tapetal cells in young anthers. A, RT-PCR analysis of KNS4 using RNA extracted from rosette leaves, inflorescence stems, roots, inflorescence apices, open flowers, and fruits (siliques) of wild-type Arabidopsis (Col-0), and inflorescence apices of kns4-2 and kns4-3 mutants. Leftmost lane shows the PCR product amplified from Col-0 genomic DNA (gDNA) with the same primer set. ACT2 was used as a control. Black and gray triangles indicate cDNA- and gDNA-derived bands, respectively. B, ProKNS4-GUS expression in flower buds. C to F, ProKNS4-GFP expression in anthers from stages 8 to 10/11 observed by CLSM. Green and magenta channels show GFP signal and chlorophyll autofluorescence, respectively. G, Expression levels of KNS4 in anthers at various flower developmental stages (stages 8 and 9E, 9M and 9L, 10/11, 12E, 12M, and 12L) measured by qRT-PCR. Bars show relative average expression values (stage 8/9E = 1) of triplicate samples ± sd. H to Q, In situ hybridization using either an antisense (H–L) or sense (M–Q) probe for KNS4. Cross sections of paraffin-embedded anthers from stages 8 to 10/11 were examined. Scale bars = 1 mm in B, 100 μm in (C–F), and 50 μm in (H–Q).
To more specifically determine the tissues/organs where KNS4 is expressed, the KNS4 promoter was separately fused to β-glucuronidase (GUS) and GFP, and the reporter constructs introduced into wild-type Arabidopsis plants. In all 14 individual T1 ProKNS4:GUS transformants, the GUS signal was specifically detected in anthers with strongest staining in young flower buds and reduced signal in older open flowers (Fig. 4B. No signal was observed in any other organs including leaves, stems, roots, and fruits, consistent with the RT-PCR analysis and the public microarray datasets (Fig. 4A). The same result was obtained for all 10 individual T1 transformants carrying the ProKNS4:GFP transgene (Fig. 4, C–F. Imaging revealed that the GFP fluorescence was specifically detected in anthers ranging from stages 8 through to 10/11, with the strongest expression at stage 9L (Fig. 4, C–F).
We also examined KNS4 mRNA levels in anthers by quantitative RT-PCR (qRT-PCR; Fig. 4G). KNS4 transcript was detected from stages 8 through to 12E, declining in stage 12M and 12L along with the degeneration of tapetal cells (Fig. 4G). These results indicated that KNS4 might have a constitutive function in tapetal cells. It should be noted that the expression of ProKNS4:GFP did not reflect the accumulation of KNS4 mRNA at stages 10/11 and 12E, possibly due to the differential stabilities of mRNAs.
To more accurately define the location of KNS4 transcripts, we performed in situ hybridization of anthers from stage 8 through to stage 10/11. While the sense probe resulted in little signal (Fig. 4, M–Q), the antisense probe produced strong signals in tapetal cells throughout these stages, with the strongest labeling detected at stage 9 (Fig. 4, H–L). The relatively high background signal by the antisense probe in young anthers possibly reflects weak KNS4 expression in tissues other than tapetum, which has been suggested in a previous study (Peiffer et al., 2008).
Enzymatic Activity of KNS4
KNS4 is predicted to encode a β-(1,3)-galactosyltransferase (GalT) that belongs to GT family 31 (CAZy; http://www.cazy.org; Strasser et al., 2007; Qu et al., 2008; Egelund et al., 2011). Among the members of this family, At1g77810, one of the closest homologs of KNS4, has been shown to encode a β-(1,3)-GalT (Qu et al., 2008). An enzyme with this activity is required for the biosynthesis of the β-(1,3)-galactan core structure of type II AGs, the glycan moieties that decorate AGPs (Ellis et al., 2010; Showalter and Basu 2016), as well as the type II AG side chains on RG-I (Mohnen, 2008). Furthermore, it was recently reported that the gene product of another close homolog, AtGALT31A (At1g32930), possesses β-(1,6)-GalT activity, which is also required for the formation of side chains in type II AGs (Geshi et al., 2013). Thus, it was thought that KNS4 may have a similar GalT enzymatic activity.
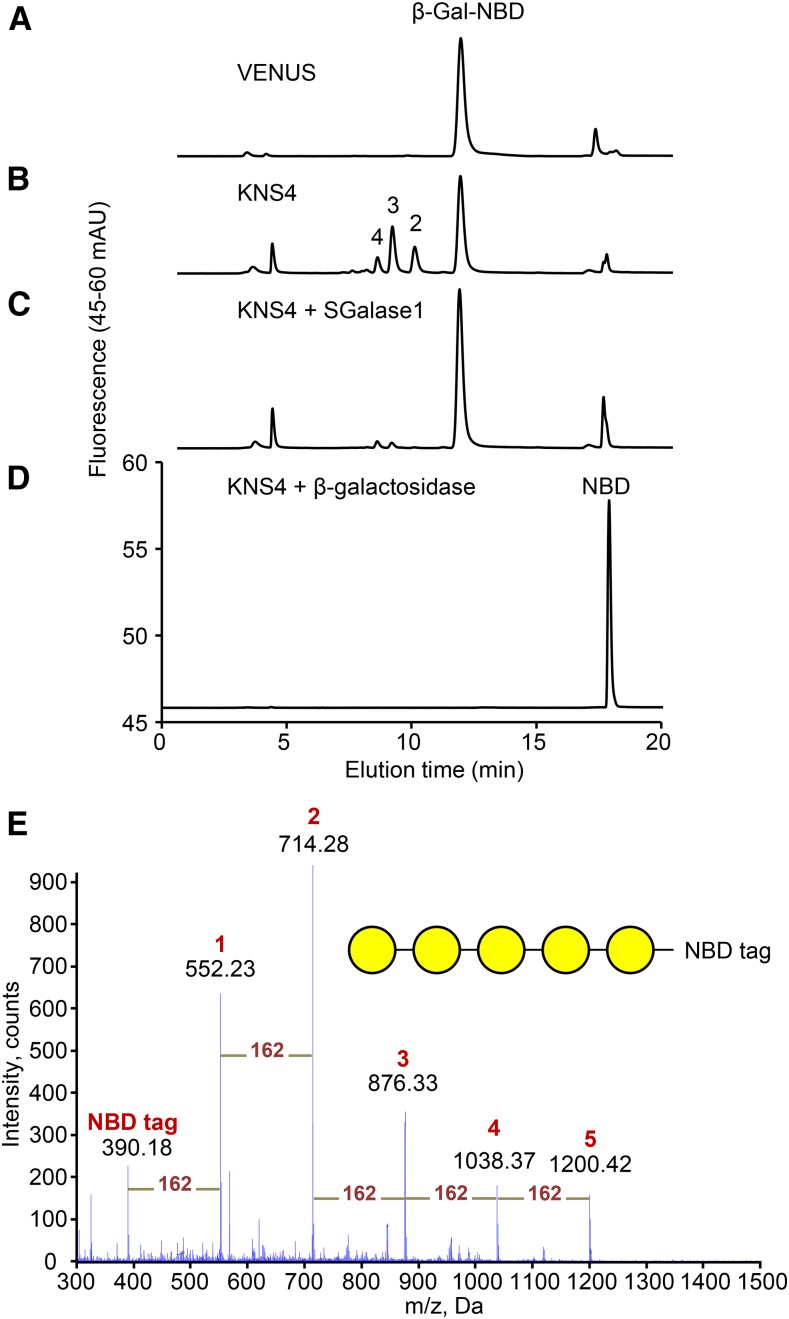
To examine this possibility, the full-length KNS4 coding sequence was transiently expressed in Nicotiana leaves. As a negative control, the yellow fluorescent protein VENUS was also transiently expressed. Microsomal membrane (MM) fractions prepared from leaves 4 d post-infiltration were used as the enzyme source in biochemical activity assays using fluorescent β-Gal-NBD as the acceptor (McGill and Williams, 2009; Fig. 5). When MMs prepared from the control plants were assayed in the presence of UDP-Gal, the reversed phase-HPLC profile was indistinguishable from the β-Gal-NBD standard, indicating the absence of detectable endogenous enzyme activity under these assay conditions (Fig. 5A). In contrast, when MMs derived from KNS4-expressing leaves were assayed under the same conditions, a series of new peaks were observed with Rt of 9.6 min (Peak 2), 8.7 min (Peak 3), and 8.1 min (Peak 4; Fig. 5B). The donor specificity was restricted to UDP-Gal as none of the other UDP-sugar donors tested (UDP-Glc, UDP-Arap, and UDP-Xyl) resulted in the formation of product using β-Gal-NBD as the acceptor (Supplemental Fig. S2).
Figure 5.
Enzymatic activity of heterologously expressed KNS4 in microsomal membranes from Nicotiana leaf. A and B, RP-HPLC profiles of GalT enzyme assay products with KNS4-expressing MMs in the reaction mixture (B) and control experiment with VENUS-expressing MMs (A). Numbers above peaks indicate the number of Gal residues. C and D, Treatment of the KNS4 reaction products with either SGalase1 (C) or β-galactosidase (D). E, ESI-MS full scan spectrum of KNS4 GalT enzyme assay products. Peaks corresponding to β-Gal1-5-NBD are labeled 1 to 5, respectively. Yellow circles represent Gal residues.
To determine the nature of the glycosidic linkage(s) in the NBD-labeled products formed by the KNS4 MM preparation, the oligosaccharide products were digested with SGalase1, an exo-β-(1,3)-d-galactanase (CAZy GH family 43) that has been shown to specifically cleave the β-(1,3)-d-galactan backbone of AGPs (Ling et al., 2012; Fig. 5C). SGalase1 digestion resulted in the hydrolysis of all the reaction products to β-Gal-NBD (Rt is approximately 11.5 min). A commercial Escherichia coli β-galactosidase, which also digests the Gal-NBD bond, hydrolyzed these same oligosaccharides in KNS4 MM preparations to NBD (Rt is approximately 17.0 min; Fig. 5D). Analysis of the assay products by electrospray ionization (ESI)-MS/MS revealed the presence of the protonated pseudo-molecular ions [M+H] of m/z: 552.2, 714.3, 876.3, 1038.4, and 1200.4 corresponding to β-Gal-NBD, Gal2-NBD, Gal3-NBD, Gal4-NBD, and Gal5-NBD, respectively (Fig. 5E). The mass difference between each of the pseudo-molecular ions is 162 dalton, indicating a hexose series (Fig. 5E). Thus, from the combined linkage-specific enzyme digestions and the MS data we can conclude that KNS4 is a β-(1,3)-GalT likely to play a role in type II AG glycan biosynthesis on either AGPs or RG-I.
Distribution of AGP Epitopes in Developing Anthers
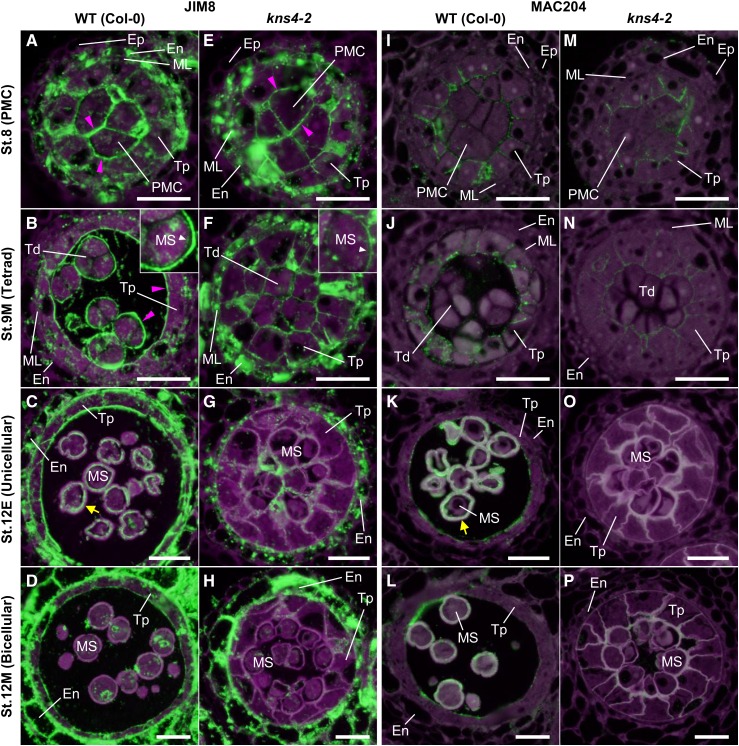
To investigate the impact of the kns4 mutant on the accumulation of AGPs in anthers, cross sections of young flower buds up to stage 12M were labeled with the JIM8 antibody that recognizes the AG epitope on AGPs (Pennell et al., 1991; Fig. 6, A–H). At stage 8, JIM8 labeling was observed in the cell walls surrounding the PMCs in both wild-type and kns4 mutant anthers (Fig. 6, A and E). Bright, punctate signals were observed within the cytoplasm of the PMCs, tapetal cells, middle layer cells, and endothecial cells, which likely corresponds to the endoplasmic reticulum/Golgi apparatus as they are known to be the intracellular sites of AGP biosynthesis. At stage 9M, JIM8 strongly labeled the outer surface (primary walls) of wild-type tetrads and the tapetal walls facing the locule (Fig. 6B). JIM8-labeled signal was also observed in primexine in tetrads (inset in Fig. 6B). In kns4 mutants, signal could still be detected, but was much weaker (Fig. 6F). At stage 12E, the cavities of the developing exine and walls of the tapetal cells were labeled in wild type, but only weak punctate signals were observed in the walls of kns4 tapetal cells (compare Fig. 6, C and G, respectively). By stage 12M in wild type, the JIM8 signal in the exine cavities and on the locular surface of tapetal walls had decreased (Fig. 6D). The signal in kns4 was similarly weaker at the same stage (Fig. 6H). In contrast, walls and cytoplasm of endothecial cells in both wild type and kns4 were strongly labeled by JIM8 throughout anther development.
Figure 6.
Distribution of JIM8- and MAC204-epitope labeling of AGPs in microsporocytes and microspores. Cross sections of resin-embedded anthers of wild type (Col-0; A–D and I–L) and kns4-2 (E–H and M–P) from stages 8 to 12M were incubated with the JIM8 (A–H) and the MAC204 (I–P) antibodies, and subsequently with Alexa Fluor 488-labeled secondary antibody. Fluorescence of Alexa Fluor 488 (green) and autofluorescence (magenta) were separately captured by epifluorescence microscopy and overlaid. Magenta arrowheads indicate the signal in the primary cell wall of the PMC, tetrad, and tapetal-cell walls facing the locule. Yellow arrow indicates the signal in the exine cavities. Insets in (B) and (F) are enlargements of a microspore in a tetrad. White arrowhead indicates the signal in primexine. En, Endothecium; Ep, epidermis; ML, middle layer cell; MS, microspore; PMC, pollen mother cell; Td, tetrad; Tp, tapetal cell. Bars = 20 μm.
Distinct immunolabeling results were obtained using another anti-AGP antibody, MAC204 (Bradley et al., 1988). This antibody specifically labeled tapetal walls and developing exine cavities in wild type (Fig. 6, I–L). In contrast to JIM8, MAC204 only faintly labeled the primary walls of PMCs and tetrads, and no labeling was observed in the endothecium. The signal in the tapetal walls at stage 12E was very weak and restricted to the locular surface, whereas fluorescence in exine cavities was bright and remained in stage 12M. In the kns4 mutant, the signals on tapetal cell walls appeared similar to wild type at early developmental stages (stage 8), but decreased in later stages (stage 9M and beyond) with very weak labeling observed in exine cavities (Fig. 6, M–P).
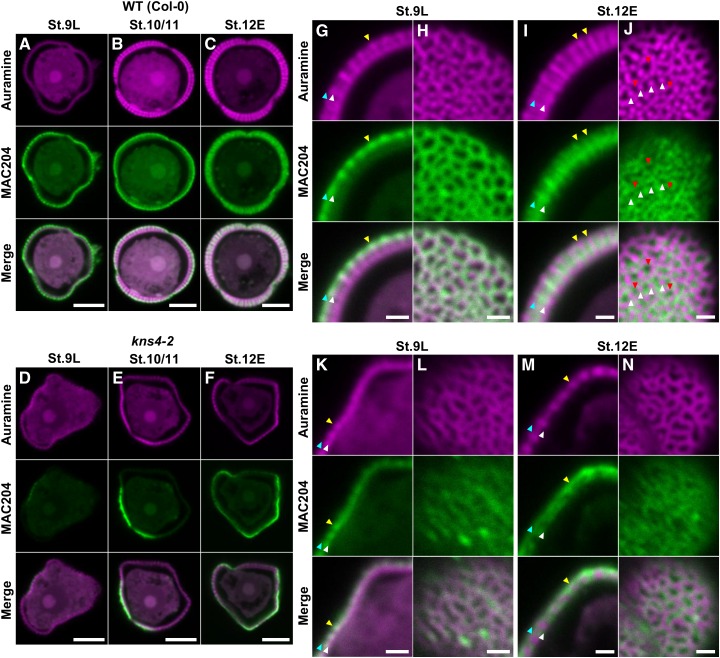
Given the demonstrated β-(1,3)-GalT activity of KNS4 (Fig. 5), the reduced labeling with two AGP antibodies (Fig. 6) and the loss of exine thickening observed in kns4 mutants (Supplemental Fig. S1), we hypothesized that AGPs play a critical role in exine development. Thus, to test whether the kns4 mutant phenotype is due to a loss of AGPs, we examined the localization of AGP epitopes using the MAC204 antibody specifically in relation to exine development at stages 9L to 12E in isolated microspores. In wild type at stage 9L, a strong signal was observed surrounding the microspores just released from tetrads (Fig. 7A). Under higher magnification, we found that a layer of AGPs overlays the developing exine (the position of the AGP layer in relation to the exine is indicated by blue and white arrowheads in Fig. 7G) and localized on the top of baculae (yellow arrowhead in Fig. 7G). An optical section near the surface of the microspore revealed a reticulate pattern of MAC204 labeling (Fig. 7H), indicating that at this developmental stage the AGPs formed a reticulate structure overlaying the tectum. At stage 10/11, while the exine gradually thickened, most AGP still overlayed the tectum but some began to be redistributed into the exine cavities (Fig. 7B). At stage 12E, when the exine reaches its maximum thickness in wild type (Supplemental Fig. S1), MAC204 signal was also strongest (Fig. 7C). AGP labeling was reduced at the exine surface but appeared in the exine cavities (in the space between baculae (yellow arrowheads in Fig. 7I)). Tangential optical sections showed the nonuniform distribution of AGPs in the cavities of the reticulate exine, specifically they were absent from both baculae and lacunae (white and red arrowheads, respectively in Fig. 7J). In kns4 microspores, MAC204 labeling in the exine was reduced across all stages, being either absent (Fig. 7D) or localized on only a part of the microspore surface (Fig. 7, E and F). The redistribution of signal from the outer layer at stage 9L to within the exine cavities at stage 12E was not as obvious in the mutant compared to wild type (compare Fig.7, K–N to Fig. 7, G–J, respectively). These results suggest an association between AGP deposition and exine formation and that the synthesis of AGPs is dependent on the function of KNS4.
Figure 7.
Distribution of MAC204-epitope labeling of AGPs on the surface of developing microspores. A to F, Median optical sections of wild type (Col-0; A–C) and kns4-2 (D–F) microspores isolated from stages 9L, 10/11, and 12E anthers after labeling with the MAC204 antibody and subsequently with Alexa Fluor 546-labeled secondary antibody. Exine was visualized by auramine O staining. Specimens were observed by CLSM in which magenta and green channels show fluorescence of auramine O and Alexa Fluor 546, respectively. G to N, Enlarged optical sections of wild-type (G–J) and kns4-2 (K–N) microspores isolated from stages 9L (G, H, K, and L) and 12E (I, J, M, and N) anthers. (G, I, K, and M) are median optical sections of microspores showing the exine in longitudinal cross section. Blue, white, and yellow arrowheads indicate the top and bottom boundary of the AGP layer, and the top of representative bacula, respectively. (H, L, and N) are tangential optical sections almost at the microspore surface. J, Tangential optical section that transversely cuts baculae. White and red arrowheads indicate the position of representative baculae and lacunae, respectively. Bars = 5 μm in (A–F), and 1 μm in (G–N).
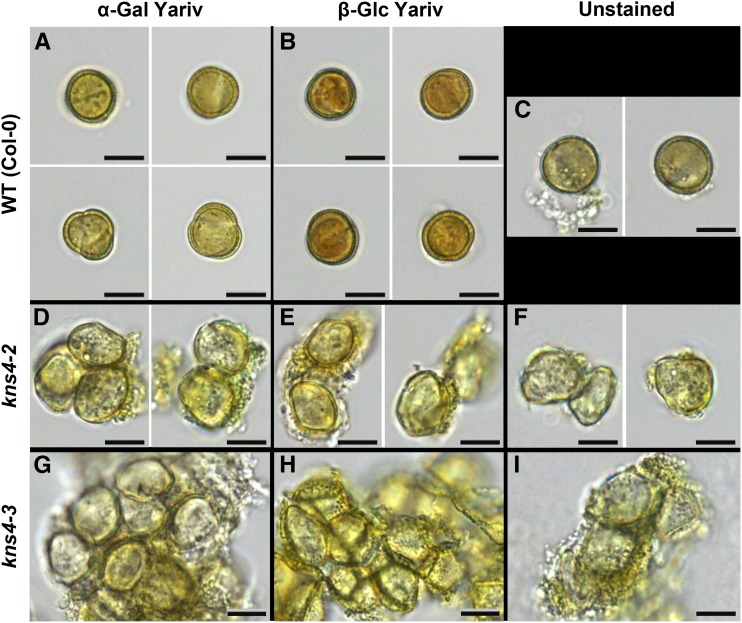
To further validate that the loss of KNS4 function leads to a reduction in AGPs in developing microspores, we used the AGP-specific β-Glc Yariv reagent to stain stage 12M microspores of both wild-type (Fig. 8B) and kns4 mutants (Fig. 8, E and H). AGPs were detected in the wild-type exine as indicated by orange/red staining of the microspore surface while minimal staining was observed in the wall of kns4 microspores. Control experiments using α-Gal Yariv reagent showed similar levels of background staining in wild-type (Fig. 8A) and kns4 mutant (Fig. 8, D and G) microspores. The β-Glc Yariv staining is consistent with more AGPs being present on the surface of developing wild-type microspores compared with the kns4-2 and kns4-3 mutants.
Figure 8.
Yariv dye reagent staining of microspores (stage 12M) isolated from flower buds of wild type and kns4 mutants. Microspores from wild type (Col-0; A–C), kns4-2 (D–F), and kns4-3 (G–I) were independently stained either with α-Gal Yariv (A, D, and G), β-Glc Yariv (B, E, and H), or left unstained (C, F, and I) and viewed with light microscopy. Positive staining is observed as a reddish-orange color. Bars = 20 µm.
Localization of AGPs in the exine cavities was examined in further detail by immuno-TEM using JIM8 and another AGP epitope antibody, JIM13 (Supplemental Fig. S3). In wild-type stage 12M microspores, most JIM13 labeling was seen in the locular space, on the exine and in exine cavities, and at the base of exine (nexine layer; Supplemental Fig. S3, B and G). A sparse but similar labeling pattern was observed with JIM8 (Supplemental Fig. S3A). In kns4 microspores, JIM8 and JIM13 labeling was generally similarly located, although more label was detected outside the exine (Supplemental Fig. S3, D, E, and G). Labeling of the sporopollenin granules was also seen (Supplemental Fig. S3E). Callose labeling was detected mainly in the nexine II layer in both the wild-type and kns4 mutants (Supplemental Fig. S3, C and F, respectively), indicating no impact on the synthesis and deposition of this polysaccharide.
Pectin and AGP Distribution Overlap in Developing Anthers
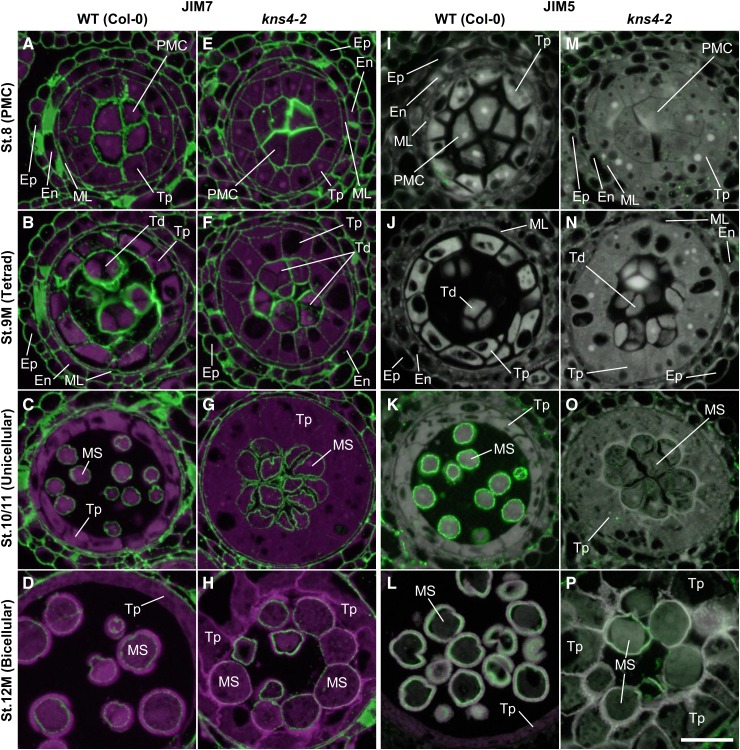
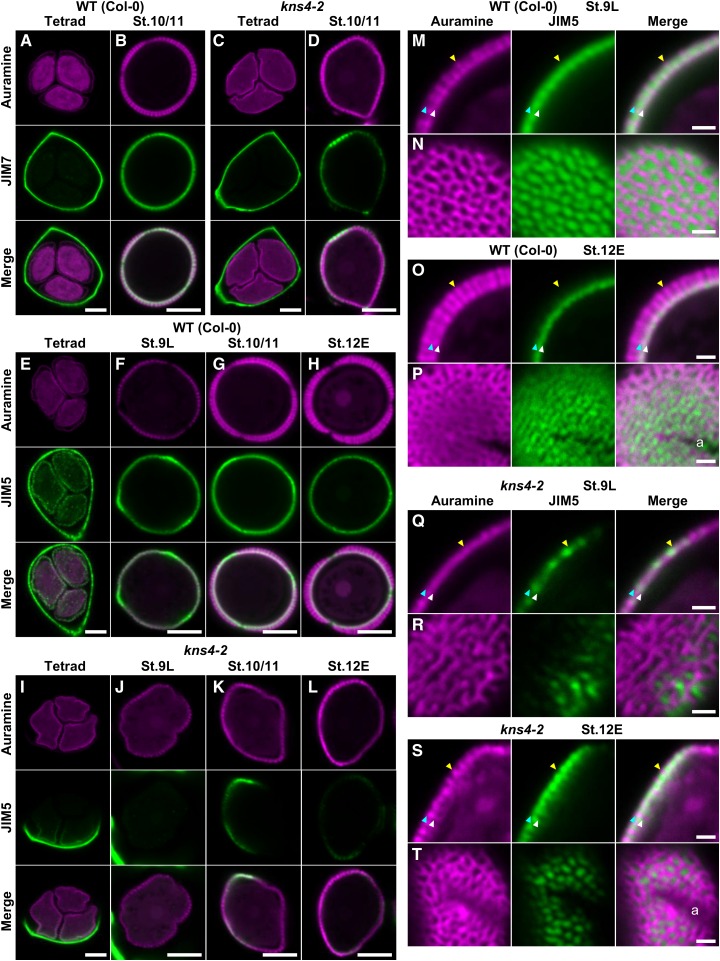
Pectins are known to be involved in cell-to-cell adhesion and cell separation in higher plants (for review, see Caffall and Mohnen, 2009). Pectins are generally composed of HGs, RG-I, and RG-II, and are synthesized in the GA; in the case of HG, the major type of pectin, as a highly methylesterified form that is then demethylesterified by pectin methylesterases after being deposited into the wall (Mohnen, 2008). Pectins are present on the surface of tetrads and defects in their degradation cause abnormal exine fusion (quartet phenotype; Rhee and Somerville, 1998). Moreover, pectins have been shown to be a component of primexine (Rhee and Somerville, 1998; Aouali et al., 2001; Majewska-Sawka et al., 2004). To test if the cell-to-cell adhesion and thin exine phenotypes in kns4 anthers were due to changes in the distribution of pectic polysaccharides and/or their methyl-esterification, two monoclonal antibodies, JIM7 and JIM5, recognizing highly methylesterified and less methylesterified pectins (HG), respectively (Knox et al., 1990), were used in labeling experiments.
In wild type at stage 8, JIM7 strongly labeled the walls separating PMCs and tapetal cells (Fig. 9A). At stage 9M, the primary walls of tetrads, which were detached from each other and from tapetal walls, were strongly labeled (Fig. 9B). After the degradation of the callosic walls, the labeling on the surface of microspores was visible (Fig. 9, C and D). A similar labeling pattern was detected in kns4 mutants when compared to the wild type (compare Fig. 9, E–H to Fig. 9, A–D, respectively). In contrast, the distribution of JIM5 labeling (Fig. 9, I–P) was distinct from that of JIM7. Specifically, no signal was detected in the primary walls of the PMC (stage 8) and tetrad (stage 9M) in both wild type (Fig. 9, I and J, respectively) and kns4 (Fig. 9, M and N, respectively). Note that the JIM5 images have higher background signal in comparison to the JIM7 images (compare Fig. 9, I–P, and Fig. 9, A–H, respectively) because the signal was weak and longer exposure times were required. After dissolution of the callosic walls, JIM5 labeled the surface of microspores (Fig. 9, I–P; for stage 10/11, see Fig. 9K). JIM5 labeling gradually decreased but was detectable until the end of stage 12M (Fig. 9L). In contrast, JIM5 labeling was largely absent in the walls of developing kns4 microspores (Fig. 9, M–P).
Figure 9.
Fluorescence micrographs of the distribution of JIM7- and JIM5-epitope labeling of pectins within anthers. Cross sections of resin-embedded anthers of wild type (Col-0; A–D and I–L) and kns4-2 (E–H and M–P) from stages 8 to 12M were incubated with either JIM7 (A to H) or JIM5 antibody (I to P), and subsequently with Alexa Fluor 488-labeled secondary antibody. Fluorescence of Alexa Fluor 488 (green) and autofluorescence (magenta) were captured separately by epifluorescence microscopy and overlaid. Note that the exposure time for capturing JIM5 signals is longer than that for JIM7, resulting in relatively higher background in JIM5 images. En, Endothecium; Ep, epidermis; ML, middle layer cell; MS, microspore; PMC, pollen mother cell; Td, tetrad; Tp, tapetal cell. Bar = 20 μm.
Developing tetrads and microspores were also isolated and immunolabeled using the JIM7 and JIM5 antibodies (Fig. 10). As in undissected anthers (Fig. 9, B and F), primary cell walls of both wild-type and kns4 tetrads were labeled with JIM7 in a relatively uniform manner (compare Fig. 10, A and C). JIM7 labeling was also observed on the surface of Fig. 9, I–P microspores through stages 9L to 12E, with maximum intensity at stage 10/11 (Fig. 10B). In general, weaker labeling appeared on the surface of kns4 microspores although some parts were as strongly labeled as wild type (Fig. 10D), suggesting HG deposition was unevenly distributed in the mutant.
Figure 10.
Fluorescence micrographs of the distribution of JIM7- and JIM5-epitope labeled pectins on the surface of isolated tetrads and developing microspores. A to D, Median optical sections of JIM7-labeled tetrads isolated from stage 9M anthers (A and C) and microspores isolated from stage 10/11 anthers (B and D), respectively. Exine was visualized by auramine O staining. Specimens were observed by CLSM in which magenta and green channels show fluorescence of auramine O and Alexa Fluor 546, respectively. Wild type (A and B) and kns4-2 (C and D) are compared. E to L, Median optical sections of JIM5-labeled tetrads (E and I) isolated from stage 9M anthers and microspores (F–H and J–L isolated from anthers at indicated stages. Wild type (Col-0; E–H) and kns4-2 (I–L) are compared. M to T, Enlarged optical sections of JIM5-labeled microspores. Median optical sections showing exine longitudinal sections (M, O, Q, and S) and tangential optical sections that cut the surface of microspore (N, P, R, and T) or that cut baculae transversely (O). Microspores were isolated from stage 9L (M, N, Q, and R) and stage 12E (O, P, S, and T) anthers. Wild type (M–P) and kns4-2 (Q–T) are compared. Blue, white, and yellow arrowheads indicate top and bottom boundary of the pectin layer, and the top of representative bacula, respectively. a, aperture. Bars = 5 μm in (A–L), and 1 μm in (M–T).
The wild-type tetrad wall also showed a relatively even labeling pattern with JIM5 (Fig. 10E), although the labeling was never observed in resin-embedded sections (Fig. 9J). In contrast, primary walls of the kns4 tetrads often displayed a partial labeling pattern (Fig. 10I), indicating that HG demethylesterification occurs only locally in the mutant. JIM5 also labeled the surface of wild-type microspores (Fig. 10, F–H). The labeling was initially observed at the end of stage 9M, when the microspores were still within tetrads (Fig. 10E), reached a maximum at stage 10/11 (Fig. 10G), and then decreased thereafter (Fig. 10H). In contrast, the kns4 microspores were labeled only weakly and partially with JIM5 (Fig. 10, J–L), similar to the labeling pattern observed with JIM7 (Fig. 10D).
Enlarged optical sections of JIM5-labeled wild-type stage 9L microspores revealed that the less methylesterified HG is localized in exine cavities, between the nexine and tectum (Fig. 10M), but in later stages it forms a distinct layer at the base of exine cavities on nexine I (stage 12E, Fig. 10O). Tangential optical sections near the surface showed that the JIM5-labeled HG at stage 9L is located in the lacunae of the reticulate exine (Fig. 10N). This location is different from that of the AGPs, which cover the outer reticulate tectum surface at the same developmental stage (Fig. 7G). At stage 12E, JIM5-labeled HG on nexine I is still distributed in a pattern that is opposite to the reticulate surface of the exine (Fig. 10P). Similar to wild type, the pectin deposited on kns4 microspores filled the shallow cavities of the exine (Fig. 10, Q–T).
kns4 Pollen Defects Lead to Reduced Fertility
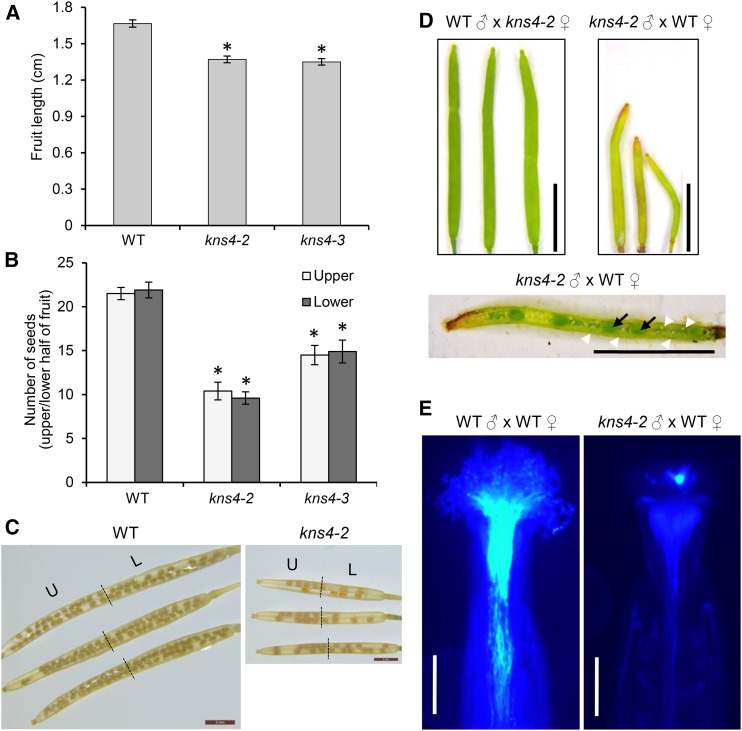
Given the AGP and pectin defects observed during microspore development in the kns4 mutant, we wanted to determine the possible function(s) of KNS4 in planta, especially during sexual reproduction. Because fruit length has previously been shown to be correlated with seed set (Barendse et al., 1986), fruits of wild type (Col-0) and kns4 mutants were collected and their lengths measured. Fruits of kns4 mutant plants were significantly shorter (kns4-2: 1.37 ± 0.03 cm; kns4-3: 1.35 ± 0.03 cm, n = 15) than those from wild type (1.67 ± 0.03 cm, n = 15, P < 0.001; Fig. 11A). Examination of the number of seeds present in each wild-type and kns4 fruit at stage 17 (fully expanded green fruit, Smyth et al., 1990) showed that the loss of KNS4 function reduced seed set by as much as 50% (wild type: 43 ± 1; kns4-2: 29 ± 2; kns4-3: 20 ± 2, n = 15, P < 0.001), although no bias toward the upper or lower half of the fruit was observed (Fig. 11, B and C).
Figure 11.
kns4 has defects in sexual reproduction. A to C, Morphological phenotypes of fully elongated fruit length (A) and seed set (number of seeds per upper and lower half of fruit; B and C) were observed and quantified. Error bars indicate SD. *P < 0.0001 in a Student’s t-test, wild type (Col-0) compared to kns4-2 and kns4-3 in (A) and (B). D, Reciprocal crosses between wild type and kns4-2. Fruits 7 d after pollination are shown. Dissection of fruits from kns4-2-pollinated wild-type pistils revealed formation of fewer seeds (black arrows) and many unfertilized ovules (white arrowheads), indicating lower probability of ovule fertilization by kns4 pollen. E, wild-type, or kns4-2 pollen tubes growing in vivo in manually pollinated wild-type pistils stained with aniline blue after 24 h. Staining appears as a bright light blue color. Scale bar = 2 mm in (C), 500 μm in (D) and 5 mm in (E). L, lower; U, upper.
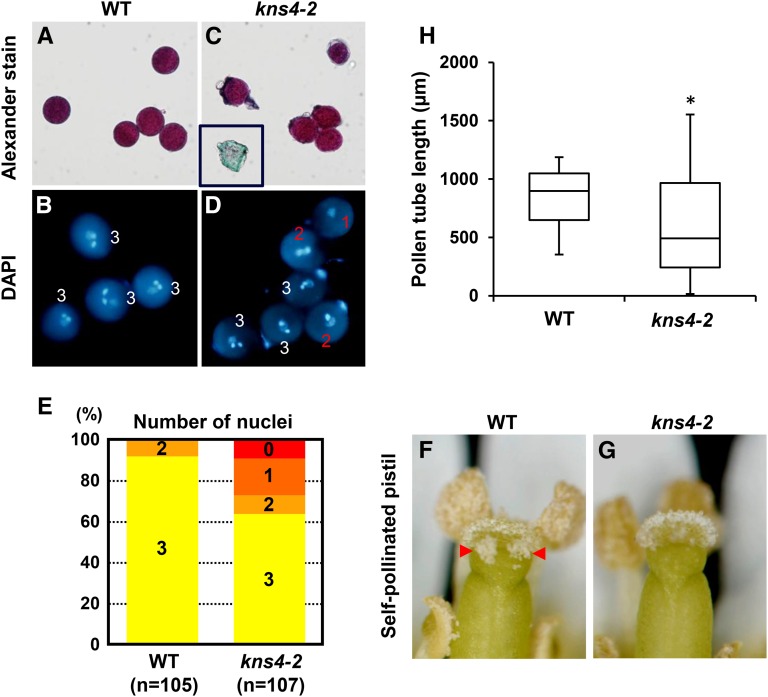
To confirm that the reduced fertility observed in kns4 mutants was a consequence of a disruption in male gametophyte development, reciprocal crosses were performed. When wild-type pollen was used to pollinate kns4-2 pistils, fruits with normal lengths (1.51 ± 0.04 cm) resulted, whereas kns4-2 pollen on wild-type pistils yielded significantly shorter fruits (0.86 ± 0.06 cm) with few fertilized ovules (Fig. 11D). These results suggested that kns4 pollen grains have reduced fertility. Alexander staining to test for pollen viability showed most of the kns4-2 pollen grains were colored purple, indicating they were viable (Fig. 12C). However, DAPI staining revealed that one-third of kns4-2 pollen grains were developmentally aborted before entering tricellular stage (Fig. 12, B, D, and E). Hence, mutations in KNS4 cause not only an exine abnormality but also a defect in pollen development.
Figure 12.
Viability of wild-type and kns4-2 pollen grains. A to E, Staining of wild-type (Col-0) and kns4-2 pollen grains with Alexander dye (A and C, respectively) and DAPI (B and D, respectively). A few aborted pollen grains are observed in the kns4-2 pollen population (inset in C). The numbers in (B) and (D) show the number of nuclei in each pollen grain; (E) shows the percentage of wild-type and kns4-2 pollen grains with 0, 1, 2, or 3 nuclei. F and G, Images of self-pollinated pistils in wild-type (Col-0; F) and kns4-2 (G) flowers. Pollen adhesion is more obvious on the wild-type stigma (red arrowheads). H, Lengths of wild-type (Col-0) and kns4-2 pollen tubes grown in vitro for 16 h as represented by a bar-and-whiskers plot (n = 28). *P < 0.01 (Student’s t-test).
Because pollen grains produced by stamens in a flower are in large excess of ovules in a pistil, developmental defects in a subset of pollen grains usually pose few, if any problems with self-pollination. However, observations of self-pollinated pistils revealed that the number of pollen grains that adhered to the stigma was far fewer in kns4-2 than in wild type (Col-0; compare Fig. 12, G and F, respectively), indicating that the pollen clumps formed in kns4 anthers affect pollen dispersal. Furthermore, it is also possible that kns4 pollen grains are functionally less active. To examine this possibility, we tested if the pollen grains of kns4 mutants could germinate, and that pollen tubes could extend down into the style. To do this we used an in vivo pollen tube growth assay, where preemasculated mature wild-type (Col-0) pistils were pollinated with either wild-type (Col-0) or kns4-2 pollen grains, and pollen tube elongation observed after 24 h (Fig. 11E). Fewer kns4 pollen tubes grew into the pistil compared to wild type, suggesting pollen tube germination and/or growth was compromised in the mutant. Deficiency in pollen tube elongation of kns4-2 pollen grains were also observed in an in vitro pollen tube growth assay (Fig. 12H). Taken together, these data suggest that loss of KNS4 function leads to multiple cell wall defects during male gametogenesis that have developmental/reproductive consequences.
DISCUSSION
We have identified and characterized the gene defective in kns4 mutants that results in a unique thin exine phenotype among 12 previously reported kns mutants (Suzuki et al., 2008). The KNS4 gene is At1g33430 and is identical to the underlying gene of the upex1 mutant [Dobritsa et al., 2011; Li et al., 2017 (accompanying article)]. KNS4 expression is restricted to tapetal cells in young anthers from stages 8 to 12E of flower development, consistent with the timing of exine development (Smyth et al., 1990). Our biochemical studies demonstrate that KNS4 is a β-(1,3)-GalT involved in the biosynthesis of the β-(1,3)-galactan backbone of type II AGs. Type II AGs are found in both pectins as side chains of RG-I and AGPs; however, because changes in both AGPs and pectins were observed at the same stage of development in the tapetum of wild-type and kns4 anthers, it is difficult to discern the exact molecular target of KNS4. That kns4 mutants display a relatively severe phenotype also indicates that KNS4 is not redundant with other GalTs that are likely expressed in tapetal cells.
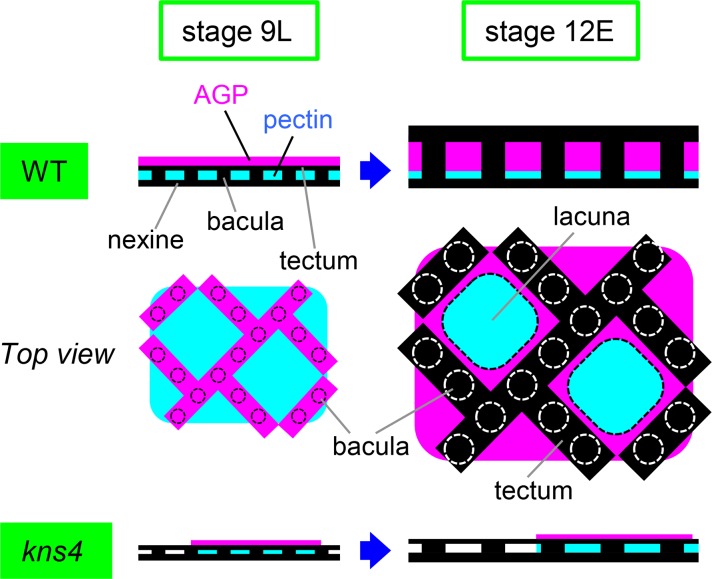
Exine formation is initiated with the construction of its basic structure composed of baculae and tectum in the primexine layer between the callosic wall and microspore plasma membrane in tetrads. We found that JIM8-labeled AGPs are a constituent of primexine in tetrads (Fig. 6B), together with previously identified pectin (HG and RG-II; Rhee and Somerville, 1998). Furthermore, JIM8- and MAC204-labeled AGPs, as well as JIM5- and JIM7-labeled HGs, form matrix layers on the surface of free microspores after being released from tetrads (Figs. 6–10). Although the basic structure of the exine has been established in tetrads, its thickness increases more than 2-fold by stage 12E (Supplemental Fig. S1A). Therefore, it is expected that the matrix of AGP and pectin functions as a framework for sporopollenin deposition on the surface of free microspores at stages 9L to 12E. Because it is proposed that sporopollenin is transferred from tapetal cells to the microspore surface as either copolymers or fine fibrillar materials (Owen and Makaroff 1995; Quilichini et al., 2010), AGPs (and pectins, and possibility other, as yet unidentified, polysaccharides/glycoproteins) may also be incorporated in the copolymers and transferred concomitantly with sporopollenin. We propose a model of how AGPs and pectins might function in the construction of exine (Fig. 13). By stage 9L, a framework of exine is made in the primexine layer that is mainly composed of pectin. AGPs are laid down over the pectin but initially, they do not enter into the exine cavities. The AGP layer is reticulate and displays a similar pattern to exine. During microspore development, sporopollenin supplied by tapetal cells is deposited onto the expanding exine. AGPs are also supplied by tapetal cells and are deposited into the AGP layer, the thickening of which elongates the baculae and raises the tectum. At stage 12E, a thin pectin layer remains at the bottom of the exine cavities and the thickened AGP layer fills the space beneath the tectum. According to this model, the thin exine phenotype of the kns4 mutants is well explained by the observed defects in pectins and/or AGPs. The uneven thickness of exine observed in the mutant is likely due to the uneven distribution of AGP and pectins and is consistent with the nonuniform labeling of AGP and pectin epitopes seen in Figures 7 and 10. It is also conceivable that other matrix phase polysaccharides act as the framework that regulate the size of the lacunae and impact on the 3D architecture of the pollen wall.
Figure 13.
Schematic model of the function of microspore primexine in exine development. On the surface of wild-type microspores just released from the tetrad (stage 9L), the tectum of developing exine is sandwiched between pectin and AGP. The top view indicates AGPs lie on the outer surface of the tectum and display a similar reticulate pattern, and pectins show an internal distribution. As microspores develop, AGPs and sporopollenin supplied by tapetal cells are deposited into the primexine and exine, respectively. By stage 12E, the AGP layer laid down over the pectins becomes thicker, lifts the tectum up, and extends the baculae, resulting in exine thickening. AGPs fill the exine cavities except for baculae and lacunae, whereas pectins remain in the lacunae. In kns4, the AGP and pectin layers are incompletely formed at stage 9L. Subsequent deposition of AGP and sporopollenin is reduced and the exine remains thin at stage 12E.
The interactions (covalent and/or noncovalent) between pectins and AGPs has been reported in numerous studies (Baldwin et al., 1993; Li et al., 1995; Jauh and Lord, 1996; Mollet et al., 2002; Majewska-Sawka and Rodriguez-Garcia, 2006) and structural analyses have provided multiple lines of evidence that AGPs are associated with pectins, especially RG-I (Immerzeel et al., 2006; Mohnen, 2008; Caffall and Mohnen, 2009) and more recently xylan (Tan et al., 2013; Voiniciuc et al., 2015). Moreover, SOS5/FLA4 (At3g46550), a fasciclin-like GPI-anchored AGP, was recently shown to be involved in the formation of either a pectin or a pectin-hemicellulose network in seed mucilage (Griffiths et al., 2014). Although the distribution of AGPs and pectins are not identical in primexine, AGPs and/or pectins and possibly xylan (Li et al., 2017) might function as a scaffold for other wall components and could potentially be cross-linked to each other.
The first developmental abnormality identified in kns4 anthers was the failure of PMC separation before meiosis (stage 9E), a defect that continues into stage 9M when PMCs become tetrads (Fig. 2). It has been proposed that pectin plays an important role in both cell adhesion and cell separation in plant organ development (Micheli, 2001; Pelloux et al., 2007; Wolf and Greiner, 2012). We detected differences between wild type and kns4 both in the deposition of AGPs (Figs. 6, E and F and 7D) and in the demethylesterification of HG (Fig. 10I) in the walls of tetrads, although changes to AGP epitope labeling were initially observed in PMC walls (stage 8). Therefore, the apparent reduction in abundance of AGPs slightly precedes the changes observed in pectin, suggesting AGPs are the more likely direct target of KNS4 activity. While highly methylesterified HG pectin appeared to be deposited in a normal manner in kns4 mutant microspores (Figs. 9, E and F and 10C), the appearance of less-methylesterified HG pectin as revealed by JIM5 labeling was patchy (Fig. 10I) unlike wild type where the labeling is relatively uniform (Fig. 10E). This suggests there is an accessibility issue of these walls to the pectin methylesterases that demethylesterify pectin. After degradation of the tetrad wall, released microspores of the kns4 mutants still adhered to each other (Fig. 2, J–L) and no additional JIM5 epitopes were revealed (Fig. 10, J–L) when normally signal would increase (Fig. 10G; stage 10/11). Hence, reduced accessibility to kns4 microspore walls persists after tetrad wall dissolution. We showed that mutant microspores at stage 12E formed a clump when they were extruded from anthers and that callosic walls abnormally remained between microspores (Fig. 3H). Furthermore, Li et al. (2017) have shown the existence of abnormally remaining xylan around microspores. From these results, it seems likely that the AGPs and/or pectins produced by the enzymatic function of KNS4 contribute to cell wall architecture in a way that impacts the accessibility of glycan hydrolytic enzymes (glycosyl hydrolases) to the locular space.
The second abnormality relates to the deposition of the sporopollenin. Sporopollenin is synthesized in tapetal cells, secreted to locules, and transferred to the microspore surface (Owen and Makaroff 1995; Quilichini et al., 2014). Besides the failure of exine formation in kns4, we revealed that the sporopollenin is abnormally deposited on the surface of tapetal cell walls, which is in contact with the developing exine of microspores (Figs. 2, R–T and 3, D–F). In wild type, tapetal walls are usually degraded and have disappeared by stage 12L when tapetal cells collapse. In contrast, kns4 mutant walls fail to degrade completely, remaining attached to the surface of microspores (Figs. 2L and 3F).
The third abnormality of kns4 mutants is related to microspore development and pollen fertility, issues that are likely consequences of abnormal pollen development. We studied the reproductive success of kns4 pollen with reciprocal crosses between wild type and the kns4-2 mutant showing reduced seed set in kns4 (Fig. 11D). Likewise, for self-pollinated mutants, the fruits were shorter compared to wild type (Fig. 11, A–C). In addition, an in vivo pollen tube growth assay indicated that fewer pollen tubes germinated and grew into the style (Fig. 11E). DAPI staining revealed that one-third of kns4 microspores abort before generative cell mitosis (Fig. 12, B–E). As a consequence, the mutants show reduced seed set (Fig. 11). These results suggest that the microspore phenotype has an impact on the reproductive success of kns4. Considerable evidence has now accumulated implicating the involvement of AGPs in pollen fertility and pollen tube growth (for review, see Nguema-Ona et al., 2012, Pereira et al., 2015). KNS4, as a β-(1,3)-GalT, is predicted to be involved in the synthesis of β-(1,3)-galactan backbone of type II AGs, which can either be attached to AGPs or pectin. It is possible that KNS4 has multiple cell wall polysaccharide targets in pollen; hence, knocking out KNS4 leads to a severe microspore phenotype, which results in reduced pollen fertility.
KNS4/UPEX1 encodes a β-(1,3)-GalT responsible for type II AG biosynthesis on either the protein backbones AGPs or the side chains of the pectic polysaccharide RG-I. This study, together with that of the accompanying article by Li et al., (2017), demonstrates that the absence of these glycans leads to severe defects in primexine and exine formation and subsequent microspore development.
MATERIALS AND METHODS
Source and Growth of Plants
The kns4-1 mutant allele in the Landsberg erecta background of Arabidopsis (Arabidopsis thaliana) was previously described in Suzuki et al. (2008). T-DNA insertion alleles in the Columbia-0 (Col-0) background, SALK_091466 (kns4-2) and SAIL_544_C02 (kns4-3), were obtained from the Arabidopsis Biological Resource Centre (Ohio State University). Arabidopsis seeds were sown directly on soil, stratified at 4°C for 3 d, and grown under a regime of 16 h light at 21°C and 8 h darkness at 17°C. Nicotiana benthamiana plants were grown in soil (Debco seed raising mix, Tyabb) at 20° to 26°C in a glasshouse under continuous cool-white lights supplemented with daylight for 5 to 6 weeks.
Electron Microscopy and Immunolabeling
For SEM observations of pollen grains and TEM observations of anther sections, the methods as described by Suzuki et al. (2008) were followed. Immunogold labeling was carried out as described in Wilson and Bacic (2012).
Immunofluorescence Labeling
Flower bud clusters at inflorescence apices were fixed [2% (v/v) formaldehyde, 2.5% (v/v) glutaraldehyde, 25 mM Na-P buffer, pH 7.5] for 24 h at 4°C, then dehydrated through an ethanol series. The ethanol was replaced with 1:1 mix of LR White resin (type medium; Electron Microscopy Sciences) and ethanol, then with pure resin. The resin-infiltrated samples were placed separately in small plastic tubes, and polymerized by heat treatment at 50°C for 12 h in a plastic bag containing oxygen absorber. Sections (1–2 µm) were cut in a microtome (RM2125RT; Leica) equipped with TC-65 blade and mounted on a slide (MAS-coated glass slide; Matsunami). Sections were treated with a solution (1% (w/v) bovine serum albumin (BSA) in PBST (5.1 mm Na2HPO4, 1.6 mm KH2PO4, 130 mm NaCl, 0.02% Tween 20) for 1 h at RT for blocking, and subsequently incubated with a 1:40 dilution of primary antibody in the same solution for 4 h at RT with gentle rocking. Rat monoclonal antibodies JIM5, JIM7, JIM8, and MAC204 (CarboSource Service) were used as primary antibodies. After washing with PBST, samples were reacted with a 1:100 dilution of secondary antibody (Alexa Fluor 488 Goat anti-Rat IgG; Thermo Fisher Scientific) for 2 h and then washed with PBST and mounted with aqua-poly/mount (Polysciences). A BX60 epifluorescence microscope equipped with a DP72 digital camera (Olympus) was used for observation. Fluorescence of Alexa Fluor 488 and background autofluorescence of the sample were captured under fluorescence mirror units NIBA and WBV, respectively. To make a merged image of these signals, the NIBA and WBV images were captured simultaneously, split into RGB channels by Photoshop (Adobe), and both the R and B channels of the NIBA image were replaced with the G channel of the WBV image.
Anthers collected from developing flower buds were immersed into fixative (3.7% formaldehyde, 50 mm Na P buffer, pH7.2), ground with a homogenizer (BioMasher II; Nippi) and put on ice for 1 h. After tissue debris were removed, tetrads and microspores were collected by centrifugation at 400g for 1 min, washed three times with PBST, treated with a blocking solution (1% (w/v) BSA in PBST), and incubated with a 1:100 dilution of primary antibody in blocking solution for 3 h at RT. Samples were washed twice with PBST, rinsed with blocking solution, and then incubated with a 1:100 dilution of secondary antibody (Alexa Fluor 546 Goat anti-Rat IgG, Thermo Fisher Scientific) for 1 h. After washing twice with PBST, samples were suspended in 0.5 × PBS (2.6 mm Na2HPO4, 0.8 mm KH2PO4, 65 mm NaCl), 0.005% (w/v) Tween 20, 18% (v/v) glycerol, 0.005% (w/v) auramine O. The sample was spread on a coverslip coated with 0.2% (v/v) polyethyleneimine solution and mounted on a glass slide. The specimens were observed by a CLSM (FV1000; Olympus) with Ex: 473 nm, Em: 485 to 545 nm for auramine O staining, and Ex: 559 nm, Em: 565 to 665 nm for Alexa Fluor 546.
Yariv Dye Staining of Arabidopsis Pollen
Floral buds at stage 12 (Smyth et al., 1990) were collected from five plants of each genotype (wild type, kns4-2, and kns4-3) and anthers dissected with a fine blade. The dissected anthers were then placed into either α-Gal Yariv reagent dye [40 µL of 2 mg/mL in 1% (w/v) NaCl] as a negative control or β-Glc Yariv reagent dye [40 µL of 2 mg/mL in 1% (w/v) NaCl] to stain for AGPs. The samples were tapped gently to release the pollen and then placed in a ThermoMixer (Eppendorf) at 800 rpm for 20 min, both to assist pollen release and dye infiltration. The samples were incubated at 4°C overnight and subsequently washed twice with 0.15 m NaCl before viewing under a light microscope.
Toluidine Blue Staining
Sections (1 µm) of resin-embedded anthers as prepared for TEM were mounted on a glass slide, stained with a toluidine blue staining solution [0.5% (w/v) toluidine blue, 0.5% (w/v) sodium borate], and the slides heated on a hot plate for 20 min. After washing out the stain, the specimen was viewed under a light microscope (BX60; Olympus).
Calcofluor Staining
Sections (8 µm) of paraffin-embedded flower bud clusters were mounted on a MAS-coated glass slide, deparaffinized by a serial treatment with Lemosole (Wako Pure Chemical), 1:1 mix of Lemosole and ethanol, then ethanol, and rehydrated by an ethanol series. The sample was incubated with 1% (w/v) Calcofluor White (Fluorescent Brightener 28; Sigma-Aldrich) for 5 min, washed five times with water, and sealed with aqua-poly/mount. Fluorescence was detected by an epifluorescence microscope (BX60; Olympus) with a WBV mirror unit.
Aniline Blue Staining of Anther Sections
Sections of resin-embedded anthers were incubated with aniline blue fluorochrome (0.1 mg/mL; Biosupplies) and sealed with aqua-poly/mount. The specimens were observed by a CLSM (FV1000; Olympus) with Ex 405 nm, Em 485-555 nm for aniline blue staining, and Ex 440 nm, Em 610–710 nm for autofluorescence of sporopollenin.
Measurement of Exine Thickness
Median optical sections of developing microspores prepared for immunofluorescence labeling were used to measure exine thickness by CLSM. Local maxima of exine thickness were measured at three positions in a microspore, and average and sd of 30 measurements (10 microspores for each developmental stage) was calculated. Intine and exine thickness were also measured using labeled and unlabeled TEM sections.
Evaluation of Seed Set Activity
The fruits from self-pollinated kns4-2, kns4-3 and wild type (Col-0) were collected and the individual fruit length measured. Fruits were decolorized with 90% acetone for 30 min, followed by 50% ethanol for 30 min, and 70% ethanol overnight to view contents. After a further 70% ethanol wash for 30 min, samples were incubated for an additional 30 min in 100% ethanol. Fruits were then viewed under a model no. M205A stereo microscope (Leica Microsystems), and seeds counted.
Reciprocal crosses were performed between wild type and kns4-2 following the protocol from Weigel and Glazebrook (2002). Ten flowers from each line were emasculated and hand-pollinated with the appropriate pollen type. Fruits were allowed to develop for a week before images were captured using a model no. M205A stereo microscope (Leica Microsystems). Two biological replicates comprising 10 flowers for each line were performed for the reciprocal crosses. The fruits were then dissected and the embryos and ovules were checked for any developmental defects.
Pollen Tube Growth
The in vivo pollen tube growth protocol of Wu et al. (2010) was followed. Imaging was carried out using a model no. DM 6000B compound light microscope utilizing a 5× objective (Leica Microsystems) under an I3 filter (BP 450-490; Excitation: LP 515 nm).
Mapping of the kns4-1 Mutation
A F2 population obtained by a cross between kns4-1 and Col-0 was used for a mapping the KNS4 locus using the method described by Suzuki et al. (2008).
Genetic Complementation
The KNS4 promoter region (ProKNS4, 2006 bp upstream of the initiation codon) was amplified by PCR from Col-0 genomic DNA using KNS4p-attB4-F (5′-GGGGACAACTTTGTATAGAAAAGTTGTATCATCTTCAACACCATCTTAGCT-3′) and KNS4p-attB1-R (5′-GGGGACTGCTTTTTTGTACAAACTTGTGAAAACAAGAATGTAGTTGCAGAA-3′) primers. The amplified fragment was cloned into pDONR P4-P1R vector by a Gateway BP reaction (Invitrogen) to make a promoter entry clone, pDONR-L4KNS4pR1. The coding region of KNS4 was PCR-amplified from Col-0 inflorescence first-strand cDNA using KNS4-attB1-F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTTTATATAAATAACATCTCCTCAACATTC-3′) and KNS4-attB2-R (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTAGACGAAGAAGCGAAAATTAGCTAGA-3′) primers, and cloned into the pDONR201 vector by a BP reaction to make a CDS entry clone, pDONR-KNS4. PrimeSTAR HS DNA polymerase (Takara Bio) was used for cloning PCRs. The promoter and CDS were joined and transferred to a binary vector, R4pGWB401 (Nakagawa et al., 2008) by a Gateway LR reaction (Invitrogen) to generate the binary plasmid pGWB-KNS4p:KNS4. This plasmid was introduced into Agrobacterium tumefaciens strain C58C1 (pMP90) by electroporation and the bacteria then used to infect kns4-1 plants by floral dip (Ishiguro et al., 2010). Transformants were identified on agar plates supplemented with 30 mg/L kanamycin.
RT-PCR
RNA was extracted from various organs using the RNeasy Mini Kit (Qiagen) and incubated with DNase I to remove residual DNA. First strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) primed with an oligo(dT)20 primer. PCRs were carried out to amplify a part of coding region using Ex Taq DNA polymerase (Takara Bio). ACT2 (At3g18780) was used as a control. Primers used for PCR were as follows: KNS4-F2 (5′-GAGAACTCACCAAGCAGTTA-3′), KNS4-R2 (5′-CCTGTTGCATGCCTAAAGTA-3′), ACT2-F (5′-GAAGAGCACCCTGTTCTTCT-3′), and ACT2-R (5′-TTCTCGATGGAAGAGCTGGT-3′).
To determine mRNA levels in developing anthers, ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo) was used for preparing first strand cDNA. Real-time qRT-PCR was performed using the SYBR Premix Ex Taq II (Takara) on a LightCycler 480 (Roche). PDF2 (At1g13320) was used as an internal control. Primers used for qRT-PCR were as follows: KNS4, 5′-GAGAACTCACCAAGCAGTAA-3′ and TTTCTGCAGACGGCTCTGGTTTTTC-3′; PDF2, 5′-TAACGTGGCCAAAATGATGC-3′ and 5′-GTTCTCCACAACCGCTTGGT-3′.
In Situ Hybridization
A DNA fragment corresponding to a half of a KNS4 mRNA sequence was prepared by PCR amplification with two primers KNS4-SacI-F (5′-CCCGAGCTCTTTTATATAAATAACATCTCCTCAACATTC-3′) and KNS4-SacI-R2 (5′-CCCGAGCTCAGAAGTACAGTCTTGTCTTTGTCGA-3′) and cloned in both directions into the SacI site of the pBS-KS(+) plasmid. The resulting two plasmids, pBS-KNS4ΔC3f and pBS-KNS4ΔC3r, contain the sense and antisense orientation of the KNS4 sequence, respectively, relative to the direction of T7 promoter. A DNA fragment was amplified from each plasmid by PCR using M460 and RV58 primers, and purified using the QIAquick PCR purification kit (Qiagen). The purified fragment derived from pBS-KNS4ΔC3f was transcribed in vitro by T7 RNA polymerase in the presence of Fluorescein labeling mix (Roche, Switzerland) to prepare a fluorescein-labeled sense probe for KNS4. An antisense probe was prepared from a fragment derived from pBS-KNS4ΔC3r in a similar manner. To remove unincorporated nucleotides, transcripts were purified by a ProbeQuant G-50 Micro Column (GE Healthcare).
Flower bud clusters were fixed in a fixative [3.7% (v/v) formaldehyde, 5% (v/v) acetic acid, 50% (v/v) ethanol] for 15 h at RT. Samples were dehydrated in a graded ethanol series, immersed in Lemosole, and embedded in paraffin (Paraplast Plus; McCormic Scientific). Serial sections of 8 µm thick were prepared by a microtome, mounted on a MAS-coated glass slide, deparaffinized by Lemosole, and rehydrated with a graded ethanol series. Samples were treated with 5 µg/mL proteinase K for 30 min at 37°C in a buffer (100 mm Tris-HCl, 50 mm EDTA, pH7.5), fixed in freshly hydrolyzed 5% (v/v) paraformaldehyde in 10 mm Na-P buffer (pH 7.2), and acetylated in 0.5% (v/v) acetic anhydride dissolved in 100 mm triethylamine-HCl buffer (pH 8.0). Samples were washed in 2× SSPE and dehydrated in a graded ethanol series. Two-hundred microliters of hybridization mix [50% (v/v) formamide, 10 mm Tris-HCl, 0.3M NaCl, 1 mm EDTA, 1× Denhardt’s solution, 10% dextran sulfate, 30 mm DTT, 1 µg/µL Escherichia coli tRNA, 1 µg/µL poly A, and 0.5 ng/µL sense or antisense probe] was placed on a slide and the slide incubated overnight at 50°C in a humid box. Samples were washed in 4× SSC at 50°C for 15 min, treated with 20 µg/mL RNase A in a buffer (10 mm Tris-HCl, 500 mm NaCl, 10 mm EDTA, pH 7.5), and washed twice in 0.5× SSC at 50°C for 20 min. The samples were then rinsed in buffer 1 (100 mm Tris-HCl, 150 mm NaCl, pH 7.5), treated with blocking solution (1% Blocking reagent (Roche) in buffer 1), and reacted with Anti-Fluorescein-AP (Roche) in buffer 2 (100 mm Tris-HCl, 150 mm NaCl, 0.2% (v/v) Tween 20, 1% (w/v) BSA, pH 7.5) for 2 h at RT. After washing in buffer 1, the slide was incubated in a stain solution (0.55 mm NBT, 0.43 mm BCIP, 100 mm Tris-HCl, 100 mm NaCl, 50 mm MgCl2, pH 9.5) for 9 h at RT. After washing in TE (10 mm Tris-HCl, 1 mm EDTA, pH 7.5), samples were sealed with aqua-poly/mount.
Construction of KNS4 Promoter-reporter Constructs
The same KNS4 promoter fragment was PCR-amplified as described above and cloned into the pDONR201 vector by a Gateway BP reaction. The resulting plasmid, pDONR-L1KNS4pL2, was recombined with pGWB433 and pGWB450 (Nakagawa et al., 2007) by a Gateway LR reaction to produce pGWB-KNS4p:GUS (binary plasmid for GUS fusion) and pGWB-KNS4p:GFP (for GFP fusion), respectively. Plant transformations were carried out as described above.
Transient Expression of KNS4 in N. benthamiana Leaves
Transient expression in N. benthamiana leaves was carried out following Lampugnani et al. (2016) with minor modifications. To generate the Pro-35S:KNS4 construct, the KNS4 CDS was amplified from cDNA of wild-type Arabidopsis floral buds using primers KNS4-Fwd (5′-TCGAGGAATTCGGTACCATGAGGGCGAAAGCTGCTTCGGG-3′) and KNS4-Rev (5′-AAGCAGGACTCTAGAGGATCCTCAGACGAAGAAGCGAAAATTAGC-3′). The PCR product was cloned into the binary vector pFUERTE (Wilson et al., 2015) by GeneArt seamless cloning according to the manufacturer’s instructions (Invitrogen). Sequence-verified constructs along with the helper plasmid pSOUP were introduced into A. tumefaciens strain AGL1 via electroporation as described in Lampugnani et al. (2012). A 35S:VENUS construct was used as a transformation control (Wilson et al., 2015).
Microsomal Membrane Preparation from N. benthamiana Leaves
Microsomal membranes (MMs) were prepared according to Song et al. (2015) with minor modifications. Briefly, 2 g of plant tissue was ground using a mortar and pestle in 10 mL of chilled extraction buffer (0.1 m HEPES/KOH pH 6.8, 0.4 m Suc, 5 mm MgCl2, 5 mm MnCl2, 1 mm PMSF; Roche EDTA-free complete protease inhibitor). MM preparations were resuspended in 250 μL extraction buffer and stored at −80°C before use.
Biochemical Assay of KNS4
MMs were pretreated on ice for 10 min with 1% v/v Triton X-100 (Sigma-Aldrich) before use in a GalT biochemical activity assay. The GalT enzyme assay was conducted in a reaction volume of 50 μL consisting of detergent-permeabilized MM (100 µg total protein as determined by the Pierce BCA protein assay, using BSA as standard), 5 μm fluorescent acceptor [β-Gal-NBD; NBD = 7-nitro-2,1,3-benzoxadiazole; McGill and Williams (2009)] kindly provided by Professor Spencer Williams (Department of Chemistry, Bio21 Institute, The University of Melbourne), and 0.4 mm UDP-sugar (Sigma-Aldrich) as donor. The reaction mixture was incubated for 1 h at 25°C and subsequently terminated by adding 1 µL glacial acetic acid and 1 µL 0.5 m EGTA. The reaction mixture was then centrifuged for 2 min at maximum speed in a bench microfuge and the supernatant filtered using a 0.22 µm filter unit (Millipore). The filtrate was transferred to an HPLC vial for Reversed Phase-HPLC (RP-HPLC) analysis.
RP-HPLC Analysis of Biochemical Assay Products
Five microliters of enzyme assay product filtrate was analyzed by RP-HPLC using a Zorbax Eclipse XDB-C18 RP column (4.6 × 150 mm, 5 µm; Agilent Technologies) with the following step gradient profile: 0 to 1.0 min 25% B; 1.1 to 12.1 min 30% B; 12.2 to 17.2 min 80% B; 17.3 to 25.0 min 100% B where A: 50 mm ammonium acetate, pH 4.3 and B: acetonitrile, and the flow rate was 0.5 mL/min. NBD-tagged enzyme assay products were detected by a model no. FP-920 Intelligent Fluorescence Detector (Ex 470 nm, Em 540 nm; JASCO).
Enzymatic Characterization of Biochemical Assay Products
The enzyme assay product (10 µL; see above) was mixed with 17.5 μL 50 mm sodium acetate, pH 3.8 and digested with 2.5 μL of recombinant SGalase1, an exo-β-(1,3)-galactanase from Streptomyces sp. prepared according to the method of Ling et al., (2012). In parallel, 10 μL of GalT enzyme assay product was mixed with 17.5 μL extraction buffer (see above) and digested with 1.5 U (approximately 2.5 μL) of E. coli β-galactosidase (Grade VIII, G5635; Sigma-Aldrich). Both reactions were incubated at 37°C for 1 h and then terminated by heating at 95°C for 10 min, spun down, and the sample reanalyzed by RP-HPLC as described above.
Mass Spectrometric Characterization of Biochemical Assay Products
For mass spectrometry analysis of the enzyme assay reaction products, the entire filtered assay mixture was injected into the RP-HPLC column. Peaks were manually collected and analyzed by direct infusion ESI-MS on a QSTAR Elite hybrid quadrupole time-of-flight mass spectrometer (Sciex). The MS was operated in positive ion mode with an ion source voltage of 2200 V using 10 µm uncoated SilicaTips (New Objective). Analyst QS 2.0 software (Sciex) was used to collect data in data-dependent acquisition mode for the three most intense ions fulfilling the following criteria: m/z between 450 and 2000; ion intensity 40 counts; and charge state between +1 and +2. After MS/MS analysis, these ions were dynamically excluded for 18 s, using a mass tolerance of 50 mDa. MS scans were accumulated for 0.5 s, and MS/MS scans collected in automatic accumulation mode for a maximum of 2 s Mass and charge state-dependent rolling collision energy was used and the MS instrument was calibrated daily with [Glu]-fibrinopeptide B (Sigma-Aldrich).
To obtain a full scan of all the possible m/z peaks for the enzyme products, a nano LC-MS was performed. The sample was loaded onto a 300 µm × 5 mm Zorbax 300SB-C18 (Agilent Technologies) RP precolumn attached to a Prominence nano-LC system (Shimadzu). The precolumn was washed with 0.1% (v/v) formic acid in 5% (v/v) acetonitrile for 15 min before being placed in-line with a 75 µm i.d. × 150 mm Zorbax 300SB-C18 RP column (Agilent Technologies). Products were eluted using a gradient of 5% to 80% (v/v) acetonitrile in 0.1% (v/v) formic acid over 1 h, at a flow rate of 0.2 µL/min. Products with a charge state of either +1 or +2, between 300 D and 1500 D were analyzed via ESI-MS on a QSTAR Elite hybrid quadrupole time-of-flight mass spectrometer (Applied Biosystems/MDS Sciex) operated in the positive ion mode, as described above.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The average exine thickness measured from CLSM and TEM images.
Supplemental Figure S2. Enzyme activity of KNS4 using different UDP-sugar donors.
Supplemental Figure S3. Transmission electron micrographs of stage 12M wild-type and kns4-2 microspores labeled with either anti-AGP (JIM8, JIM13) or anti-callose gold-labeled antibodies.
Supplementary Material
Acknowledgments
We thank Professors Hitoshi Sakakibara and Kenzo Nakamura for helpful discussions and Rie Ishiguro and Fumika Ezuka for excellent technical assistance.
Footnotes
This work was supported by KAKENHI grant no. 20000304 (to T.S.); grants no. 21024004, 23370018, and 15H01229 (to S.I.) from the Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science (MEXT/JSPS), Japan; grant no. CE1101007 (to W.Z., E.R.L., A.v.d.M., M.S.D., and A.B.) from the Australia Research Council (ARC) to the ARC Centre of Excellence in Plant Cell Walls; and the University of Melbourne under the Melbourne International Research Scholarship (MRS) and Melbourne International Fee Remission Scholarship (MIFRS) (to J.O.N.).
Articles can be viewed without a subscription.
References
- Aarts MGM, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Aouali N, Laporte P, Clément C (2001) Pectin secretion and distribution in the anther during pollen development in Lilium. Planta 213: 71–79 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Kawanabe T, Hatakeyama K, Sato S, Kato T, Tabata S, Toriyama K (2008) Ultrastructural characterization of exine development of the transient defective exine 1 mutant suggests the existence of a factor involved in constructing reticulate exine architecture from sporopollenin aggregates. Plant Cell Physiol 49: 58–67 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Baldwin TC, McCann MC, Roberts K (1993) A novel hydroxyproline-deficient arabinogalactan protein secreted by suspension-cultured cells of Daucus carota (purification and partial characterization). Plant Physiol 103: 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendse GWM, Kepczynski J, Karssen CM, Koornneef M (1986) The role of endogenous gibberellins during fruit and seed development: studies on gibberellin-deficient genotypes of Arabidopsis thaliana. Physiol Plant 67: 315–319 [Google Scholar]
- Basu D, Liang Y, Liu X, Himmeldirk K, Faik A, Kieliszewski M, Held M, Showalter AM (2013) Functional identification of a hydroxyproline-o-galactosyltransferase specific for arabinogalactan protein biosynthesis in Arabidopsis. J Biol Chem 288: 10132–10143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D, Wang W, Ma S, DeBrosse T, Poirier E, Emch K, Soukup E, Tian L, Showalter AM (2015) Two hydroxyproline galactosyltransferases, GALT5 and GALT2, function in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. PLoS One 10: e0125624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley DJ, Wood EA, Larkins AP, Galfre G, Butcher GW, Brewin NJ (1988) Isolation of monoclonal antibodies reacting with peribacteriod membranes and other components of pea root nodules containing Rhizobium leguminosarum. Planta 173: 149–160 [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Chang HS, Zhang C, Chang YH, Zhu J, Xu XF, Shi ZH, Zhang XL, Xu L, Huang H, Zhang S, Yang ZN (2012) No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol 158: 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Jin JY, Choi S, Hwang JU, Kim YY, Suh MC, Lee Y (2011) An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J 65: 181–193 [DOI] [PubMed] [Google Scholar]
- Coimbra S, Almeida J, Junqueira V, Costa ML, Pereira LG (2007) Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. J Exp Bot 58: 4027–4035 [DOI] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG (2009) Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J Exp Bot 60: 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Mendes MA, Pereira AM, Pinto J, Pereira LG (2010) Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex Plant Reprod 23: 199–205 [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ (2009) A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Geanconteri A, Shrestha J, Carlson A, Kooyers N, Coerper D, Urbanczyk-Wochniak E, Bench BJ, Sumner LW, Swanson R, Preuss D (2011) A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol 157: 947–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Nishikawa S, Preuss D, Urbanczyk-Wochniak E, Sumner LW, Hammond A, Carlson AL, Swanson RJ (2009a) LAP3, a novel plant protein required for pollen development, is essential for proper exine formation. Sex Plant Reprod 22: 167–177 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D (2009b) CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DP (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Dou XY, Yang KZ, Zhang Y, Wang W, Liu XL, Chen LQ, Zhang XQ, Ye D (2011) WBC27, an adenosine tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J Integr Plant Biol 53: 74–88 [DOI] [PubMed] [Google Scholar]
- Egelund J, Ellis M, Doblin M, Qu Y, Bacic A (2011) Genes and enzymes of the GT31 family: towards unravelling the function(s) of the plant glycosyl transferase family members. Ann Plant Res 41: 213–234 [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB, Stone BA, Clarke AE (1983) Arabinogalactan-proteins: structure, biosynthesis, and function. Annu Rev Plant Physiol 34: 47–70 [Google Scholar]
- Geshi N, Johansen JN, Dilokpimol A, Rolland A, Belcram K, Verger S, Kotake T, Tsumuraya Y, Kaneko S, Tryfona T, Dupree P, Scheller HV, et al. (2013) A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J 76: 128–137 [DOI] [PubMed] [Google Scholar]
- Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza CdeA, Heitz T, Douglas CJ, Legrand M (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JS, Tsai AY, Xue H, Voiniciuc C, Šola K, Seifert GJ, Mansfield SD, Haughn GW (2014) SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol 165: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1968) Pollen wall development. The succession of events in the growth of intricately patterned pollen walls is described and discussed. Science 161: 230–237 [DOI] [PubMed] [Google Scholar]
- Hoedemaekers K, Derksen J, Hoogstrate SW, Wolters-Arts M, Oh SA, Twell D, Mariani C, Rieu I (2015) BURSTING POLLEN is required to organize the pollen germination plaque and pollen tube tip in Arabidopsis thaliana. New Phytol 206: 255–267 [DOI] [PubMed] [Google Scholar]
- Hu J, Wang Z, Zhang L, Sun MX (2014) The Arabidopsis Exine Formation Defect (EFD) gene is required for primexine patterning and is critical for pollen fertility. New Phytol 203: 140–154 [DOI] [PubMed] [Google Scholar]
- Immerzeel P, Eppink MM, de Vries SC, Schols HA, Voragen AGJ (2006) Carrot arabinogalactan proteins are interlinked with pectins. Physiol Plant 128: 18–28 [Google Scholar]
- Ishiguro S, Nishimori Y, Yamada M, Saito H, Suzuki T, Nakagawa T, Miyake H, Okada K, Nakamura K (2010) The Arabidopsis FLAKY POLLEN1 gene encodes a 3-hydroxy-3-methylglutaryl-coenzyme A synthase required for development of tapetum-specific organelles and fertility of pollen grains. Plant Cell Physiol 51: 896–911 [DOI] [PubMed] [Google Scholar]
- Jauh YG, Lord EM (1996) Localization of pectins and arabinogalactan-proteins in lily (Lilium longiflorum L.) pollen tube and style, and their possible roles in pollination. Planta 199: 251–261 [Google Scholar]
- Jia QS, Zhu J, Xu XF, Lou Y, Zhang ZL, Zhang ZP, Yang ZN (2015) Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for Nexine formation. Mol Plant 8: 251–260 [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang Z, Cao J (2013) Pollen wall development: the associated enzymes and metabolic pathways. Plant Biol (Stuttg) 15: 249–263 [DOI] [PubMed] [Google Scholar]
- Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza CdeA, Geoffroy P, Heintz D, Krahn D, Kaiser M, Kombrink E, Heitz T, et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K (1990) Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Ito T, Sugimoto E, Shinozaki K (2011) Arabidopsis mutant of AtABCG26, an ABC transporter gene, is defective in pollen maturation. J Plant Physiol 168: 2001–2005 [DOI] [PubMed] [Google Scholar]
- Lampugnani ER, Ho YY, Moller IE, Koh PL, Golz JF, Bacic A, Newbigin E (2016) A glycosyltransferase from Nicotiana alata pollen mediates synthesis of a linear (1,5)-α-L-arabinan when expressed in Arabidopsis. Plant Physiol 170: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Kilinc A, Smyth DR (2012) PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J 71: 724–735 [DOI] [PubMed] [Google Scholar]
- Li WL, Liu Y, Douglas CJ (2017) Role of glycosyl transferases in pollen wall primexine formation and exine patterning. Plant Physiol 173: 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-Q, Faleri C, Geitmann A, Zhang H-Q, Cresti M (1995) Immunogold localization of arabinogalactan proteins, unesterified and esterified pectins in pollen grains and pollen tubes of Nicotiana tabacum L. Protoplasma 189: 26–36 [Google Scholar]
- Ling NXY, Lee J, Ellis M, Liao M-L, Mau S-L, Guest D, Janssen PH, Kováč P, Bacic A, Pettolino FA (2012) An exo-β-(1→3)-D-galactanase from Streptomyces sp. provides insights into type II arabinogalactan structure. Carbohydr Res 352: 70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A, Münster A, Wisniewska E (2004) Temporal and spatial distribution of pectin epitopes in differentiating anthers and microspores of fertile and sterile sugar beet. Plant Cell Physiol 45: 560–572 [DOI] [PubMed] [Google Scholar]
- Majewska-Sawka A, Rodriguez-Garcia MI (2006) Immunodetection of pectin and arabinogalactan protein epitopes during pollen exine formation of Beta vulgaris L. Protoplasma 228: 41–47 [DOI] [PubMed] [Google Scholar]
- McGill NW, Williams SJ (2009) 2,6-Disubstituted benzoates as neighboring groups for enhanced diastereoselectivity in β-galactosylation reactions: synthesis of β-1,3-linked oligogalactosides related to arabinogalactan proteins. J Org Chem 74: 9388–9398 [DOI] [PubMed] [Google Scholar]
- Micheli F. (2001) Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci 6: 414–419 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Mollet JC, Kim S, Jauh GY, Lord EM (2002) Arabinogalactan proteins, pollen tube growth, and the reversible effects of Yariv phenylglycoside. Protoplasma 219: 89–98 [DOI] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Nakamura S, Tanaka K, Kawamukai M, Suzuki T, Nakamura K, Kimura T, Ishiguro S (2008) Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci Biotechnol Biochem 72: 624–629 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Newbigin E, Bacic A, Read S (2009) Callose and its role in pollen and embryo-sac development in flowering plants. In Bacic A, Fincher G B, Stone B A, eds, Chemistry, Biochemistry and Biology of (1,3)-β-Glucans and Related Polysaccharides. Academic Press, Elsevier, New York, pp 465–498 [Google Scholar]
- Nguema-Ona E, Coimbra S, Vicré-Gibouin M, Mollet J-C, Driouich A (2012) Arabinogalactan proteins in root and pollen-tube cells: distribution and functional aspects. Ann Bot (Lond) 110: 383–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D (2005) Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsubayashi Y (2015) Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J 81: 736–746 [DOI] [PubMed] [Google Scholar]
- Owen HA, Makaroff CA (1995) Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Owen HA, Makaroff CA (1997) A comparative ultrastructural analysis of exine pattern development in wild-type Arabidopsis and a mutant defective in pattern formation. Protoplasma 198: 53–65 [Google Scholar]
- Peiffer JA, Kaushik S, Sakai H, Arteaga-Vazquez M, Sanchez-Leon N, Ghazal H, Vielle-Calzada J-P, Meyers BC (2008) A spatial dissection of the Arabidopsis floral transcriptome by MPSS. BMC Plant Biol 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ (2007) New insights into pectin methylesterase structure and function. Trends Plant Sci 12: 267–277 [DOI] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K (1991) Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AM, Pereira LG, Coimbra S (2015) Arabinogalactan proteins: rising attention from plant biologists. Plant Reprod 28: 1–15 [DOI] [PubMed] [Google Scholar]
- Qu Y, Egelund J, Gilson PR, Houghton F, Gleeson PA, Schultz CJ, Bacic A (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol Biol 68: 43–59 [DOI] [PubMed] [Google Scholar]
- Quilichini TD, Douglas CJ, Samuels AL (2014) New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana. Ann Bot (Lond) 114: 1189–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ (2010) ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Grienenberger E, Douglas CJ (2015) The biosynthesis, composition and assembly of the outer pollen wall: a tough case to crack. Phytochemistry 113: 170–182 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR (1998) Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J 15: 79–88 [DOI] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Showalter AM, Basu D (2016) Extensin and arabinogalactan-protein biosynthesis: glycosyltransferases, research challenges, and biosensors. Front Plant Sci 7: 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zeng W, Wu A, Picard K, Lampugnani ER, Cheetamun R, Beahan C, Cassin A, Lonsdale A, Doblin MS, Bacic A (2015) Asparagus spears as a model to study heteroxylan biosynthesis during secondary wall development. PLoS One 10: e0123878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glössl J, Léonard R, Stadlmann J, Altmann F, Steinkellner H, Mach L (2007) A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MX, Huang XY, Yang J, Guan YF, Yang ZN (2013) Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod 26: 83–91 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Masaoka K, Nishi M, Nakamura K, Ishiguro S (2008) Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol 49: 1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, Baldwin D, Pham C, et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Showalter AM, Egelund J, Hernandez-Sanchez A, Doblin MS, Bacic A (2012) Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front Plant Sci 3: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LK, Chu H, Yip WK, Yeung EC, Lo C (2009) An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol 181: 576–587 [DOI] [PubMed] [Google Scholar]
- van Aelst AC, van Went JL (1992) Ultrastructural immuno-localization of pectins and glycoproteins in Arabidopsis thaliana pollen grains. Protoplasma 168: 14–19 [Google Scholar]
- Voiniciuc C, Günl M, Schmidt MH, Usadel B (2015) Highly branched xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol 169: 2481–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S, Fleming A, Wellman CH, Beerling DJ (2011) Evolutionary development of the plant and spore wall. AoB Plants 2011: plr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Wilson SM, Bacic A (2012) Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat Protoc 7: 1716–1727 [DOI] [PubMed] [Google Scholar]
- Wilson SM, Ho YY, Lampugnani ER, van de Meene AML, Bain MP, Bacic A, Doblin MS (2015) Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-D-glucan in grasses. Plant Cell 27: 754–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Greiner S (2012) Growth control by cell wall pectins. Protoplasma 249(Suppl 2): S169–S175 [DOI] [PubMed] [Google Scholar]
- Wu Y, Yan J, Zhang R, Qu X, Ren S, Chen N, Huang S (2010) Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell 22: 3745–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhu J, Cui Y-L, Yang Z-N (2015) Ultrastructure analysis reveals sporopollenin deposition and nexine formation at early stage of pollen wall development in Arabidopsis. Science Bulletin 62: 273–276 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.