Arabidopsis CRK28 is synthesized upon immune perception, associates with the FLS2 receptor complex, and silencing multiple related CRKs enhances disease susceptibility.
Abstract
Membrane-localized proteins perceive and respond to biotic and abiotic stresses. We performed quantitative proteomics on plasma membrane-enriched samples from Arabidopsis (Arabidopsis thaliana) treated with bacterial flagellin. We identified multiple receptor-like protein kinases changing in abundance, including cysteine (Cys)-rich receptor-like kinases (CRKs) that are up-regulated upon the perception of flagellin. CRKs possess extracellular Cys-rich domains and constitute a gene family consisting of 46 members in Arabidopsis. The single transfer DNA insertion lines CRK28 and CRK29, two CRKs induced in response to flagellin perception, did not exhibit robust alterations in immune responses. In contrast, silencing of multiple bacterial flagellin-induced CRKs resulted in enhanced susceptibility to pathogenic Pseudomonas syringae, indicating functional redundancy in this large gene family. Enhanced expression of CRK28 in Arabidopsis increased disease resistance to P. syringae. Expression of CRK28 in Nicotiana benthamiana induced cell death, which required intact extracellular Cys residues and a conserved kinase active site. CRK28-mediated cell death required the common receptor-like protein kinase coreceptor BAK1. CRK28 associated with BAK1 as well as the activated FLAGELLIN-SENSING2 (FLS2) immune receptor complex. CRK28 self-associated as well as associated with the closely related CRK29. These data support a model where Arabidopsis CRKs are synthesized upon pathogen perception, associate with the FLS2 complex, and coordinately act to enhance plant immune responses.
Plants are in close contact with a variety of microorganisms, including pathogens. Plants possess physical barriers to pathogen ingress, including a waxy cuticle and cell wall. There also are large numbers of germline-encoded extracellular and intracellular innate immune receptors that actively perceive pathogens and induce cellular reprogramming for defense (Zipfel, 2014; Chiang and Coaker, 2015). Surface-localized receptors with extracellular domains can perceive conserved pathogen-associated molecular patterns (PAMPs)/microbe-associated molecular patterns (MAMPs), such as bacterial flagellin or fungal chitin, and induce pattern-triggered immunity (PTI; Zipfel, 2014). Common PTI responses include Ca2+ influx, an extracellular reactive oxygen species (ROS) burst, activation of mitogen-activated protein kinases (MAPKs), transcriptional reprogramming, and callose deposition (Boller and Felix, 2009). While Ca2+ influx, the oxidative burst, and MAPK activation occur within minutes of PAMP perception, callose deposition is a later response occurring several hours post perception (Boller and Felix, 2009). Thus, adapted pathogens must overcome PTI in order to grow and to cause disease. A common mechanism for PTI suppression is the secretion of pathogen proteins, called effectors, into the apoplast or inside host cells (Dou and Zhou, 2012; Sánchez-Vallet et al., 2013). Plants also can recognize specific intracellular pathogen effectors using resistance proteins and induce effector-triggered immunity (ETI; Chiang and Coaker, 2015). Although there is significant overlap in PTI- and ETI-based responses, ETI typically induces a stronger response and culminates in programmed cell death at the site of infection (Chiang and Coaker, 2015).
PTI receptors are receptor-like kinases (RLKs) or receptor-like proteins (RLPs) possessing extracellular ligand-binding domains (Zipfel, 2014). The Arabidopsis (Arabidopsis thaliana) FLAGELLIN-SENSING2 (FLS2) is a well-studied RLK and perceives a 22-amino acid epitope of bacterial flagellin called flg22 (Chinchilla et al., 2006). Upon flg22 perception, FLS2 rapidly heterodimerizes with its RLK coreceptor BAK1 and its closest homolog SOMATIC EMBRYOGENESIS-RELATED KINASE4 (SERK4), leading to the activation of PTI responses (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). BAK1, also known as SERK3, is a member of the SERK family. BAK1 also heterodimerizes with other immune-related RLKs and RLPs in a ligand-dependent manner (Liebrand et al., 2014). Some immune receptors require additional SERK family members for defense signaling (Roux et al., 2011). Thus, members of the SERK family play crucial roles in plant immunity. RLKs can function not only in defense but also in development and abiotic stress responses (Huffaker and Ryan, 2007; Osakabe et al., 2013; Zipfel, 2014).
Cys-rich receptor-like kinases (CRKs) are a large subfamily of Arabidopsis RLKs. Like most RLKs, CRKs possess an extracellular domain, a single-pass transmembrane domain, and an intracellular Ser/Thr protein kinase domain (Chen, 2001). In their extracellular domain, most CRKs possess two copies of DOMAIN OF UNKNOWN FUNCTION26 (DUF26). The DUF26 domain possesses a conserved C-X8-C-X2-C motif (Ohtake et al., 2000; Chen, 2001). The Cys residues in each DUF26 domain are predicted to form two Cys bridges, which are hypothesized to be targeted for apoplastic redox modification (Bourdais et al., 2015). In the Arabidopsis genome, there are 46 predicted CRKs, eight plasmodesmata-located proteins (PDLPs), and more than 50 Cys-rich repeat secretory proteins that possess DUF26 domains (Chen, 2001; Amari et al., 2010; Bourdais et al., 2015). PDLPs are RLPs with similarity to CRK extracellular domains (Chen, 2001). PDLPs are involved in the regulation of plant cell-to-cell communication, viral cell-to-cell movement, and plant immunity (Amari et al., 2010; Lee et al., 2011; Caillaud et al., 2014).
Previous studies have demonstrated that Arabidopsis CRKs are transcriptionally induced in response to abiotic stresses, including salicylic acid, ozone, UV light, salt, and drought treatments (Chen et al., 2003, 2004; Wrzaczek et al., 2010; Bourdais et al., 2015; Yeh et al., 2015). A subset of CRKs is strongly induced in response to pathogens and PAMP treatment (Wrzaczek et al., 2010; Bourdais et al., 2015). The coordinated transcriptional induction of CRKs during immune responses suggests that they cooperate to enhance defense signaling. Recently, large-scale phenotyping of T-DNA knockout lines for 41 Arabidopsis CRKs was performed (Bourdais et al., 2015). Individual T-DNA knockout lines in most CRKs lack strong immune-related phenotypes (Bourdais et al., 2015). In contrast, overexpression of some CRKs robustly enhances plant immune responses. Overexpression of CRK4, CRK5, CRK6, CRK13, and CRK36 in Arabidopsis resulted in enhanced resistance to the bacterial pathogen Pseudomonas syringae pv tomato (Pst; Chen et al., 2003; Acharya et al., 2007; Yeh et al., 2015). Furthermore, overexpression of CRK4, CRK6, and CRK36 enhanced the activation of early and late PTI responses (Yeh et al., 2015). Overexpression of CRK4, CRK5, CRK13, CRK19, and CRK20 in Arabidopsis also induces cell death (Chen et al., 2003; Acharya et al., 2007).
A mechanistic understanding of CRK function for plant immune responses, including the identification of CRK-interacting proteins, the role of the extracellular Cys residues, and the importance of kinase activity, remains largely unknown. Quantitative proteomics analyses were performed upon the perception of flg22, identifying multiple CRKs whose protein abundance is increased significantly. Here, we report the functional characterization of CRK28 in plant immunity. Overexpression of CRK28 in Arabidopsis enhanced resistance to Pst. Transient expression of CRK28 in Nicotiana benthamiana induced cell death. CRK28’s extracellular Cys residues and the conserved Lys in its kinase domain are required for cell death induction. Furthermore, we show that CRK28 associates with BAK1 and is present in the FLS2-BAK1 immune complex. Lastly, using immunoprecipitation (IP), we demonstrate that CRK28 self-associates and also associates with the closely related CRK29.
RESULTS
Quantitative Proteomics Identifies Multiple RLKs Induced upon Flagellin Perception
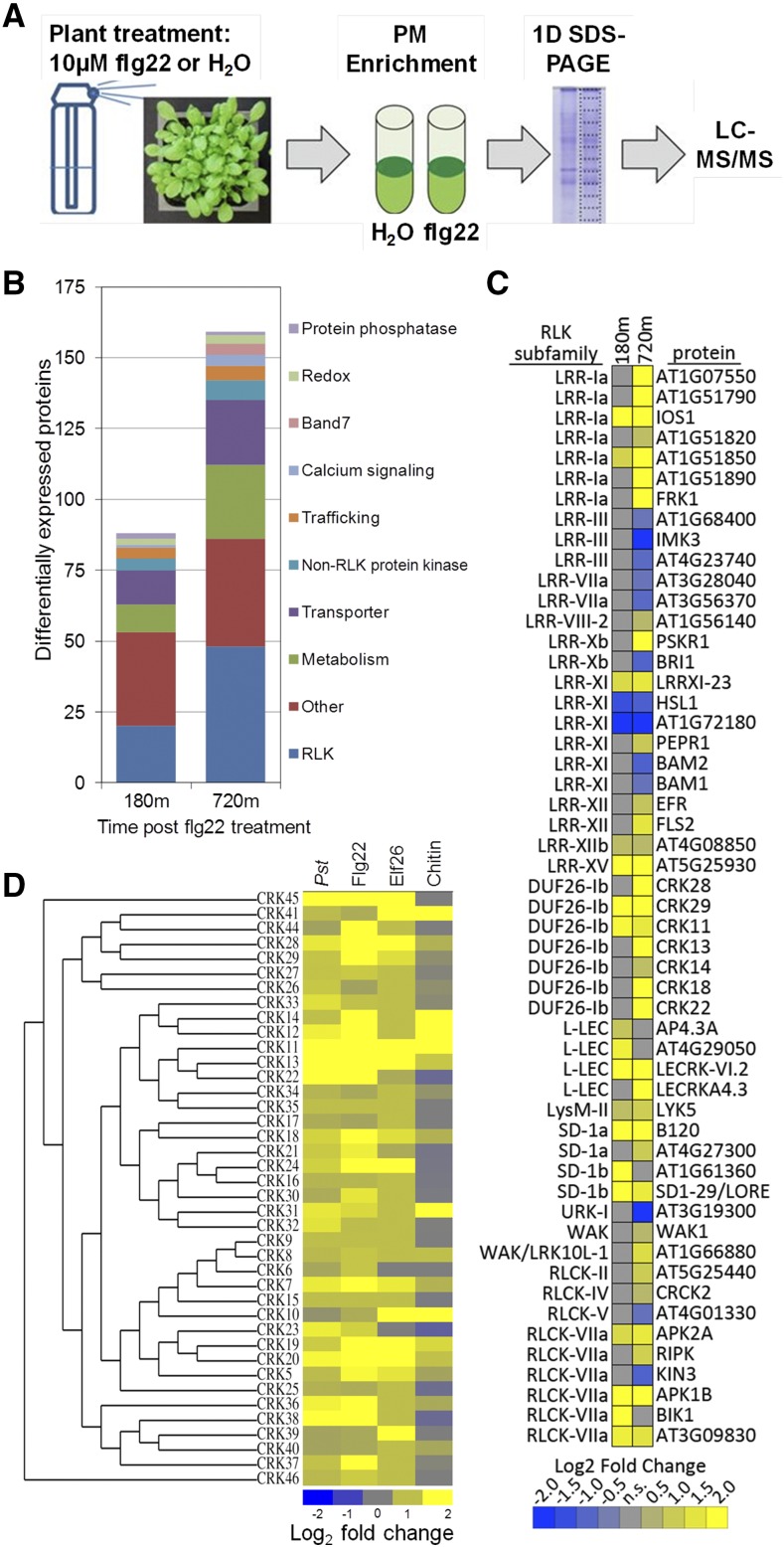
In order to identify proteins whose expression levels change dynamically upon the perception of flg22, 4-week-old Arabidopsis plants were sprayed with 10 µm flg22 peptide or water. Rosette tissue was harvested at 180 and 720 min for plasma membrane enrichment followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses (Fig. 1A). A total of 3,827 proteins were identified across the entire experiment (Supplemental Table S1). For a given time point, proteins identified in all three biological replicates of one condition (water or flg22) were used for differential abundance analyses. The edgeR negative bionomial model was used to detect proteins exhibiting differential abundance based on spectral count data (Robinson et al., 2010). Differential abundance was detected for 200 distinct proteins: 88 at 180 min and 159 at 720 min post flg22 treatment. Many plasma membrane proteins or protein classes known to be involved in immune signaling changed in abundance, including protein phosphatases, calcium-signaling proteins, protein kinases, and RLKs (Fig. 1B; Supplemental Table S1).
Figure 1.
Plasma membrane proteomics identified multiple differentially expressed RLKs upon the perception of bacterial flagellin. A, General overview of the experimental setup of the plasma membrane proteome analyses. Four-week-old Arabidopsis Columbia-0 (Col-0) plants grown in soil were sprayed with 10 μm flg22 peptide or water. Rosette leaf tissue was harvested for plasma membrane (PM) protein enrichment followed by mass spectrometry analyses at 180 and 720 min. B, Number of differentially expressed proteins in particular protein families at 180 and 720 min post flg22 treatment. C, Heat map showing differentially expressed RLKs post flg22 treatment. Blue indicates down-regulated proteins and yellow indicates up-regulated proteins, and the values are log2 fold change compared with the respective water-treated sample. At left are RLK subfamilies, and at right are gene names or gene identifiers. D, Unrooted phylogenetic tree of all CRKs located on chromosome 4 based on their full-length amino acid sequences. The heat map shows corresponding CRK expression patterns after pathogen and PAMP treatment. Transcript expression data were obtained from the Bio-Analytic Resource (BAR) database and published microarray data. Plants were treated with Pst for 120 min, 1 µm flg22 for 240 min, 10 µm elf26 for 60 min, and 1 µm chitin for 30 min. The heat map was generated using log2 fold change compared with the respective untreated control plants.
We reproducibly identified 391 RLKs by tandem mass spectrometry (MS/MS), representing 63% of all RLKs encoded in the Arabidopsis genome. Furthermore, 53 of the identified RLKs were differentially expressed over the course of the experiment (Fig. 1C). Differential protein abundance of Leucine-rich repeat (LRR)-RLKs was detected primarily in the LRR-Ia and LRR-XI subfamilies (Fig. 1C). Several LRR-XI subfamily members with known or putative roles in development were down-regulated (e.g. BARELY ANY MERISTEM1/2, HAESA-LIKE1, and the Arabidopsis C-TERMINALLY ENCODED PEPTIDE RECEPTOR2; DeYoung et al., 2006; Stenvik et al., 2008; Tabata et al., 2014), consistent with the antagonism between plant growth and defense. The immune receptors ELONGATION FACTOR TU RECEPTOR (EFR) and FLS2 also were up-regulated at 720 min (Fig. 1C). EFR is the Arabidopsis receptor for the bacterial PAMP elongation factor Tu (Zipfel et al., 2006). Activated FLS2 is ubiquitinated and endocytosed after the perception of flg22 (Lu et al., 2011; Beck et al., 2012). However, after FLS2 degradation occurring approximately 1 h post flg22 perception, de novo protein synthesis results in an increase in FLS2 protein levels between 3 and 24 h post perception, which is consistent with our results (Smith et al., 2014). The major chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (Cao et al., 2014), the lipopolysaccharide receptor LECTIN S-DOMAIN1 RECEPTOR-LIKE KINASE (Ranf et al., 2015), the damage-associated molecular pattern receptor WALL-ASSOCIATED RECEPTOR KINASE1 (Brutus et al., 2010), the danger peptide receptor PEP RECEPTOR1 (PEPR1; Huffaker and Ryan, 2007), and the positive regulator of PTI signaling IMPAIRED OOMYCETE SUSCEPTIBILITY1 (IOS1; Yeh et al., 2016) were up-regulated upon flg22 perception (Fig. 1C). These results indicate that additional RLKs are induced to enhance immune responses or pathogen detection after initial flg22 perception.
We also detected an increase in abundance of seven CRKs upon flg22 perception (CRK11, CRK13, CRK14, CRK18, CRK22, CRK28, and CRK29; Fig. 1C). In the Arabidopsis genome, 40 of the 46 predicted CRKs are located on chromosome 4, with the largest cluster comprising 20 CRKs in a tandem array (Supplemental Fig. S1A). To investigate the transcript expression of CRKs during plant immune responses, we mined available Arabidopsis expression data generated upon treatment with the bacterial pathogen Pst, the bacterial PAMPs flg22 and elf26, as well as fungal chitin. Elf26 is an immunogenic 26-amino acid epitope of the bacterial PAMP elongation factor Tu (Kunze et al., 2004). Treatment with Pst, flg22, or elf26 strongly induces the transcription of most CRKs on chromosome 4 (Fig. 1D). Taken together, these data demonstrate that CRKs are highly expressed upon PAMP perception, suggesting that multiple CRKs are collectively involved in plant immune responses.
Genetic Analyses of CRK-Mediated Defense
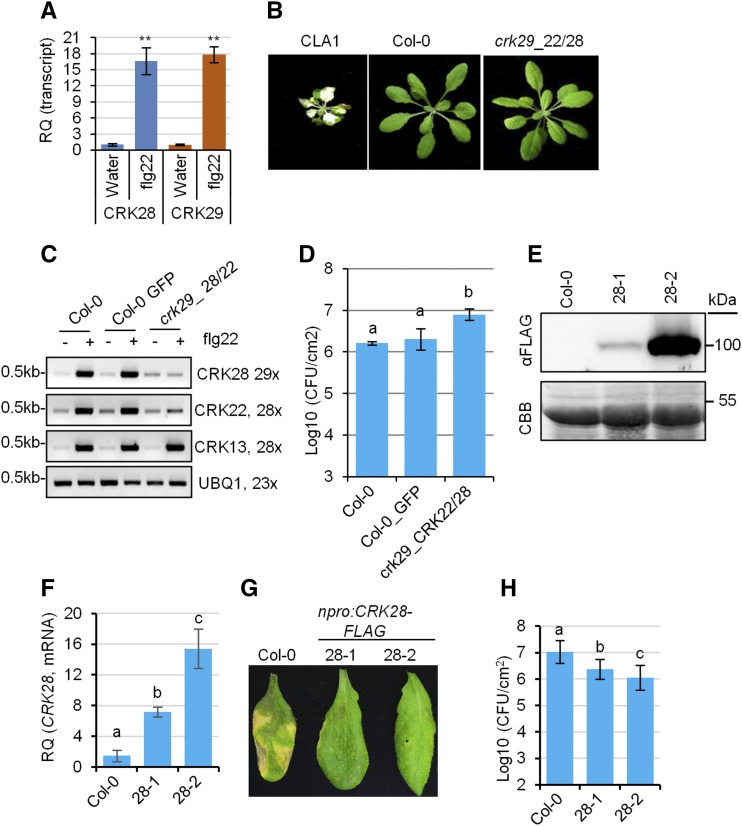
We investigated the expression levels of CRK28 (At4g21400) and CRK29 (At4g21410), two closely related CRKs that are induced upon PTI perception in our proteomics analyses. In order to investigate their transcriptional regulation, 4-week-old Col-0 plants were sprayed with either flg22 or water and leaf samples were subjected to quantitative real-time PCR (qPCR) analyses. Both CRK28 and CRK29 were strongly induced 3 h post flg22 elicitation (approximately 18-fold) compared with water treatment (Fig. 2A). We obtained T-DNA insertion lines for CRK28 (crk28-1) and CRK29 (crk29-1; Supplemental Fig. S2A). While crk29-1 is a true knockout, crk28-1 is a knockdown (Supplemental Fig. S2, B and C). crk28-1 and crk29-1 were challenged by syringe infiltration with the virulent Pst or spray inoculated with Pst ∆hrcC, which is unable to deliver effectors, but we did not observe a significant difference in bacterial titer between the mutants and Col-0 (Supplemental Fig. S2, D and E). Flg22 pretreatment protects Arabidopsis plants from subsequent infection by virulent Pst (Zipfel et al., 2004). We did not observe a significant difference in flg22-mediated protection in crk28-1 and crk29-1 compared with Col-0 (Supplemental Fig. S2F).
Figure 2.
Genetic investigation of CRK-mediated responses to P. syringae. A, qPCR analyses of CRK28 and CRK29 transcripts after treatment with the bacterial elicitor flg22. RQ, Relative quantification. Col-0 was treated with flg22 or water, and rosette leaves were collected 3 h later for RNA extraction and cDNA synthesis. Expression was normalized to Arabidopsis ELONGATION FACTOR1α (EF-1α; AT5G60390). This experiment was repeated three times with similar results. Error bars indicate sd (n = 4). Statistical differences were detected by Fisher’s lsd, α = 0.01. Asterisks indicate statistically significant differences. B, Phenotypes of 4-week-old VIGS-silenced Arabidopsis lines. Cloroplastos alterados 1 (CLA1)-silenced plants were used as a control for silencing efficiency. C, RT-PCR analyses of VIGS efficiency. Expression data were normalized to Arabidopsis UBIQUITIN EXTENSION PROTEIN1 (AT3G52590). Leaves of 4-week-old VIGS-silenced plants were treated with or without flg22 for 1 h before RNA extraction and RT-PCR analyses. D, Quantification of bacterial titers in crk29-1 plants silenced for CRK22 and CRK28 3 d post syringe infiltration with Pst DC3000. Col-0 plants infiltrated with VIGS constructs carrying GFP were included as a control. Error bars indicate sd; n = 3. Statistical differences were detected by Fisher’s lsd, α = 0.05, and letters indicate significant differences. This experiment was repeated twice with similar results. E, Anti-FLAG immunoblot showing protein level expression of CRK28 in two lines expressing CRK28 genomic DNA (npro:CRK28-FLAG 28-1 and 28-2) in the crk28-1 T-DNA mutant. The bottom gel shows the membrane stained with Coomassie Brilliant Blue (CBB) to indicate protein loading. F, qPCR analyses of CRK28 expression in Col-0, 28-1, and 28-2. Data were analyzed as described in A. G, Increased expression of CRK28 enhances Arabidopsis resistance to Pst strain DC3000. Four-week-old Col-0 and npro:CRK28-FLAG expression lines were syringe infiltrated with Pst DC3000, and leaves were photographed 4 d post infiltration. H, Bacterial titers in Col-0 and npro:CRK28-FLAG expression lines 4 d post syringe infiltration with Pst DC3000 at the indicated levels (colony-forming units [CFU] mL−1). Error bars indicate se; n = 6. Statistical differences were detected by Fisher’s lsd, α = 0.01, and letters indicate significant differences. This experiment was repeated four times with similar results.
The seven induced CRKs in the proteomics analyses phylogenetically cluster into group II and IV clades (Supplemental Fig. S1B; Bourdais et al., 2015). CRK22 is present in group IV, while CRK28 and CRK29 are present in group II (Supplemental Fig. S1B). To investigate the cumulative role of pathogen-responsive CRKs, we silenced CRK22 and CRK28 in the crk29-1 knockout using virus-induced gene silencing (VIGS; Fig. 2, B and C). Plants silenced with Cloroplastos alterados 1 exhibited an albino phenotype and were used as a visual marker for VIGS efficiency (Fig. 2B). Wild-type Col-0 plants silenced with VIGS constructs containing GFP or crk29-1 plants silenced for CRK22 and CRK28 did not exhibit alterations in plant growth (Fig. 2B). Reverse transcription (RT)-PCR demonstrated silencing of CRK22 and CRK28 in crk29-1 lines but not of CRK13 (Fig. 2C). As has been reported previously, silencing Arabidopsis Col-0 plants with VIGS constructs containing GFP does not affect Pst growth (Fig. 2D; de Oliveira et al., 2016). Silencing of CRK22 and CRK28 in the crk29-1 knockout resulted in significantly higher bacterial titers compared with Col-0 control plants 3 d post inoculation (Fig. 2D). These data indicate that pathogen-induced CRKs are important in inhibiting bacterial growth.
Increased Expression of CRK28 Enhances Resistance to P. syringae
Transgenic lines were generated expressing CRK28 genomic DNA under the control of its native promoter with a C-terminal fusion to the 3×FLAG epitope (npro:CRK28-FLAG) in the crk28-1 background. Two independent T3 homozygous lines (28-1 and 28-2) were obtained. Anti-FLAG immunoblotting revealed that CRK28 expression in npro:CRK28-FLAG line 28-1 is significantly lower than in line 28-2 (Fig. 2E). Quantitative PCR analyses revealed that both npro:CRK28-FLAG lines exhibited significantly higher CRK28 transcript accumulation than Col-0 (Fig. 2F). No obvious developmental phenotypes were observed in npro:CRK28-FLAG lines. The npro:CRK28-FLAG lines were infiltrated with Pst strain DC3000, and disease progression was monitored over time. Col-0 exhibited strong disease symptoms, whereas npro:CRK28-FLAG lines did not display any obvious Pst symptoms 4 d post infiltration (Fig. 2G). Quantification of the bacterial titers correlated with visual disease symptoms. Col-0 harbored significantly higher bacterial titers than npro:CRK28-FLAG lines 28-1 and 28-2 (Fig. 2H). Bacterial titers negatively correlated with the level of CRK28 expression: npro:CRK28-FLAG line 28-2 exhibited approximately 10-fold lower bacterial growth, and line 28-1 exhibited approximately 5-fold lower bacterial growth, than Col-0 (Fig. 2H). Taken together, these results demonstrate that CRK28 is rapidly induced upon PAMP perception and the level of expression correlates with the heighted resistance to Pst.
Extracellular Cys Residues Are Required for CRK28-Induced Cell Death in N. benthamiana
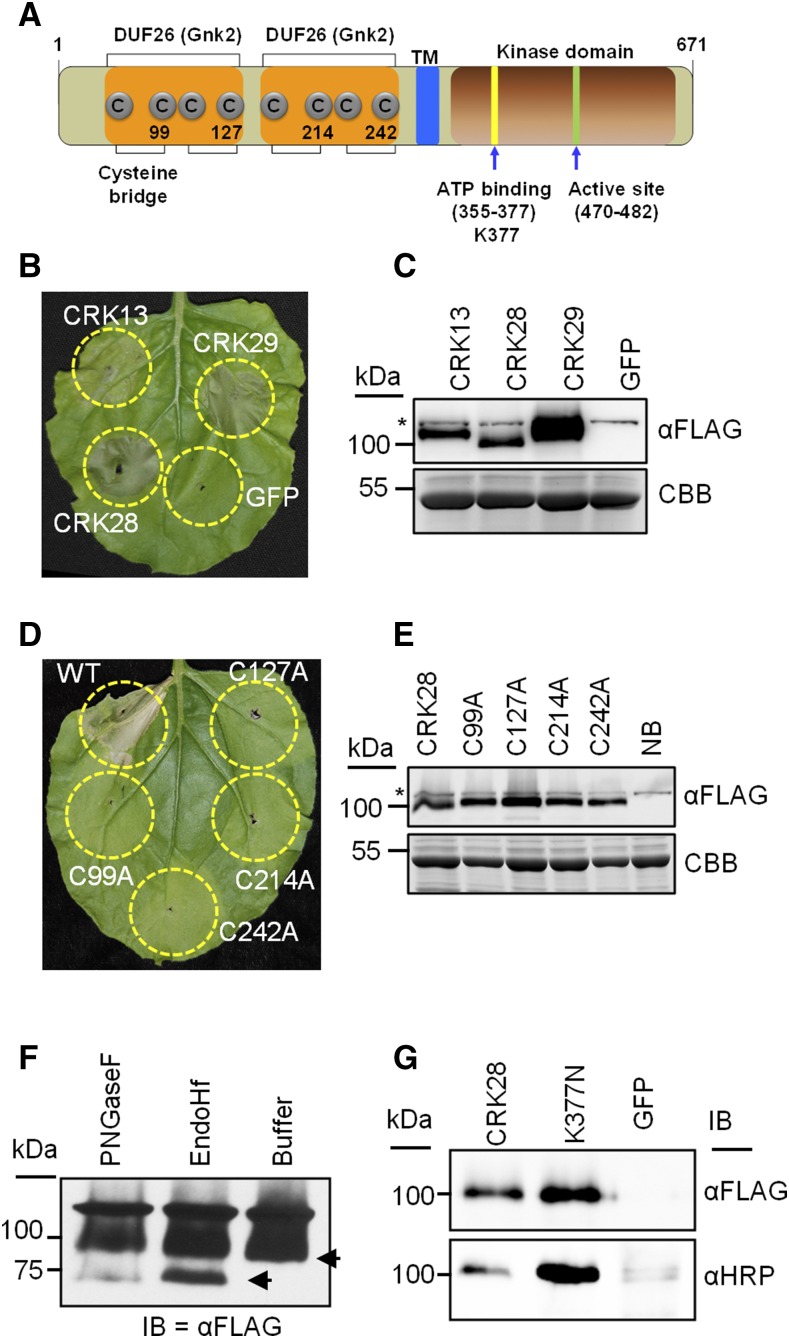
Previous studies have demonstrated that overexpression of CRK4, CRK5, CRK13, CRK19, and CRK20 in Arabidopsis induces cell death (Chen et al., 2003, 2004; Acharya et al., 2007). In order to identify a strong CRK-related phenotype for functional analyses, we investigated the ability of CRK13, CRK28, and CRK29 to elicit cell death upon transient expression in N. benthamiana. CRK13 was included as a control because overexpression in Arabidopsis has been shown previously to induce cell death (Acharya et al., 2007). When transiently expressed in N. benthamiana, 35S:CRK13-FLAG, 35S:CRK28-FLAG, and 35S:CRK29-FLAG elicited cell death 24 h post inoculation (hpi; Fig. 3B). Leaves infiltrated with the negative control 35S:GFP did not exhibit cell death (Fig. 3B). Expression of 35S:CRK28-FLAG and 35S:CRK29-FLAG induced stronger and faster cell death than expression of 35S:CRK13-FLAG (Fig. 3B). Using anti-FLAG immunoblotting, all three constructs were expressed in N. benthamiana (Fig. 3C). CRK28 and CRK29 are closely related, strongly induced in response to PAMP treatment, and elicit similar phenotypes when expressed in N. benthamiana (Figs. 1–3). Therefore, we focused on investigating CRK28 in subsequent experiments. When amplifying cDNA corresponding to the CRK28 transcript, we found that it does not match the gene model present in The Arabidopsis Information Resource (TAIR; version 10) database and contains seven exons (Supplemental Figs. S2A and S3). However, the PCR-amplified CRK28 transcript did match with the gene model (At4G21400.2) present in the Arabidopsis Information Portal (version 11) database (Cheng et al., 2016).
Figure 3.
CRK28’s extracellular Cys residues are required for cell death induction in N. benthamiana. A, Diagram showing the general domain architecture of Arabidopsis CRK28. The N terminus includes a secretion signal with two predicted extracellular DUF26 domains (orange) and an intracellular kinase domain (brown). Predicted extracellular Cys residues and subdomains important for ATP binding and activity in other kinases are highlighted. TM, Transmembrane. Numbers correspond to amino acid residues. B, Cell death induced by transient expression of CRK13, CRK28, and CRK29 in N. benthamiana. A. tumefaciens containing 35S:CRK13-FLAG, 35S:CRK28-FLAG, 35S:CRK29-FLAG, and 35S:GFP was infiltrated into N. benthamiana leaves and photographed 48 hpi. C, Anti-FLAG immunoblot showing the expression of CRK13, CRK28, CRK29, and GFP 20 hpi in N. benthamiana. The bottom gel represents Coomassie Brilliant Blue (CBB) stain to show protein loading. The asterisk shows an anti-FLAG cross-reacting band. D, Transient expression of 35S:CRK28-FLAG and four Cys mutants (C99A, C127A, C214A, and C242A) in N. benthamiana. Cell death was observed for wild-type CRK28, whereas no cell death was observed for Cys mutants. The leaf was photographed 48 hpi. E, Anti-FLAG immunoblot demonstrating the expression of CRK28 and the Cys mutants 20 hpi. NB, N. benthamiana (uninfiltrated). F, 35S:CRK28-FLAG was transiently expressed in N. benthamiana, and the detergent-soluble supernatant was used for PNGase F and Endo Hf enzymatic deglycosylation assays. Although the predicted molecular mass of CRK28 is 74.46 kD, by SDS-PAGE it is detected at 100 kD. G, Anti-HRP immunoblot (IB) demonstrating that CRK28 carries the complex N-linked glycan β-(1,2)-Xyl or α-(1,3)-Fuc. 35S:CRK28-FLAG and 35S:CRK28K377N-FLAG were expressed transiently in N. benthamiana and subjected to anti-FLAG IP. Mature glycosylation (in the Golgi) was detected using anti-HRP antibody. Anti-FLAG immunoblot analysis was performed to show the amount of IP loaded.
In both Arabidopsis and N. benthamiana, CRK28 was detected at a molecular mass of approximately 100 kD by western blotting (Figs. 2E and 3C). However, CRK28’s molecular mass is predicted to be 74.46 kD. Similarly, CRK13 and CRK29 also ran higher than their predicted molecular masses of 75.29 and 75.72 kD, respectively (Fig. 3C). Other transmembrane proteins, such as FLS2 and EFR, also run higher than their predicted molecular masses due to complex N-linked glycosylation (Coppinger et al., 2004; Häweker et al., 2010). Incubation of the CRK28 protein extract from N. benthamiana with both N-linked glycosylases PNGase F and Endo Hf resulted in the detection of some of CRK28 at its predicted molecular mass (Fig. 3F). A significant amount of CRK28 was resistant to glycosylase treatment, likely due to either incomplete digestion or the presence of more complex N-linked glycans. N-Glycosylation is further modified in the Golgi apparatus by the addition of β-(1,2)-Xyl or α-(1,3)-Fuc (Henquet et al., 2008). These complex N-linked glycans often are resistant to a single enzymatic digestion. To investigate if CRK28 carries complex N-linked glycans, we transiently expressed 35S:CRK28-FLAG and 35S:GFP in N. benthamiana and performed anti-FLAG IP. Anti-horseradish peroxidase (HRP) antibody has been used previously to detect complex N-linked glycans (Henquet et al., 2008). Anti-HRP immunoblotting showed that, in addition to endoplasmic reticulum N-glycosylation, CRK28 is further glycosylated in the Golgi (Fig. 3G). Together, these results show that CRK28 is a complex glycosylated transmembrane protein.
The majority of the Arabidopsis CRKs possess two copies of DUF26 in their extracellular region. The four Cys residues in each DUF26 domain are predicted to form two Cys bridges (Fig. 3A). Disulfide bonds are ROS sensitive and are important for protein folding, structure, and stability (Wedemeyer et al., 2000; Feige and Hendershot, 2011; Waszczak et al., 2015). Thus, we investigated the role of extracellular Cys residues predicted to be involved in disulfide bond formation for CRK28-mediated cell death induction in N. benthamiana. Cys bridges were disrupted by mutating one Cys residue from each pair into Ala. Wild-type CRK28 (35S:CRK28-FLAG) and the Cys mutants (35S:CRK28C99A-FLAG, 35S:CRK28C127A-FLAG, 35S:CRK28C214A-FLAG, and 35S:CRK28C242A-FLAG) were transiently expressed in N. benthamiana using Agrobacterium tumefaciens. As expected, N. benthamiana leaves infiltrated with 35S:CRK28-FLAG exhibited cell death starting at 24 hpi (Fig. 3D). However, none of the Cys mutants induced cell death (Fig. 3D), suggesting that the extracellular Cys residues are required for cell death induction. Mutating the Cys residues did not affect protein stability, as all the Cys mutants were expressed to the same level as the wild type by anti-FLAG immunoblotting (Fig. 3E).
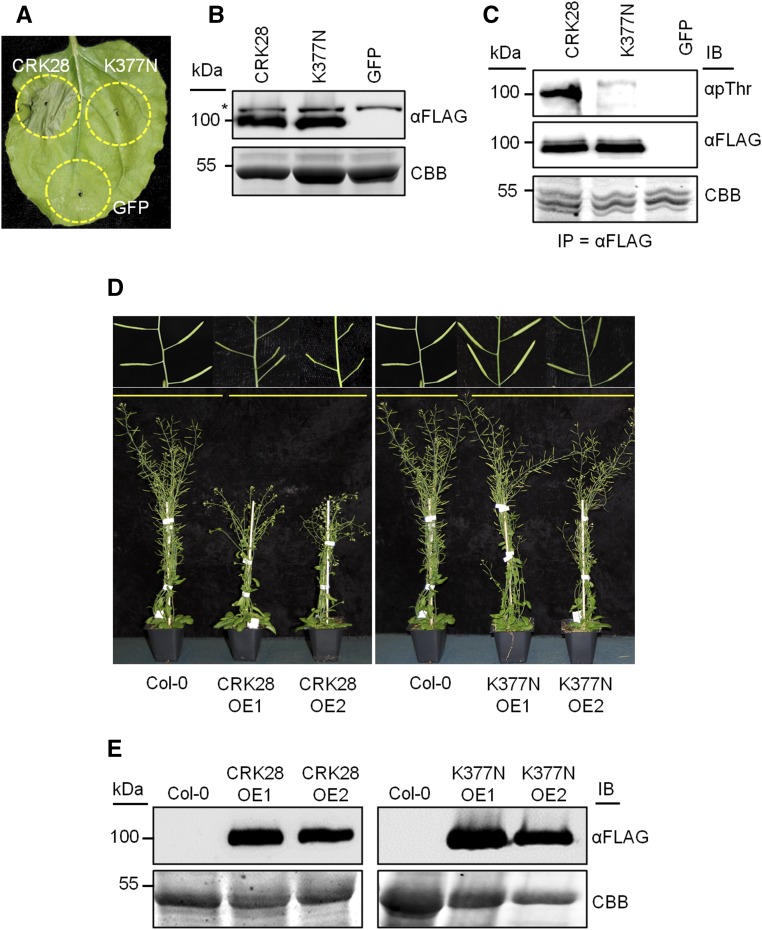
CRK28’s Kinase Active Site Is Required for Cell Death Induction in N. benthamiana and Arabidopsis Developmental Phenotypes
CRKs possess a Ser/Thr kinase domain with all the predicted consensus features (Fig. 3A; Hanks et al., 1988). However, it is still unknown whether kinase activity is required for CRK function or if CRKs are phosphorylated in planta. We tested the importance of CRK28’s active site for inducing cell death in N. benthamiana. The highly conserved Lys (K377) residue in CRK28’s ATP-binding motif was mutated to Asn (N; 35S:CRK28K377N-FLAG). Mutating this conserved Lys residue in Ser/Thr protein kinases has been shown to block activity (Schulze et al., 2010; Lin et al., 2015). None of the leaves infiltrated with 35S:CRK28K377N-FLAG exhibited cell death, but all leaves infiltrated with wild-type 35S:CRK28-FLAG exhibited cell death (Fig. 4A). Anti-FLAG immunoblotting demonstrated equal expression of CRK28-FLAG and CRK28K377N-FLAG (Fig. 4B). Overall, these results suggest that the invariant Lys residue is required for CRK28-induced cell death.
Figure 4.
The CRK28 intracellular ATP-binding Lys (Lys-377) residue is required for CRK28-mediated function, and CRK28 is phosphorylated in planta. A, Transient expression of 35S:CRK28-FLAG, 35S:CRK28K377N-FLAG, and 35S:GFP in N. benthamiana. No cell death was observed for the K377N mutant. The leaf was photographed 48 hpi. B, Anti-FLAG immunoblot demonstrating the expression of CRK28 and CRK28K377N 20 hpi. The asterisk shows an anti-FLAG cross-reacting band. The bottom gel shows the membrane stained with Coomassie Brilliant Blue (CBB) to indicate protein loading. C, In planta phosphorylation of CRK28 and CRK28K377N. 35S:CRK28-FLAG, 35S:CRK28K377N-FLAG, and 35S:GFP were expressed transiently in N. benthamiana and subjected to anti-FLAG IP. Twenty microliters of the resuspended beads was used for a kinase reaction containing 100 µm ATP. After incubating the reaction at 30°C for 1 h, the proteins were separated by SDS-PAGE and immunoblotted (IB) with anti-phospho-Thr antibody and anti-FLAG antibody. D, Overexpression of CRK28 in Arabidopsis affects flowering and silique development. 35S:CRK28-FLAG and 35S:CRK28K377N-FLAG constructs were transformed into Col-0. Transgenic T1 seeds were selected on kanamycin plates, and transformants were transferred into soil along with Col-0 grown on Murashige and Skoog plates without kanamycin. Representative images show transgenic T1 plants expressing 35S:CRK28-FLAG and 35S:CRK28K377N-FLAG 7 weeks post transplanting onto soil. The same Col-0 plant was used in both images, as the T1 plants were transplanted at the same time and grown in the same conditions. E, Anti-FLAG immunoblots showing the protein level expression of CRK28-FLAG and 35S:CRK28K377N-FLAG. The bottom gel shows Coomassie Brilliant Blue stain indicating total protein loading.
The kinase domains of CRK28 and CRK28K377N were expressed in Escherichia coli as MBP fusion proteins. However, no kinase activity was detected with recombinant proteins (Supplemental Fig. S4). However, kinase activity was observed for the positive control RIPK (Liu et al., 2011). To investigate if CRK28 is phosphorylated in planta, CRK28-FLAG, CRK28K377N-FLAG, and GFP were expressed in N. benthamiana and immunoprecipitated using anti-FLAG agarose beads. Subsequently, the IP was used for anti-phospho immunoblotting. We were able to detect phosphorylation of wild-type CRK28, but not CRK28K377N, using anti-phospho immunoblotting after IP (Fig. 4C). Thus, CRK28 is phosphorylated in planta.
The importance of CRK28’s conserved Lys residue also was investigated in Arabidopsis. 35S:CRK28-FLAG and 35S:CRK28K377N-FLAG constructs were transformed into Col-0. Few transformants were obtained from Col-0 transformed with 35S:CRK28-FLAG compared with 35S:CRK28K377N-FLAG. Furthermore, the surviving 35S:CRK28-FLAG T1 transgenics exhibited shorter stature, increased inflorescence branch numbers, shorter siliques, and a lack of seed set (Fig. 4D). Plants transformed with 35S:CRK28K377N-FLAG exhibited wild-type growth (Fig. 4D). Protein expression levels were similar between CRK28-FLAG and CRK28K377N-FLAG transformants (Fig. 4E), further confirming that an intact kinase domain is required for the phenotypes observed.
Increased CRK28 Expression Enhances the Extracellular ROS Burst, and CRK28 Associates with FLS2 and BAK1
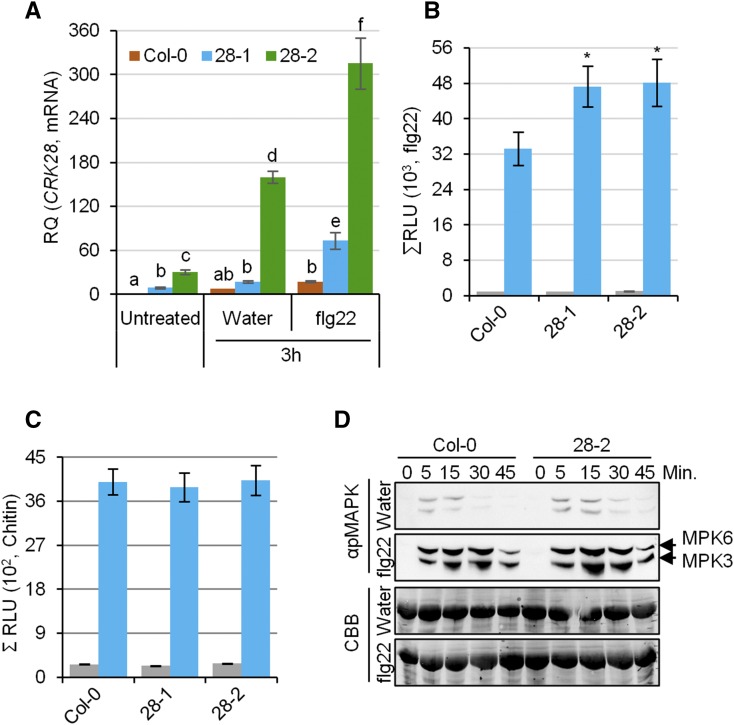
There is a positive correlation between CRK28’s basal expression level and resistance to Pst (Fig. 2). In order to investigate the expression of CRK28 in Col-0 and npro:CRK28-FLAG transgenic lines, we treated with 10 µm flg22 or water and leaf samples were collected at 0 and 3 h post treatment for qPCR. CRK28 transcripts were strongly induced in response to flg22 treatment and were significantly higher in npro:CRK28-FLAG lines 28-1 and 28-2 compared with Col-0 (Fig. 5A). Next, we examined the induction of the extracellular ROS burst and MAPK activation, two common PTI markers (Boller and Felix, 2009). To measure the ROS burst, 2-week-old Col-0 and npro:CRK28-FLAG lines 28-1 and 28-2 were treated with flg22 and the fungal elicitor chitin. While flg22 treatment significantly increased ROS accumulation in npro:CRK28-FLAG lines 28-1 and 28-2 compared with Col-0, treatment with chitin did not alter ROS accumulation (Fig. 5, B and C). Consistent with this observation, transcript analyses of CRK expression upon elicitor treatment revealed that many CRKs, including CRK28, are induced upon flg22 treatment (Fig. 1D). In contrast, fewer CRKs are induced in response to chitin treatment (Fig. 1D). We also examined if MAPK activation is altered in npro:CRK28-FLAG expressing line 28-2. Increased expression of CRK28 did not significantly alter MAPK activation compared with Col-0 after flg22 treatment (Fig. 5D). These results suggest that heightened expression of CRK28 results in an increase of a subset of PTI responses after the perception of specific microbial features.
Figure 5.
CRK28 expression is induced in response to flg22, and heighted expression of CRK28 enhances the flg22-triggered ROS burst. A, qPCR analyses of CRK28 expression in response to treatment with flg22. Four-week-old Col-0 and npro:CRK28-FLAG lines 28-1 and 28-2 were treated with flg22 or water, and leaf samples were collected for RNA extraction at the indicated time points. Expression was normalized to Arabidopsis EF-1α. RQ, Relative quantification. Error bars indicate se; n = 3. Statistical differences were detected by Fisher’s lsd, α = 0.05, and different letters indicate statistical significance. The experiment was repeated twice with similar results. B and C, ROS burst in Col-0, 28-1, and 28-2 after treatment with flg22 and chitin, respectively. Gray and blue bars indicate water and flg22/chitin treatment, respectively. ROS was quantified using a luminol-based assay. The graphs depict total relative light units (ΣRLU) detected over a 30-min period. Error bars indicate se; n = 16. Statistical differences after flg22 or chitin treatment were detected by Fisher’s lsd, α = 0.05. These experiments were repeated three times with similar results. D, MAPK3/6 activation assay in Col-0 and 28-2 after flg22 or water treatment. Three-week-old plants were sprayed with flg22 or water, and tissues were collected at the indicated time points and processed. Phosphorylated MAPK3 and MAPK6 were detected with anti-p44/42 MAPK antibody. The bottom two gels show membranes stained with Coomassie Brilliant Blue (CBB) to indicate protein loading. These experiments were repeated three times with similar results.
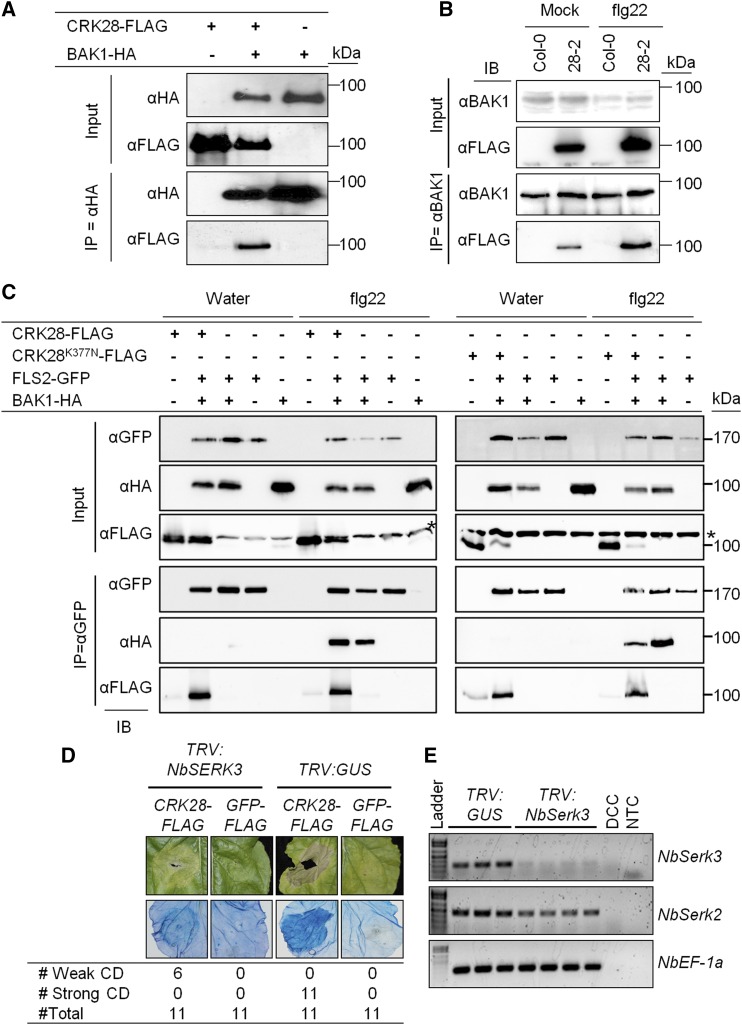
Bacterial flagellin is perceived by the RLK FLS2, whose signaling is dependent on the coreceptor BAK1 (Chinchilla et al., 2007; Boller and Felix, 2009; Zipfel, 2014). Chitin perception in Arabidopsis does not require BAK1 (Shan et al., 2008; Gimenez-Ibanez et al., 2009). The heightened expression of CRK28 positively affects the flg22-induced ROS burst but not the chitin-induced ROS burst (Fig. 5C). Therefore, we sought to examine the role of BAK1 for CRK28-mediated immune responses. Using coimmunoprecipitation (Co-IP), we examined if BAK1 can associate with CRK28 in N. benthamiana. We transiently expressed 35S:CRK28-FLAG and 35S:BAK1-HA (for hemagglutinin) in N. benthamiana, and IP was performed with anti-HA agarose beads. Immunoblotting with the anti-FLAG antibody demonstrated that CRK28 can Co-IP with BAK1 in N. benthamiana (Fig. 6A). To further confirm this association, 4-week-old Col-0 and npro:CRK28-FLAG line 28-2 were sprayed with 10 µm flg22 or water to induce CRK28 expression, and leaves were sampled after 3 h. IP with the anti-BAK1 antibody showed that CRK28-FLAG can Co-IP with BAK1 (Fig. 6B).
Figure 6.
BAK1 associates with CRK28 in Arabidopsis and N. benthamiana, and silencing NbSerk3 reduces CRK28-mediated cell death. A, CRK28 Co-IPs with BAK1 in N. benthamiana. 35S:CRK28-FLAG and 35S:AtBAK1-HA were coexpressed in N. benthamiana and subjected to anti-HA IP. Proteins were then subjected to anti-HA and anti-FLAG immunoblotting. B, CRK28 Co-IPs with BAK1 in Arabidopsis. Col-0 and the npro:CRK28-FLAG expression line 28-2 were sprayed with flg22 or water and subjected to anti-BAK1 IP. Proteins were separated by SDS-PAGE and immunoblotted with anti-BAK1 and anti-FLAG antibodies. C, CRK28 and CRK28K377N associate with the FLS2/BAK1 immune complex in N. benthamiana. 35S:FLS2-GFP, 35S:BAK1-HA, 35S:CRK28-FLAG, and 35S:CRK28K377N-FLAG were coexpressed in N. benthamiana and subjected to anti-GFP IP. Proteins were then subjected to anti-GFP, anti-FLAG, and anti-HA immunoblotting (IB). The asterisk shows an anti-FLAG cross-reacting band. D, CRK28-mediated cell death in NbSerk3-silenced and control plants (top row), and Trypan Blue staining of dead tissue (bottom row). The table represents the number of leaves that showed weak or strong cell death (CD) per total number of leaves infiltrated. The leaf image with CRK28-FLAG expression after NbSerk3 silencing is representative of the weak cell death observed. E, RT-PCR showing the expression of NbSerk3 and NbSerk2 in silenced plants 3 weeks post infiltration with VIGS constructs. NbEF-1α was used as an endogenous control. DCC, DNA contamination control; NTC, no template control. All experiments were repeated at least three times with similar results.
BAK1 forms a ligand-dependent immune complex with multiple pattern recognition receptors (PRRs), including FLS2 in Arabidopsis (Chinchilla et al., 2007; Zipfel, 2014). Recently, Yeh et al. (2015) reported that CRK4, CRK6, and CRK36 associate with FLS2 in a flg22-independent manner in Arabidopsis protoplasts. Thus, we were interested to know whether CRK28 is present in the activated FLS2/BAK1 immune complex. 35S:CRK28-FLAG, 35S:FLS2-GFP, and 35S:BAK1-HA were expressed transiently in N. benthamiana. Twenty hours later, leaf tissue was collected 5 min after elicitation with 1 µm flg22 or water for IPs. As expected, the anti-HA immunoblot showed that BAK1 can Co-IP with FLS2 in a ligand-dependent manner (Fig. 6C; Chinchilla et al., 2007). Similar to Yeh et al. (2015), we observed that CRK28 can Co-IP with FLS2 in a ligand-independent manner (Fig. 6C). However, the Co-IP of CRK28, BAK1, and FLS2 was observed only in a flg22-dependent manner (Fig. 6C).
Mutating the conserved Lys residue in an ATP-binding motif (K377N) abrogates CRK28-mediated cell death in N. benthamiana (Fig. 4A). We investigated the importance of Lys-377 for association with the activated FLS2/BAK1 immune complex. 35S:CRK28K377N-FLAG, 35S:FLS2-GFP, and 35S:BAK1-HA were expressed transiently in N. benthamiana and treated with either water or flg22, and samples were collected 5 min later for IP. Similar to wild-type CRK28, CRK28K377N also can Co-IP with FLS2-GFP in a flg22-independent manner (Fig. 6C). Previous work also has demonstrated for FLS2/BAK1 that kinase activity is not required for immune complex formation (Schulze et al., 2010). These data indicate that kinase activity also is not required for CRK28’s association with the FLS2/BAK1 immune complex.
AtBAK1 is a member of the larger SERK family, and the closest N. benthamiana homolog of BAK1 is NbSerk3 (Heese et al., 2007). In N. benthamiana, NbSerk3 is required for NbFLS2-dependent responses and silencing of NbSerk3 significantly affects the flg22-triggered ROS burst (Heese et al., 2007). We used a VIGS approach to silence NbSerk3. TRV:GUS-infiltrated plants were used as a negative control. As reported previously, NbSerk3-silenced plants displayed slight stunting and aberrant leaf morphology (Heese et al., 2007). RT-PCR demonstrated that the transcript level of NbSerk3 is reduced significantly in NbSerk3-silenced plants compared with TRV:GUS-infiltrated plants (Fig. 6E). We transiently expressed 35S:CRK28-FLAG and 35S:GFP in NbSerk3- and TRV:GUS-infiltrated N. benthamiana leaves. Strong cell death was observed when 35S:CRK28-FLAG was expressed in TRV:GUS-infiltrated N. benthamiana leaves (Fig. 6D). CRK28-induced cell death was either abolished completely or reduced significantly in all of the NbSerk3-silenced leaves (Fig. 6D). We also confirmed the cell death reduction by Trypan Blue staining (Fig. 6D). As expected, the expression of 35S:GFP did not induce cell death (Fig. 6D). Anti-FLAG immunoblotting confirmed that CRK28 was expressed at similar levels in both NbSerk3- and TRV:GUS-infiltrated plants (Supplemental Fig. S5A).
To verify that the reduction of CRK28-induced cell death in NbSerk3-silenced plants is specific to CRK28, we infiltrated 35S:RPS2-FLAG, an Arabidopsis resistance protein known to trigger cell death in N. benthamiana (Day et al., 2005). 35S:RPS2-FLAG induces cell death to the same level in both NbSerk3- and TRV:GUS-infiltrated leaves (Supplemental Fig. S5B). Anti-FLAG immunoblotting confirmed that RPS2-FLAG was expressed at similar levels in both NbSerk3-silenced and TRV:GUS-infiltrated leaves (Supplemental Fig. S5C). Overall, these results indicate that NbSerk3 is required for CRK28-induced cell death in N. benthamiana. Furthermore, silencing NbSerk3 specifically affects CRK28-induced cell death but not cell death induced by RPS2.
CRK28 Self-Associates and Associates with CRK29
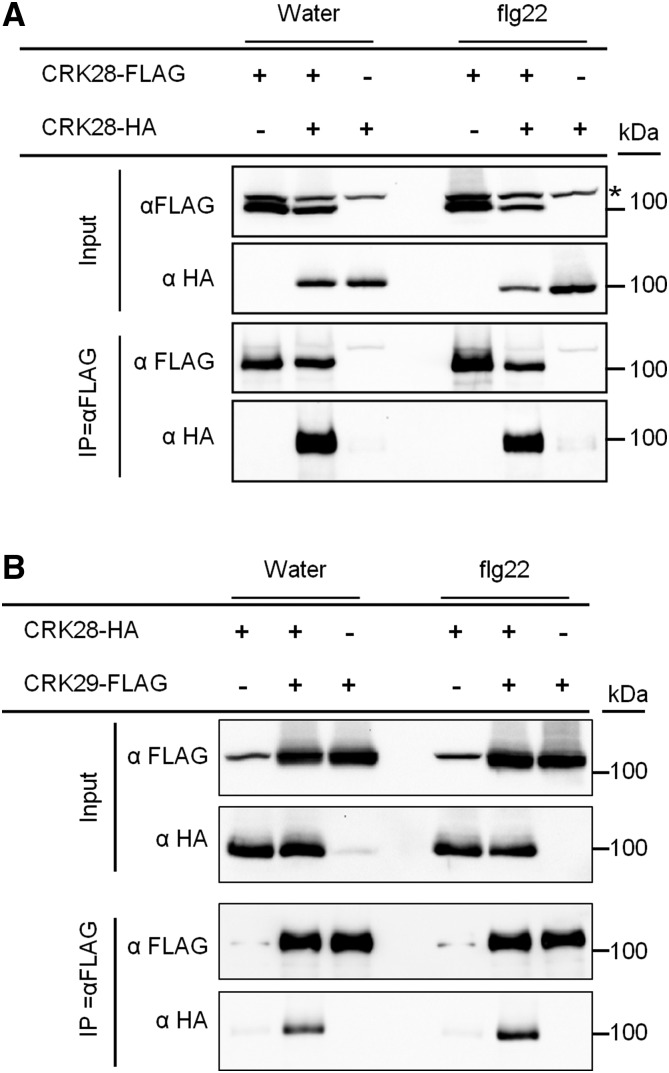
Multiple studies have demonstrated that receptors assemble into ligand-induced immune complexes to initiate defense signaling (Zipfel, 2014). Thus, we investigated the ability of CRK28 to self-associate in the presence and absence of immune perception. 35S:CRK28-FLAG and 35S:CRK28-HA were expressed transiently in N. benthamiana, 20 h later leaf tissue was collected after elicitation with 1 µm flg22 or water, and IPs were performed with FLAG antibody beads. Anti-HA immunoblotting demonstrated that CRK28-HA can Co-IP with CRK28-FLAG, indicating the self-association of CRK28 in N. benthamiana (Fig. 7A). CRK28 also self-associates after flg22 treatment (Fig. 7A).
Figure 7.
CRK28 self-associates in N. benthamiana and also associates with CRK29. A, Immunoblot showing the Co-IP of CRK28-HA with CRK28-FLAG in the presence or absence of flg22. 35S:CRK28-HA and 35S:CRK28-FLAG were coinfiltrated into N. benthamiana leaves, and 20 hpi, the leaves were infiltrated with either flg22 or water and leaf samples were collected 5 min later for IP. The asterisk shows an anti-FLAG cross-reacting band. B, Co-IP of CRK28-HA along with CRK29-FLAG in N. benthamiana. The experiments were performed as described in A. An anti-FLAG cross-reacting band is present in the first and fifth lanes. These experiments were repeated three times with similar results.
Next, we examined CRK28’s ability to associate with CRK29, a closely related and genetically linked CRK (Supplemental Figs. S1 and S2). 35S:CRK28-HA and CRK29-FLAG were transiently expressed in N. benthamiana for 20 h. At this point, leaves were collected 5 min after elicitation with flg22 or water, and IP was performed with anti-FLAG beads. Anti-HA immunoblotting revealed that CRK28-HA can Co-IP with CRK29-FLAG in both flg22- and water-treated samples, indicating that CRK28 associates with CRK29 (Fig. 7B). Taken together, these results demonstrate that CRK28 self-associates and also can associate with CRK29.
DISCUSSION
Some of the largest gene families in plants encode RLKs with similarity to characterized immune receptors (Shiu and Bleecker, 2003). Many of these proteins localize to the plasma membrane and can control pathogen perception as well as propagate signals downstream of pathogen recognition (Huffaker and Ryan, 2007; Zipfel, 2014). We identified 391 RLKs by MS/MS, revealing a large dynamic range in estimated LRR-RLK abundance (Supplemental Fig. S1). Most known PRRs are present at relatively low levels in plasma membrane fractions (Supplemental Figs. S1 and S2). PRR abundance is likely tightly controlled in order to avoid inappropriate activation. Pretreatment of plants with flg22 primes the plant immune system to elicit robust defense responses upon pathogen challenge (Boller and Felix, 2009). The up-regulation of many known PRRs (Supplemental Figs. S1 and S2) around 720 min post flg22 treatment supports a model where the primary recognition event stimulates an increase in pathogen receptors at the plasma membrane, leading to stronger activation of defense responses against subsequent or sustained pathogen attack. The increase in PRRs recognizing different microbial ligands may allow the plant to more rapidly achieve the signaling threshold required for the activation of defense. Moreover, the expression patterns of known PRRs strongly implicate uncharacterized RLKs with similar expression profiles as additional plant immune receptors or regulatory proteins.
The tandem duplication of many CRKs on chromosome 4 in Arabidopsis is reminiscent of resistance gene clusters in plant genomes and may facilitate the adaptive evolution of novel specificity, subfunctionalization, or coordinated gene expression (Bergelson et al., 2001). Large-scale phenotyping of CRK T-DNA insertion lines revealed robust phenotypes related to plant growth and stress adaptation for knockouts in CRK2 and CRK5, respectively (Bourdais et al., 2015). However, most individual CRK T-DNA insertions exhibited more subtle alterations in plant growth, stomatal responses, and abiotic/biotic responses (Bourdais et al., 2015). For example, the crk28-1 knockdown exhibited a slight increase in visual Pst disease symptoms in young plants, which typically exhibit enhanced susceptibility (Kus et al., 2002; Bourdais et al., 2015). We detected a significant range in CRK protein abundance at a resting state (Supplemental Table S1). However, the seven CRK proteins that were robustly induced by PTI perception exhibited low-level protein expression at a resting state (Supplemental Table S1). We hypothesize that the subset of CRKs that exhibited a strong increase in protein abundance after PRR perception are coordinately involved in immune signaling after initial pathogen perception. Consistent with this hypothesis, silencing CRK22, CRK28, and CRK29 resulted in enhanced susceptibility to Pst. Heightened expression of CRK28 in Arabidopsis resulted in increased resistance to Pst and an increased ROS burst upon flg22 perception. Previous studies have demonstrated that the PEPR1/2 RLKs amplify immune signaling by perceiving endogenous Pep epitopes whose corresponding PROPEP transcripts are induced rapidly upon the perception of PAMPs (Huffaker and Ryan, 2007; Krol et al., 2010; Yamaguchi et al., 2010). The IOS1 RLK also acts as an important mediator of PTI responses and associates with BAK1, EFR, and FLS2 in a ligand-independent manner (Yeh et al., 2016). Similarly, we observed a CRK28 association with BAK1 and FLS2 in a ligand-independent manner. The ios1 knockout exhibited enhanced susceptibility to Pst and defective PTI responses, while IOS overexpression lines were more resistant to Pst and exhibited primed PTI responses (Yeh et al., 2016). PEPR1 and IOS1 protein abundances are increased significantly after flg22 perception (Fig. 1C). Future research is necessary to determine if CRKs coordinately function in a similar manner to enhance defense responses.
Pathogen perception induces an extracellular ROS burst (Boller and Felix, 2009), and oxidative stress facilitates the formation of multiple intradisulfide and interdisulfide bonds between Cys residues (Feige and Hendershot, 2011; Nagahara, 2011). Mutating CRK28’s Cys residues predicted to be involved in disulfide bond formation into Ala completely abolished CRK28-mediated cell death in N. benthamiana but did not affect protein stability (Fig. 3, D and E). Earlier reports have indicated that redox-sensitive Cys residues and disulfide bonds can serve as a switch to modulate protein function (Feige and Hendershot, 2011; Nagahara, 2011). Alterations in apoplastic ROS upon pathogen perception could enhance disulfide bond formation of CRK28 and act as a redox switch to activate this RLK (Bourdais et al., 2015).
Previous studies have demonstrated that posttranslational modifications play various roles in immune receptor function. We have demonstrated that CRK28 is both glycosylated and phosphorylated in planta. Recently, CRK4 was demonstrated to be glycosylated, and STAUROSPORIN AND TEMPERATURE SENSITIVE3 N-glycosylation was required for CRK4-induced cell death and protein accumulation in Arabidopsis (de Oliveira et al., 2016). Glycosylation also has been shown to be important for the function of multiple immune receptors, including FLS2, EFR, and Cf proteins (van der Hoorn et al., 2005; Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010; Liebrand et al., 2012). Mutation of the N-linked glycosylation sites in EFR reduced the level of mature EFR protein, decreased ligand binding, and decreased ligand-elicited ROS burst (Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010).
RLK-mediated immune signaling is regulated in a phosphorylation-dependent manner. For example, FLS2-mediated flg22 perception and signaling requires transphosphorylation events that occur in the receptor complex (Macho et al., 2015). In addition, the transmembrane protein RBOHD, an NADPH oxidase responsible for the production of extracellular ROS, is activated through transphosphorylation by the receptor-like cytoplasmic kinase BIK1 and calcium-dependent protein kinases (Kadota et al., 2015). Mutation of the conserved Lys residue in the kinase domain of the Ser/Thr protein kinases is known to abolish kinase activity and thereby signaling (Schulze et al., 2010; Lin et al., 2015). In vitro kinase activities have been demonstrated for CRK6, CRK7, and CRK36 (Tanaka et al., 2012; Idänheimo et al., 2014). The strength of autophosphorylation and cofactor preference varied between different CRKs (Tanaka et al., 2012; Idänheimo et al., 2014). Although we were unable to detect in vitro kinase activity for CRK28, we demonstrated that mutating the Lys residue in the kinase domain of CRK28 abolished its ability to induce cell death in N. benthamiana (Fig. 4A). Overexpression of CRK28 in Arabidopsis leads to altered developmental phenotypes, but overexpression of CRK28K377N does not (Fig. 4D). Furthermore, CRK28 is phosphorylated in planta. It is possible that CRK28 is an active kinase but that our purified protein was not properly folded or CRK28 requires additional proteins or cofactors for activity. CRK28 also could be transphosphorylated by other plant kinases. Taken together, our results suggest that an intact kinase domain is required for CRK28-mediated signaling and function.
The BAK1 RLK has been identified as a coreceptor for both primary immune receptors, such as FLS2, as well as a signal amplifying RLKs such as PEPR1/2 (Zipfel, 2014). Here, we found that BAK1 also is required for CRK28-mediated cell death induction. Silencing the homolog of AtBAK1 in N. benthamiana, NbSerk3, significantly reduced CRK28-induced cell death (Fig. 6D). Using IP, we demonstrated that BAK1 associates with CRK28. Consistent with the requirement of BAK1 for CRK28-induced cell death, increased expression of CRK28 resulted in a higher flg22-triggered ROS burst but did not affect the chitin-triggered ROS burst (Fig. 5, B and C). While FLS2 perception of flg22 requires BAK1, the perception of chitin occurs in a BAK1-independent manner (Shan et al., 2008; Gimenez-Ibanez et al., 2009). In addition, we identified CRK28 in the FLS2/BAK1 immune complex. CRK28’s expression is very weak at a resting state. Thus, during natural infection, substantial CRK28 protein synthesis would likely first require initial pathogen perception mediated by other PRRs such as FLS2.
FLS2 as well as other RLKs were demonstrated to be recruited to detergent-resistant microdomains upon flg22 perception (Keinath et al., 2010). The mammalian Toll-like receptor TLR2 also facilitates the formation of different receptor clusters at the membrane in response to PAMP perception to enhance PRR signaling intensity (Triantafilou et al., 2006; Inoue and Shinohara, 2014). In plants, the BAK1 coreceptor could act in a similar manner to recruit multiple PRRs in close proximity to one another upon pathogen perception, including FLS2 and CRK28. Signaling components downstream of PAMP perception are quite conserved (Boller and Felix, 2009). Thus, clusters of PRRs in the membrane could enable the rapid recruitment of signaling components, facilitating more robust defense responses. Future research focusing on the role of additional suites of CRKs to enhance plant defense responses as well as elucidating the ligand(s) perceived by these RLKs could facilitate targeted engineering for increased disease resistance.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a controlled-environment chamber at 23°C, 75% relative humidity, and a 10-h/14-h light/dark photoperiod with light intensity of 100 µE m−2 s−1. All seeds were stratified for 3 to 4 d at 4°C before sowing into soil (Sunshine Mix #1). Nicotiana benthamiana plants were grown in a controlled-environment chamber at 25°C, 85% relative humidity, and a 16-h/8-h light/dark photoperiod with light intensity of 180 µE m−2 s−1.
For the plasma membrane proteome experiments, 4- to 5-week-old plants were used. Three hours after the onset of light, Col-0 plants were sprayed with 10 μm flg22 peptide (greater than 91.6% purity; GenScript) in 18.2 MΩ cm−1 water containing 0.025% Silwet L-77 surfactant using a Preval-267 compressed air sprayer. Col-0 plants sprayed with 18.2 MΩ cm−1 water containing 0.025% Silwet L-77 were used as a negative control. Plants were incubated for 180 min and 720 min before harvesting tissue for protein isolations. Three biological replicates of plants grown and harvested at different times were performed. Water- and flg22-treated samples were processed in parallel for all replications.
T-DNA insertion lines in CRK28 (crk28-1; SALK_085178) and CRK29 (crk29-1; SALK_069665) in the Col-0 background were obtained from the Arabidopsis Biological Resource Center; the lines were genotyped by PCR to identify homozygous insertion lines, and transcript expression was analyzed by RT-PCR. Primers used in all experiments are shown in Supplemental Table S2.
Plasma Membrane Enrichment and Processing
Plasma membrane enrichment was performed on 30 to 40 g of leaf tissue using three rounds of aqueous two-phase partitioning as described previously (Elmore et al., 2012) with minor modifications. Homogenization buffer was supplemented with 50 mm sodium pyrophosphate, 25 mm sodium fluoride, 1 mm sodium molybdate, 1 mm sodium orthovanadate, and 25 mm β-glycerophosphate. The final upper phase fraction containing enriched plasma membrane vesicles was incubated with 0.02% Brij-58 detergent on ice for 10 min to invert vesicles and release cytosolic proteins (Johansson et al., 1995). Samples were diluted 20 times with water and centrifuged at 90,000g for 60 min to pellet plasma membrane vesicles. Membrane pellets were frozen in liquid nitrogen and stored at −80°C. Protein samples were solubilized in 2× Laemmli buffer with 6 m urea and quantified using the RCDC Protein Assay (Bio-Rad). Samples (300 µg of protein) were fractionated by one-dimensional SDS-PAGE using an 8% to 16% Precise Protein Gradient Gel (Thermo Scientific). The entire sample lane was excised and cut into 15 pieces of equal size using a disposable grid cutter (The Gel Company). In-gel digestions were performed with trypsin (Shevchenko et al., 2006). Digested peptides were dried using a vacuum concentrator and then solubilized in 60 μL of 2% acetonitrile/0.1% trifluoroacetic acid and frozen at −80C.
LC-MS/MS
The LC-MS/MS system configuration consisted of a CTC Pal autosampler (LEAP Technologies) and Paradigm HPLC device (Michrom BioResources) coupled to a QExactive hybrid quadrupole Orbitrap mass spectrometer (Thermo Scientific) with a CaptiveSpray ionization source (Michrom BioResources). Liquid chromatography was performed by injecting 20 μL of each peptide sample onto a Zorbax 300SB-C18 trap column (5 μm, 5 × 0.3 mm; Agilent Technologies) and desalted online. Peptides were eluted from the trap and separated on a reverse-phase Michrom Magic C18AQ (200 μm × 150 mm) capillary column at a flow rate of 2 μL min−1 using a 120-min gradient (2%–35% buffer B in 85 min, 35%–80% buffer B in 25 min, and 2% buffer B in 10 min; buffer A = 0.01% formic acid in water and buffer B = 100% acetonitrile). The mass spectrometer was operated in data-dependent acquisition mode using a standard top-15 method. The 30 protein fractions for each biological replicate were analyzed on the same column in blocks of seven and eight, alternating between flg22- and water-treated samples separated with multiple column flushes.
Protein Identification
Tandem mass spectra were extracted to mzML format using Proteome Discoverer and analyzed with the X!Tandem GPM-XE Cyclone version 2013.02.01.2 spectrum modeler (Craig and Beavis, 2004) using the TAIR 10 Arabidopsis complete proteome (TAIR10_pep_20101214.fasta; 35,386 entries) with a list of common contaminants (112 entries). A reversed and concatenated database served as a decoy sequence database to determine peptide and protein false discovery rates (FDRs; Käll et al., 2008). X!Tandem was configured to allow parent ion mass error of 20 ppm and fragment mass error of 20 ppm. Data were searched using fixed modification of +57 (carbamidomethyl) Cys residues and the following variable modifications: −18 on N-terminal E (Glu → pyro-Glu), −17 on N-terminal C (ammonia loss), −17 on N-terminal Q (Gln → pyro-Glu), +1 on NQ (Asn, Gln deamidated), +16 on MW (Met, Trp oxidation), +32 on MW (Met, Trp dioxidation), +42 on K (acetyl), and +80 on STY (Ser, Thr, Tyr phospho) while allowing one missed cleavage. X!Tandem search results were imported into Scaffold 4.0.3 (Proteome Software) with all MS/MS runs corresponding to the same sample merged. Protein identifications required two unique peptides, 95% peptide probability and 80% protein probability, resulting in a 0.05% peptide decoy FDR and 0.8% protein decoy FDR. Shared spectral count distributions were performed within Scaffold similar to Zhang et al. (2010) using each protein’s cumulative unique peptide identification probability as the distribution factor. Spectral counts of protein isoforms mapping to the same genomic locus were summed. Proteins with one or more spectral count in three of three biological replicates of a treatment condition (water or flg22) were used for differential expression analysis. Raw MS/MS data were uploaded to the MassIVE repository (accession no. MSV000079639; data will be released upon publication).
Protein Quantitative Analysis
The edgeR statistical framework was used to generate a list of high-confidence proteins exhibiting differential abundance (Robinson et al., 2010). Spectral counts are modeled from a negative binomial distribution using a generalized linear model with replicate and treatment as model factors (Robinson et al., 2010). Proteins with FDR ≤ 0.05 and an estimated log2 fold change > 0.58 (50%) were classified as differentially expressed (Supplemental Table S1).
Phylogenetic Analyses
To construct the phylogenetic tree of CRKs located on chromosome 4, the full-length amino acid sequences were downloaded from TAIR 10. The protein sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), a phylogenetic tree was generated using the neighbor-joining method, and the tree was visualized using FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Molecular Cloning
To generate a native promoter CRK28 expression construct, the 745-bp upstream region of CRK28’s start codon was amplified along with the genomic DNA of CRK28 with the CRK28_promoter_F and CRK28_Rev primer pair. The resulting PCR fragment was cloned into pENTR/D-TOPO vector (Invitrogen) and moved into the binary vector pGWB1-3xFLAG using Gateway technology (Invitrogen), generating npro:CRK28-3xFLAG. The binary vector pGWB1-3xFLAG was modified from pGWB1 to contain C-terminal 3×FLAG. To modify pGWB1, the Gateway cassette (GW) with 3×FLAG tag was amplified from the Gateway-compatible binary vector pTA700-3xFLAG with a forward and reverse primer pair containing HindIII and SacI restriction sites, respectively. Then, the GW cassette from pGWB1 was removed by HindIII/SacI digestion and replaced with the PCR product containing GW-3xFLAG, generating pGWB1-3xFLAG.
To generate a CRK28 overexpression construct driven by the cauliflower mosaic virus 35S promoter, the CRK28 cDNA was amplified with the CRK28_F and CRK28_R primer pair. The PCR fragment was then cloned into pENTR/D-TOPO vector. The ATP-binding site (Lys-377) in the kinase domain of CRK28 and the extracellular Cys residues (Cys-99, Cys-127, Cys-214, and Cys-242) were mutated into Asn and Ala by site-directed mutagenesis in pENTR/D-TOPO vector (Stratagene), respectively. All the constructs were moved into the Gateway-compatible binary vector pMD1-3xFLAG to generate 35S:CRK28-3xFLAG, 35S:CRK28K377N-3xFLAG, 35S:CRK28C99A-3xFLAG, 35S:CRK28C127A-3xFLAG, 35S:CRK28C214A-3xFLAG, and 35S:CRK28C242A-3xFLAG.
Plant Transformation
The npro:CRK28-3xFLAG construct was transformed into the crk28-1 mutant. 35S:CRK28-3xFLAG and 35S:CRK28K377N-3xFLAG were transformed into Col-0. All the constructs were transformed into Arabidopsis following the floral dip protocol with Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998).
RT-PCR and qPCR
To confirm that crk28-1 and crk29-1 were true knockouts by RT-PCR, the mutant and Col-0 rosette leaves were infiltrated with 1 µm flg22, and 3 h later, the leaves were sampled in liquid nitrogen. Total RNA was extracted from rosette leaves following a Trizol-based RNA extraction protocol. One microgram of RNA, in a total reaction volume of 20 µL, was used to synthesize cDNA using Moloney murine leukemia virus reverse transcriptase (Promega). Specific primer pairs for each T-DNA insertion line or target genes of VIGS were used for RT-PCR, and the PCR was run for 25 cycles. In the case of qPCR, the CFX96 real-time PCR detection system (Bio-Rad) was used to quantify expression of the genes. The Arabidopsis ACTIN2, UBIQUITIN1 (UBQ1), and EF-1α genes were used as endogenous controls in RT-PCR and qPCR. In N. benthamiana, the NbEF-1α gene was used as an endogenous control. All the primers used in qPCR and RT-PCR in this study are presented in Supplemental Table S1.
Pathogen Assays
Four-week-old Arabidopsis plants grown as described above were used for pathogen assays. Pseudomonas syringae pv tomato strain DC3000 and Pst DC3000 ∆hrcC were used for plant inoculations. Strains were grown on plates containing 100 µg mL−1 rifampicin and 25 µg mL−1 kanamycin overnight at 28°C. Inoculum was prepared in 5 and 10 mm MgCl2 for infiltration (3 × 105 CFU mL−1) and spray inoculation (1 × 109 CFU mL−1), respectively. For flg22 protection assays, leaves were infiltrated with 1 µm flg22, and 24 h later, the same leaves were infiltrated with Pst DC3000. After inoculation, the plants were covered for 24 h to maintain high humidity, and symptoms were monitored over time. Four days post inoculation, six leaves were sampled from four plants and surface sterilized with 70% ethanol for 30 s. Bacterial titers were determined as described previously (Liu et al., 2009).
ROS Burst Assay
Leaf discs were collected using a cork borer (5 mm diameter) from 3-week-old Arabidopsis plants and floated overnight in demineralized water. The next day, the water was replaced with an assay solution containing 17 mg mL−1 luminol (Sigma), 10 mg mL−1 horseradish peroxidase (Sigma), 100 nm flg22 (GenScript), or 50 µg mL−1 chitin (Sigma). Luminescence was measured using a Tristar multimode reader (Berthold Technology). Statistical differences were calculated with Fisher’s lsd, α = 0.05. At least 24 leaf discs were used in each replication, and the experiment was repeated more than three times with similar results.
MAPK Assay
Four-week-old Col-0 and the CRK28-expressing line (28-2) were sprayed with 10 μm flg22 or water (control) with 0.025% Silwet L-77. Leaf samples were collected at 0 (before spray), 5, 15, 30, and 45 min. Samples were ground in liquid nitrogen and resuspended in 200 µL of extraction buffer (50 mm HEPES [pH 7.5], 50 mm NaCl, 10 mm EDTA, 0.2% Triton X-100, 1× protease inhibitor cocktail [Sigma], and 1× Halt phosphatase inhibitor cocktail [Thermo Scientific]). After centrifugation for 10 min at 10,000 rpm, the supernatants were transferred into new tubes and the total protein concentration was quantified using the Pierce 660-nm protein assay (Thermo Scientific) following the manufacturer’s protocol. Equal amounts of protein samples were separated by 12% SDS-PAGE and immunoblotted with anti-p44/42 MAPK antibody (Cell Signaling Technology).
VIGS in Arabidopsis
The DNA fragments of CRK28 and CRK22 (approximately 400 bp of each starting from the ATG start codon) were amplified from Col-0 cDNA. CRK28 was digested with EcoRI and KpnI and cloned into pTRV-RNA2 (pYL156), yielding pYL156-CRK28. The CRK22 fragment was digested with NcoI and KpnI and cloned into pYL156-CRK28 via the same restriction sites. The construct pYL156-CRK28/22 contains two fragments targeting CRK28 and CRK22, respectively. The binary TRV vectors, pTRV-RNA1, pYL156-GFP, pYL156-Cla1, and pYL156-CRK28/22, were electroporated into A. tumefaciens strain GV3101, and VIGS was performed as described previously on 2-week-old plants (de Oliveira et al., 2016). The pYL156-Cla1-silenced plants exhibit a leaf-bleaching phenotype, which was used as a visual marker for silencing efficiency (de Oliveira et al., 2016). One pair of fully expanded leaves on the 4-week-old plants was inoculated with Pst DC3000, and bacterial titers were determined 3 d post infiltration.
The leaves of 4-week-old VIGS plants were treated with 100 nm flg22 for 1 h (or water as a control) in order to induce CRK expression. Trizol reagents (Invitrogen) were used for RNA extraction. After DNase I treatment (37°C for 30 min), the RNAs were reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase (New England Biolabs). RT-PCR with the primers listed in Supplemental Table S2 was used to detect target transcript levels after VIGS. For CRK genes, 30 PCR cycles (98°C for 10 s, 58°C for 15 s, and 72°C for 30 s) were used. For UBQ1, 22 PCR cycles were performed.
IP
For IP in N. benthamiana, constructs cloned into binary vectors (35S:CRK28-3xFLAG, 35S:BAK1-3xHA, 35S:CBL-GFP-3xFLAG/35S:GFP, 35S:CRK29-3xFLAG, and 35S:FLS2-GFP) were transformed into A. tumefaciens strain C58C1 and infiltrated into leaves at a concentration of 2 × 108 CFU mL−1 for 35S:CRK28-3xFLAG and 35S:BAK1-3xHA and 6 × 108 CFU mL−1 for 35S:FLS2-GFP. One gram of leaf tissue was collected at 20 hpi and frozen in liquid nitrogen. In the case of induction by elicitor, 1 µm flg22 or water was infiltrated into leaves before the leaves start visually showing cell death (approximately 20 hpi), and samples were collected 5 min later.
To perform IPs in N. benthamiana tissue, the samples were ground by mortar and pestle in liquid nitrogen and homogenized in IP buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.5% Triton X-100, 6 mm 2-mercaptoethanol, and 1× complete protease inhibitor). The homogenate was precleared by centrifugation at 14,000 rpm for 20 min and further filtered using two layers of cheesecloth. Thirty microliters of anti-FLAG (Sigma) and anti-HA (Thermo Fisher Scientific), and 25 µL of anti-GFP (Chromotek), agarose beads were used for each sample. Beads were washed twice with IP buffer, added to the homogenate, and incubated at 4°C rotating end to end for 4 h. Beads were pelleted by centrifugation at 3,000 rpm for 3 min, washed three times with 1 mL of IP buffer without 2-mercaptoethanol, and resuspended in 3× Laemmli buffer. After boiling for 5 min, the protein samples were separated by SDS-PAGE and subsequently immunoblotted with the respective antibodies. The concentrations of antibodies used in this study were as follows: anti-FLAG HRP (Sigma; 1:3000), anti-GFP HRP (Miltenyi Biotec; 1:3,000), anti-HA HRP (Roche; 1:2,000), and anti-BAK1 (Agrisera; 1:5,000).
In Arabidopsis, 4-week-old plants were sprayed with 10 µm flg22 or water containing 0.025% Silwet L-77. Three hours later, 1 g of leaf tissue was collected in liquid nitrogen. Sample processing and IP were performed as described for N. benthamiana.
Self-Association Assays
In order to assay CRK28 self-association, 35S:CRK28-3xFLAG and 35S:CRK28-3xHA were coexpressed in N. benthamiana leaves. 35S:CRK28-3xFLAG and 35S:CRK28-3xHA also were expressed individually in N. benthamiana at the same time. In addition, 35S:CRK28-3xHA was coexpressed with 35S:CRK29-3xFLAG to investigate if CRK28 also heterodimerizes with CRK29. Before sampling for IP, the leaves were infiltrated with 1 µm flg22 or water, and 1 g of leaf tissue was collected 5 min after treatment. Sample processing and IP were performed as described above.
Purification of CRK28’s Kinase Domain from Escherichia coli
The kinase domain of CRK28 and its kinase dead variant (K377N; amino acids 349–623) were cloned into the E. coli expression vector pMAL-C4x (Liu et al., 2011) with an N-terminal fusion to MBP. Briefly, the kinase domain of CRK28 and its kinase dead variant were PCR amplified with the KDBamHI-F and KDPstI_R primer pair containing BamHI and PstI restriction sites, respectively. The PCR products were digested and cloned into pMAL-C4x using BamHI and PstI restriction enzymes.
For protein expression and purification, the constructs were transformed into the E. coli strain Rosetta DE3. A 500-mL culture was grown at 37°C until the cell density reached optical density at 600 nm (OD600) = 0.5. The culture was then transferred to a 16°C incubator for 1 h, and then 0.3 mm isopropylthio-β-galactoside was added to induce protein expression for 7 h. The E. coli culture was harvested by centrifugation at 12,000 rpm for 10 min. The pellet was then resuspended in column buffer (20 mm Tris-HCl, 200 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 µm leupeptin, and 10 µg mL−1 lysozyme), and the proteins were purified using amylose resin (New England Biolabs). RIPK was purified as described previously (Liu et al., 2011).
Kinase Activity Assay
To perform an in vitro kinase activity assay with recombinant proteins, 3 µg of CRK28, CRK28K377N, and RIPK (AT2G05940; positive control) proteins were mixed with 100 µm ATP and 3 µg of the kinase substrate myelin basic protein in a kinase buffer (50 mm HEPES, pH 7.5, 10 mm MgCl2, 50 mm NaCl, and 1 mm dithiothreitol) and incubated at 30°C for 1 h. The reactions were stopped by adding 3× Laemmli buffer, boiled for 5 min, and separated by 10% SDS-PAGE. The proteins were transferred onto polyvinylidene difluoride membrane (Millipore), blocked by 5% bovine serum albumin for 1 h at room temperature, and incubated overnight 4°C with anti-phospho-Thr antibody in 5% bovine serum albumin. Membranes were developed using the chemiDoc Touch imaging system (Bio-Rad). A radioactive kinase activity assay also was performed by adding 10 μCi of [32P]γ-ATP (Perkin-Elmer) and incubating at 37°C for 1 h. The reaction was stopped by adding 5× Laemmli buffer, incubated at 60°C for 5 min, run on a 12% Precise Tris-HEPES SDS-PAGE device (Thermo Scientific), and visualized by autoradiography.
To assess kinase activity and phosphorylation in planta, 35S:CRK28-3xFLAG and 35S:CRK28K377N-3xFLAG were transiently expressed in N. benthamiana, and at 20 hpi, 2 g of infiltrated leaves was collected in liquid nitrogen. The samples were ground in 3 mL of IP buffer, and IP was performed with anti-FLAG-conjugated agarose beads as described above. After the third wash with IP buffer, the beads were washed once with kinase buffer (50 mm HEPES, pH 7.5, 10 mm MgCl2, 10 mm MnCl2, 0.1 mm CaCl2, and 1 mm dithiothreitol), and finally the beads were resuspended in 50 µL of kinase buffer. Twenty microliters of the resuspended beads was used in the kinase assay as described above for the in vitro kinase assays.
VIGS of NbSerk3
All the VIGS constructs targeting NbSerk3 (pTRV2-NbSerk3), GUS (pTRV2-GUS), and PHYTOENE DESATURASE (PDS; pTRV2-PDS) including pTRV1 were obtained from Gregory B. Martin (Velásquez et al., 2009). Bacterial growth cultures and preparation for infiltration were performed as described previously (Liu et al., 2004) with slight modifications. Briefly, overnight A. tumefaciens cultures (strain GV3101) at OD600 = 2 carrying pTRV2-NbSerk3, pTRV2-GUS, and pTRV2-PDS were mixed with GV3101 carrying pTRV1 (OD600 = 2) in a 1:1 ratio and infiltrated into cotyledons of 10-d-old N. benthamiana seedlings. Three weeks later, A. tumefaciens cultures carrying 35S:CRK28-3xFLAG (OD600 = 0.3), 35S:RPS2-3xFLAG (OD600 = 0.3), and 35S:CBL-GFP-3xFLAG (OD600 = 0.3) were infiltrated into NbSerk3- and TRV:GUS-infiltrated N. benthamiana plants, and cell death was monitored over time.
Enzymatic Deglycosylation Assay
To enzymatically deglycosylate CRK28, 35S:CRK28-3xFLAG was expressed transiently in N. benthamiana. At 25 hpi, 0.5 g of infiltrated leaves was collected in liquid nitrogen. Sample processing and the PNGase F and Endo Hf enzymatic deglycosylation assays were performed as described (Coppinger et al., 2004). Ten microliters of the reaction was separated by 10% SDS-PAGE and immunoblotted with anti-FLAG antibody (Sigma; 1:3,000). To investigate if CRK28 carries the N-linked glycan complexes, 35S:CRK28-3xFLAG, 35S:CRK28K377N-3xFLAG, and 35S:GFP were expressed transiently in N. benthamiana. IP was performed with anti-FLAG antibody and immunoblotted with anti-HRP antibody (Jackson ImmunoResearch) as described previously (Liebrand et al., 2012).
Accession Numbers
All mass spectrometry raw data files have been uploaded to the MassIVE ftp repository (accession no. MSV000079639).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Chromosomal locations and phylogenetic tree of CRKs in the Arabidopsis genome.
Supplemental Figure S2. Genotyping and disease phenotyping of CRK28 and CRK29 T-DNA knockout lines.
Supplemental Figure S3. CRK28’s cDNA sequence from Col-0.
Supplemental Figure S4. In vitro CRK28 kinase activity assay.
Supplemental Figure S5. Silencing NbSerk3 does not inhibit cell death elicited by the resistance protein RPS2.
Supplemental Table S1. MS/MS spectral count data and differential expression analyses.
Supplemental Table S2. Sequences of primers used in this study.
Supplementary Material
Acknowledgments
We thank Tania Y. Toruño, Thomas W.H. Liebrand, and DongHyuk Lee for comments on the article.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
pattern-triggered immunity
- ROS
reactive oxygen species
- ETI
effector-triggered immunity
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MS/MS
tandem mass spectrometry
- qPCR
quantitative real-time PCR
- Col-0
Columbia-0
- VIGS
virus-induced gene silencing
- RT
reverse transcription
- hpi
hours post inoculation
- TAIR
The Arabidopsis Information Resource
- IP
immunoprecipitation
- Co-IP
coimmunoprecipitation
- PRR
pattern recognition receptor
- FDR
false discovery rate
- CFU
colony-forming units
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1054298 to G.C. and grant no. 1252539 to P.H.), the National Institutes of Health (grant no. R01–GM092772 to G.C.), and the Ministerio de Ciencia e Innovación (MICINN), Spain (Formación de Personal Investigador [FPI] fellowship to J.S.R.).
Articles can be viewed without a subscription.
References
- Acharya BR, Raina S, Maqbool SB, Jagadeeswaran G, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R (2007) Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J 50: 488–499 [DOI] [PubMed] [Google Scholar]
- Amari K, Boutant E, Hofmann C, Schmitt-Keichinger C, Fernandez-Calvino L, Didier P, Lerich A, Mutterer J, Thomas CL, Heinlein M, et al. (2010) A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog 6: e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S (2012) Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell 24: 4205–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D (2001) Evolutionary dynamics of plant R-genes. Science 292: 2281–2285 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bourdais G, Burdiak P, Gauthier A, Nitsch L, Salojärvi J, Rayapuram C, Idänheimo N, Hunter K, Kimura S, Merilo E, et al. (2015) Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet 11: e1005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud MC, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJM, Jones AME, Robatzek S, Jones JDG, Faulkner C (2014) The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Du L, Chen Z (2003) Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol 53: 61–74 [DOI] [PubMed] [Google Scholar]
- Chen K, Fan B, Du L, Chen Z (2004) Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol Biol 56: 271–283 [DOI] [PubMed] [Google Scholar]
- Chen Z. (2001) A superfamily of proteins with novel cysteine-rich repeats. Plant Physiol 126: 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan A, Schobel S, Town CD (2016) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. bioRxiv 047308. [DOI] [PubMed] [Google Scholar]
- Chiang YH, Coaker G (2015) Effector triggered immunity: NLR immune perception and downstream defense responses. The Arabidopsis Book 13: e0183 doi/10.1199/tab.0183 [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ (2004) Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J 40: 225–237 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ (2005) Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira MVV, Xu G, Li B, de Souza Vespoli L, Meng X, Chen X, Yu X, de Souza SA, Intorne AC, de A Manhães AM, et al. (2016) Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nat Plants 2: 15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE (2006) The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 45: 1–16 [DOI] [PubMed] [Google Scholar]
- Dou D, Zhou JM (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B, Coaker G (2012) Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics 11: 014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM (2011) Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol 23: 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Häweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, Robatzek S, von Schaewen A (2010) Pattern recognition receptors require N-glycosylation to mediate plant immunity. J Biol Chem 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henquet M, Lehle L, Schreuder M, Rouwendal G, Molthoff J, Helsper J, van der Krol S, Bosch D (2008) Identification of the gene encoding the α1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream n-glycan processing. Plant Cell 20: 1652–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA (2007) Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104: 10732–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idänheimo N, Gauthier A, Salojärvi J, Siligato R, Brosché M, Kollist H, Mähönen AP, Kangasjärvi J, Wrzaczek M (2014) The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem Biophys Res Commun 445: 457–462 [DOI] [PubMed] [Google Scholar]
- Inoue M, Shinohara ML (2014) Clustering of pattern recognition receptors for fungal detection. PLoS Pathog 10: e1003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson F, Olbe M, Sommarin M, Larsson C (1995) Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness: a new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J 7: 165–173 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56: 1472–1480 [DOI] [PubMed] [Google Scholar]
- Käll L, Storey JD, MacCoss MJ, Noble WS (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 7: 29–34 [DOI] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KAS, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kus JV, Zaton K, Sarkar R, Cameron RK (2002) Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. Plant Cell 14: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Wang X, Cui W, Sager R, Modla S, Czymmek K, Zybaliov B, van Wijk K, Zhang C, Lu H, et al. (2011) A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, Smit P, Abd-El-Haliem A, de Jonge R, Cordewener JHG, America AHP, Sklenar J, Jones AME, Robatzek S, Thomma BPHJ, et al. (2012) Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol 159: 1819–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, van den Burg HA, Joosten MHAJ (2014) Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci 19: 123–132 [DOI] [PubMed] [Google Scholar]
- Lin ZJD, Liebrand TWH, Yadeta KA, Coaker G (2015) PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol 169: 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJD, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nakayama N, Schiff M, Litt A, Irish VF, Dinesh-Kumar SP (2004) Virus induced gene silencing of a DEFICIENS ortholog in Nicotiana benthamiana. Plant Mol Biol 54: 701–711 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Lozano-Durán R, Zipfel C (2015) Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci 20: 269–272 [DOI] [PubMed] [Google Scholar]
- Nagahara N. (2011) Intermolecular disulfide bond to modulate protein function as a redox-sensing switch. Amino Acids 41: 59–72 [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Takahashi T, Komeda Y (2000) Salicylic acid induces the expression of a number of receptor-like kinase genes in Arabidopsis thaliana. Plant Cell Physiol 41: 1038–1044 [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP (2013) Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot 64: 445–458 [DOI] [PubMed] [Google Scholar]
- Ranf S, Gisch N, Schäffer M, Illig T, Westphal L, Knirel YA, Sánchez-Carballo PM, Zähringer U, Hückelhoven R, Lee J, et al. (2015) A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat Immunol 16: 426–433 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A, Saleem-Batcha R, Kombrink A, Hansen G, Valkenburg DJ, Thomma BP, Mesters JR (2013) Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife 2: e00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Salamango DJ, Leslie ME, Collins CA, Heese A (2014) Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol 164: 440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Osakabe Y, Katsura S, Mizuno S, Maruyama K, Kusakabe K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J 70: 599–613 [DOI] [PubMed] [Google Scholar]
- Triantafilou M, Gamper FGJ, Haston RM, Mouratis MA, Morath S, Hartung T, Triantafilou K (2006) Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem 281: 31002–31011 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, Wulff BBH, Rivas S, Durrant MC, van der Ploeg A, de Wit PJGM, Jones JDG (2005) Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17: 1000–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez AC, Chakravarthy S, Martin GB (2009) Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J Vis Exp 28: 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Jacques S, Huang J, Messens J, Van Breusegem F (2015) Oxidative post-translational modifications of cysteine residues in plant signal transduction. J Exp Bot 66: 2923–2934 [DOI] [PubMed] [Google Scholar]
- Wedemeyer WJ, Welker E, Narayan M, Scheraga HA (2000) Disulfide bonds and protein folding. Biochemistry 39: 4207–4216 [DOI] [PubMed] [Google Scholar]
- Wrzaczek M, Brosché M, Salojärvi J, Kangasjärvi S, Idänheimo N, Mersmann S, Robatzek S, Karpiński S, Karpińska B, Kangasjärvi J (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YH, Chang YH, Huang PY, Huang JB, Zimmerli L (2015) Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front Plant Sci 6: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YH, Panzeri D, Kadota Y, Huang YC, Huang PY, Tao CN, Roux M, Chien HC, Chin TC, Chu PW, et al. (2016) The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28: 1701–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem 82: 2272–2281 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.