Abstract
A comparison of eudicot and monocot model plants explores recent advances and open questions on gene regulatory networks during zygote development, parental influences on early embryogenesis, zygotic genome activation, and cell fate determination.
The flowering plant life cycle alternates between a diploid sporophytic and a haploid gametophytic generation. The sporophytic generation is initiated with the double fertilization event that results in the formation of a diploid zygote and a triploid endosperm. The unique zygote or fertilized egg cell represents the starting point of a new generation and undergoes an asymmetrical division, giving rise to a smaller apical and a larger basal cell with distinct developmental fates. Subsequently, apical and basal cells will collaborate to complete the whole process of embryogenesis and generate a multitude of different cell types (ten Hove et al., 2015). In most eudicots, the smaller apical cell will contribute to the major compartments of a mature embryo, establishing an apical-basal axis and a radial pattern through several elaborate developmental processes. The larger basal cell usually undergoes limited division to form a suspensor composed of a few cells. The uppermost suspensor cell in eudicots differentiates into the hypophysis and eventually becomes part of the primary root meristem. In monocots, apical and basal cell lineages are usually incorporated into a pear-shaped proembryo and are difficult to distinguish from each other. Over the last two decades, great efforts have been made to elucidate the molecular mechanisms underlying the early events of embryogenesis (for review, see Jenik et al., 2007; Lau et al., 2012; ten Hove et al., 2015). Despite the well-described morphological dynamics occurring during early embryogenesis and many advances in the identification of molecular players regulating embryo pattern formation in the eudicot model Arabidopsis (Arabidopsis thaliana; see Advances), there still exists a lack of knowledge about the gene regulatory network in other plant species, especially in the monocots. Moreover, there are still numerous open questions, such as the regulation of egg cell activation, establishment of zygote polarity, asymmetric zygote division, apical and basal cell fate determination, etc., which are summarized in Outstanding Questions. By comparing eudicot and monocot model plants, we will focus in this Update article on recent advances and open questions on gene regulatory networks during zygote development, parental influences on early embryogenesis, zygotic genome activation, and cell fate determination (Box 1; Rademacher et al., 2012; Zhao et al., 2011; Del Toro-De Leon et al., 2014).
TIMING OF ZYGOTIC GENOME ACTIVATION
The zygote is the starting point for embryogenesis (Fig. 1) and will develop into a mature embryo upon a series of elaborate developmental events. In animals, early embryogenesis is regulated by maternal genetic information deposited before fertilization in the egg cell and later by de novo-synthesized zygotic factors, a process known as maternal-to-zygotic transition (Tadros and Lipshitz, 2009; T. Lee et al., 2014; Baroux and Grossniklaus, 2015; Zhao and Sun, 2015). This process combines two interrelated events: (1) degradation of maternal factors and (2) onset of zygotic genome transcription, a process known as zygotic genome activation (ZGA; Tadros and Lipshitz, 2009). In plants, these processes are still poorly understood mainly because of technical limitations (Zhao and Sun, 2015).
Figure 1.
Egg cell maturation and zygote development in flowering plants. A, Egg cell maturation in the eudicot model Arabidopsis. The smaller immature egg cell will develop into a larger mature egg cell for fertilization and subsequent embryogenesis, which requires GCD1 deposited in the egg cell. After gamete fusion, the fertilized egg cell or zygote elongates rapidly along its apical-basal axis, during which zygotic polarity is established and the zygotic genome commence to transcribe. A number of genes required for zygote development and morphological changes are indicated. B, In grasses as monocot models, immature egg cells experience an evident increase in size, characterized by the formation of a high number of vacuoles distributed in the mature egg cell periphery. After gamete fusion, egg cell nucleus migration takes place, culminating in karyogamy and further movement toward the chalazal pole. In contrast to Arabidopsis, zygote elongation and increase in cell size do not take place. De novo expression of genes associated to ZGA and down-regulation of a few example genes are indicated.
Although a clear picture about the contribution of de novo zygotic transcripts to early embryogenesis could not be drawn at the present stage, after more than a decade of intense research, a common perspective in both eudicots and monocots is that de novo transcription already occurs at the zygote stage. In the eudicot model plant tobacco (Nicotiana tabacum), several de novo transcripts lacking in sperm and egg cells were detected through reverse transcription PCR in zygotes, indicating that the zygotic genome is not quiescent and instead has started to transcribe and generate novel transcripts (Ning et al., 2006). Moreover, zygotic division and elongation were strongly affected by transcription inhibitors actinomycin D and cordycepin, which could efficiently block de novo transcription, suggesting that de novo zygotic transcripts are likely required for elongation and division of the zygote (Zhao et al., 2011). In zygotes of the monocot model maize (Zea mays), de novo transcripts of three different cyclin genes, ZmCycA1;1, ZmCycB1;2, and ZmCycB2, were already detected during early zygote formation. Notably, CycA1;1 transcripts were degraded within the first 3 h after in vitro fertilization and reaccumulated 17 h after fertilization, indicating de novo transcription (Sauter et al., 1998). Comparative transcript analysis of egg cells and zygotes in maize revealed the presence of de novo transcripts for ribosomal proteins (S21A, L39) and MCM DNA replication factors in the zygote shortly after fertilization (Dresselhaus et al., 1999, 2006). Furthermore, using a paternal mRNA reporter line, translation activity was observed in early zygotes 6 h after in vitro fertilization, indicating that decondensation of the male chromatin already occurs during the initial stages of embryogenesis, allowing accessibility to the transcriptional machinery of at least a part of the male genome (Scholten et al., 2002).
Based on transcriptome analysis of sperm cells, egg cells, and zygotes, several fertilization-induced genes were identified in the zygote of rice (Oryza sativa). These include OsMAPK5, OsMET1, OsWRKY, and histone H2A, which are up-regulated after fertilization (Abiko et al., 2013). These genes have already been reported in Arabidopsis and other species as key regulators of polarity establishment and elongation. Three putative H2A.Z genes were reported being parentally expressed (Anderson et al., 2013). The transcripts of two genes (Os03g06670 and Os03g53190) are enriched in egg cells, while the third gene (Os10g28230) showed sperm cell enrichment, suggesting that at least a subset of paternal genes are poised for expression in the zygote. Similarly in wheat (Triticum aestivum), transcriptome analyses indicated that transcriptional changes take place considerably and soon after fertilization (Sprunck et al., 2005), as shown by comparative analyses of expressed sequence tags from egg cells and in vitro-developed two-celled proembryos. Novel transcripts, including those for histone H2A and ribosomal proteins, were highly represented, suggesting that similar genes are likely activated soon after fertilization in both monocots and eudicots. Furthermore, two additional genes (TaTDL1 and TaMAB2) were shown to be de novo induced after fertilization. The former encodes a Cys-rich protein and was only detected in two-celled proembryos. The latter encodes an E3-ligase component polarity protein and is likely involved in asymmetric cell divisions like its maize homolog ZmMAB1 (Juranič et al., 2012). TaMAB2 is specifically expressed in wheat zygotes and proembryos, indicating early ZGA also in this species (Leljak-Levanić et al., 2013). Related studies in Arabidopsis also support the idea that ZGA occurs at the zygote stage. Transcript comparisons between gametes and zygotes as performed in monocots have not yet been done in Arabidopsis, possibly due to the difficulties of manipulating small gametic cells and zygotes. However, genetic experiments of zygote-arrest mutants (Xu et al., 2005; Ronceret et al., 2008; Guo et al., 2016) and transcription activity analyses in zygotes using a LhG4/pOp transactivation system (Nodine and Bartel, 2012) suggested that the zygotic genome of Arabidopsis is not silenced and both paternal and maternal alleles are active in the zygote and required for zygote development and embryogenesis, respectively.
In summary, our current knowledge indicates that genome integration from male and female gametic cells commence to transcribe before zygote division, or in other words, the onset of ZGA already occurs shortly after fertilization. De novo-transcribed genetic information is likely required for zygote division and early development of embryos, although more detailed studies are urgently needed. Genome-wide transcriptional studies of zygotic genomes at different stages and functional analysis of de novo transcripts during zygote polarity, zygotic division, and apical/basal cell fate determination are now required and would allow more precise conclusions about ZGA in higher plants.
ESTABLISHMENT OF ZYGOTE POLARITY
Cellular polarity is a key aspect during plant development. Before fertilization, egg cells of eudicots such as Arabidopsis display evident polarity with the nucleus located toward its chalazal pole and a large vacuole located toward its micropylar end (Fig. 1A). After fertilization and karyogamy, the zygote elongates and the large vacuole divides into many small vacuoles, giving rise to a transiently symmetric appearance of the elongated cell lacking obvious visible polarity. Thereafter, it will undergo a series of further elaborated cytological changes, including nucleus migration toward the chalazal pole and formation of a large vacuole at the micropylar pole to rebuild cell polarity during further zygote development (Souter and Lindsey, 2000), which could be visualized by the relative position of nucleus and vacuole in the zygote.
In monocots such as the grasses, egg cell polarity is visually less distinguishable. However, due to the larger number of peripheral vacuoles at the micropylar pole, the nucleus appears to be slightly shifted toward the chalazal pole (Fig. 1B). After fertilization, the egg nucleus is surrounded by endoplasmic reticulum and other organelles and moves toward the chalazal pole where karyogamy is executed. Further movement determines the future asymmetric cell division plane. Compared with many eudicots, zygote elongation or growth does not take place (Mol et al., 1994; Sato et al., 2010), indicating that polarity establishment of egg cells and zygotes in eudicots and monocots is differentially regulated.
For Arabidopsis, it was suggested that regulatory mechanisms underlying zygote polarity establishment may be independent to that in the egg cell. This notion could be drawn, for example, from studies on the zinc-finger transcription factor WRKY2 (Ueda et al.., 2011). Expression pattern analysis revealed that WRKY2 is expressed both in the egg cell and zygote. However, in homozygous wrky2 mutants, egg cells display normal polarity but the process of zygote repolarization from the transient symmetric state failed, resulting in dispersed nucleus and vacuoles throughout the zygote. As a consequence, cell division occurred symmetrically (Ueda et al., 2011). This excellent example points out the idea that polarity establishment in egg cell and zygote is independent, and not intimately linked together. Very interestingly, zygote repolarization defects in homozygous wrky2 mutants could be restored either by paternal- or maternal-derived WRKY2 in the reciprocal crosses (Ueda et al., 2011). It is not clear yet whether WRKY2 expression in zygotes is driven by gametic cells during fertilization or de novo produced from the zygotic genome. However, the findings revealed that either male or female WRKY2 is sufficient for zygote repolarization, suggesting that both paternal- and maternal-derived WRKY2 are involved in zygote polarity establishment.
Zygote elongation occurs rapidly in eudicots along its apical-basal axis during the processes of zygote polarization. In Arabidopsis, zygote elongation and asymmetric division are regulated by the GDP/GTP exchange factor for small G-proteins of the ARF class (ARF-GEF) GNOM (GN). In GN knock-out plants, zygote elongation and asymmetric division were compromised (Mayer et al., 1993). The involvement of mitogen-activated protein kinases (MPK3 and MPK6), short suspensor (SSP), and Yoda (YDA) as regulators of zygote polarity and elongation was also reported (Wang et al., 2007; Bayer et al., 2009), but the exact mechanism remained unclear. See Table I for an overview of zygote polarity factors. It will remain a difficult task for future studies to elucidate the molecular mechanisms regulating egg cell and zygote polarity in more detail, but the identification of additional players will also help to understand the establishment of cell polarity during other developmental processes in both eudicots and monocots.
Table I. Genes involved in early embryogenesis regulation in eudicots.
| Abbreviation | Full Name | Organism | AGI No. | Protein Family/Conserved Domains | Function | Reference(s) |
|---|---|---|---|---|---|---|
| BDL | Bodenlos | Arabidopsis | At1g04550 | AUX/IAA family (IAA12) | Apical-basal embryonic pattern formation | Hamann et al. (1999, 2002) |
| FAC1 | Embryonic Factor 1 | Arabidopsis | At2g38280 | AMP deaminase | Initiation of zygotic division | Xu et al. (2005) |
| FAC19 | Embryonic Factor 19 | Arabidopsis | At1g13800 | Pentatricopeptide repeat protein | Initiation of zygotic division | Yu et al. (2012) |
| FS | FASS | Arabidopsis | At5g18580 | Ser/Thr-protein phosphatase 2A regulatory subunit B | Cell division plane and morphogenesis | Torres-Ruiz and Jürgens (1994) |
| GCD1 | Gamete Cell Defective 1 | Arabidopsis | At5g62270 | Unknown | Gamete maturation and initiation of zygote division | Wu et al. (2012) |
| GN | GNOM | Arabidopsis | At1g13980 | GDP/GTP exchange factor | Zygote elongation and asymmetric division | Mayer et al. (1993) |
| GRD | Grounded | Arabidopsis | At5g53040 | RWP-RK family | Zygote elongation and basal cell fate determination | Jeong et al. (2011) |
| MP | Monopteros | Arabidopsis | At1g19850 | Auxin response factor | Apical-basal embryonic pattern formation | Hardtke and Berleth (1998); Hamann et al. (2002) |
| NtCYS | Cystatin | Nicotiana tabacum | KF113570 | Cystatin | Prevention of precious cell death of basal cell lineage | Zhao et al. (2013) |
| PIN7 | Pin-Formed 7 | Arabidopsis | At1g23080 | Auxin transporter | Establishment of apical-basal auxin gradients | Friml et al. (2003) |
| SSP | Short Suspensor | Arabidopsis | At2g17090 | IL-1 receptor-associated kinase | Zygote elongation/zygote asymmetric division | Bayer et al. (2009) |
| WRKY2 | WRKY DNA-Binding Protein 2 | Arabidopsis | At5g56270 | Zinc-finger domain transcription factor | Zygote polarization | Ueda et al. (2011) |
| WOX2 | Wuschel Related Homeobox Protein 2 | Arabidopsis | At5g59340 | WUSCHEL-related homeobox protein | Apical-basal axis formation | Breuninger et al. (2008) |
| WOX8 | Wuschel Related Homeobox Protein 8 | Arabidopsis | At5g45980 | WUSCHEL-related homeobox protein | Apical-basal axis formation | Breuninger et al. (2008) |
| YDA | YODA | Arabidopsis | At1g63700 | MAPKK kinase | Basal cell fate determination | Lukowitz et al. (2004) |
| ZYG1 | Zygote Arrest 1 | Arabidopsis | At3g05870 | Anaphase-promoting complex/cyclosome subunit 11 | Initiation of zygotic division | Guo et al. (2016) |
| ZEU1 | Zygote Stage Zeus 1 | Arabidopsis | At5g59440 | Thymidylate kinase | Initiation of zygotic division | Ronceret et al. (2008) |
| ESF1 | Embryo Surrounding Factor 1 | Arabidopsis | At1g10747, At1g10745, At1g10717 | Cys-rich peptide | Embryonic pattern and suspensor formation | Costa et al. (2014) |
ASYMMETRIC ZYGOTE DIVISION AND ITS INFLUENCE ON EMBRYOGENESIS
Zygotic division is the first and likely the most critical cell division event during the process of early embryogenesis. The zygote usually divides transversely and asymmetrically, giving rise to an apical and a basal cell with different division patterns and distinct developmental fates. The small apical cell develops into the main body of the embryo proper, whereas the larger basal vacuolated cell continues to expand longitudinally in eudicots and divides transversely to form a suspensor composed of a single file of cells (Fig. 2A). In the grasses, the basal cell inherits most vacuoles of the zygote and divides less regularly, resulting in the formation of a basal cell lineage containing transverse, longitudinal, and intermediate cell division planes (Fig. 2B). But how does the zygote initiate its first division and how does the asymmetrical division influence apical and basal cell fate determination and embryo pattern formation?
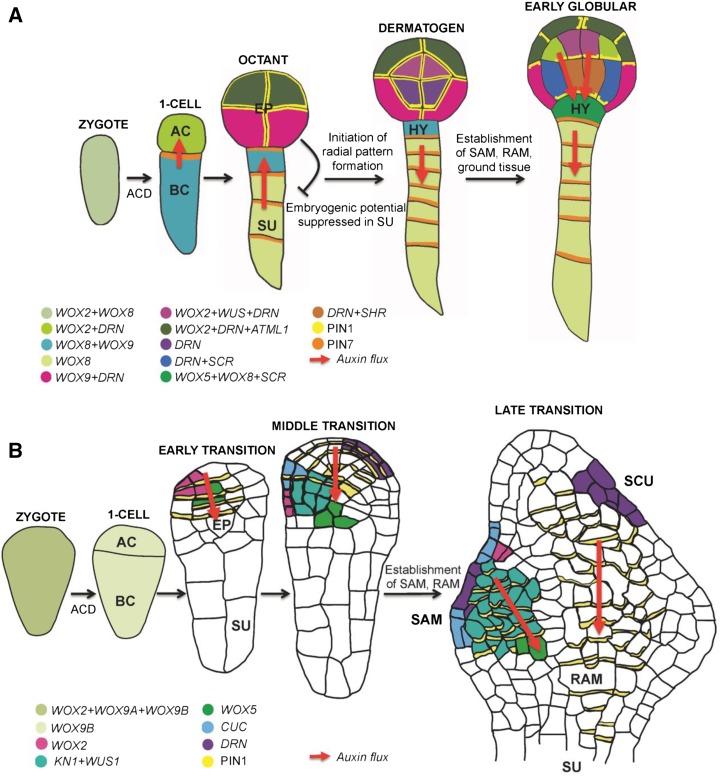
Figure 2.
A comparison of gene expression pattern and auxin fluxes regulating cell fate determination during early embryogenesis in Arabidopsis (A) and maize (B), respectively. Colors represent expression of different genes as indicated in the legends. Auxin fluxes are shown by red arrows. Transcripts are written in italic and proteins in normal letter code. Stages and major differentiation mechanisms are indicated. Abbreviations: AC, apical cell; ACD, asymmetric cell division; BC, basal cell; EP, embryo proper; HY, hypophysis; RAM, root apical meristem; SAM, shoot apical meristem; SCU, scutellum; SU, suspensor.
Over the past decade, first insights about these essential questions have been obtained. Zygote-lethal phenotypes of Arabidopsis mutants have been identified as valuable tools to understand the mechanisms regulating the initiation of the zygotic division. EMBRYONIC FACTOR1 (FAC1), encoding an AMP deaminase, was identified through a screening of ethyl methanesulfonate-mutagenized plants. Homozygous fac1 mutants are arrested at the zygote elongation stage, resulting in the failure of onset of asymmetric cell division. In addition, either maternal or paternal FAC1 could restore the zygote-lethal phenotype through reciprocal crosses with wild-type plants, suggesting that independent of the parental origin, FAC1 could support the initiation of zygote division (Xu et al., 2005). Several additional recessive mutants with the zygote-lethal phenotype include the genes FAC19, AtZYG1, and ZEU1 (Ronceret et al., 2008; Yu et al., 2012; Guo et al., 2016). AtZYG1 encodes an anaphase-promoting complex (APC) subunit 11 (APC11) protein, a ubiquitin E3 ligase that is known to target cyclins (Peters, 2006). Protein sequence analysis revealed high similarity to APC11 proteins from rice (OsAPC11) and maize (ZmAPC11; Guo et al., 2016). It needs to be shown whether knock-out of APC11 from monocots also leads to zygote lethality that could suggest a conserved pathway in both monocots and eudicots. Another example of zygote lethality was shown in the gcd1 mutant. GCD1 encodes a mitochondria-located protein, and its enrichment in female gametophytes is essential for final maturation of female gametes. GCD1 mutation leads to immature egg cells, which could fuse with sperm cells and develop into an elongated zygote, but could not initiate zygote division. Expression pattern analysis revealed that both paternal and maternal GCD1 transcript could be detected in the zygote, but paternal GCD1 could not rescue the phenotype attributed by the female gcd1 mutation, suggesting that egg-deposited GCD1 is required for the onset of zygotic division (Wu et al., 2012). Thus, its role in embryogenesis could not be replaced by its paternal counterpart. Taken together, it now becomes clear that both maternal and paternal genetic information are involved in the initiation of zygote division in plants, and parental factors may have specific roles in triggering zygote division. Moreover, rapid elongation of the zygote seems to be independent of the initiation of zygote division but appears associated with asymmetric division.
During zygote division, the control of the position and orientation of the cell division plane is a critical issue for early embryogenesis. Recently, the receptor kinase ZAR1 was found to regulate the proper position of zygote division in Arabidopsis. Homozygous zar1 leads to embryo abortion, but does not affect male or female gametogenesis. Phenotype analysis revealed that zar1 zygote elongation is normal, but its division becomes symmetrical, resulting in the failure of embryogenesis (Yu et al., 2016). NtDRP, encoding a dynamin-related protein, was found to control zygote division orientation in tobacco. Down-regulation of NtDRP caused zygotic cell division to occur in an incorrect orientation, resulting in the formation of an embryo-like structure without a typical suspensor (Zhao et al., 2016). This observation suggests that NtDRP-dependent zygotic division plane orientation is essential for the differentiation of both apical and basal cell lineages, especially for suspensor formation. In conclusion, asymmetrical cell division of the zygote appears critical for the establishment of the future embryonic pattern. However, more solid evidence in other plant species is still required to draw a final conclusion.
In rice, transcript for DNA methyltransferase 1 (OsMET1), which functions in maintaining CG DNA methylation (Kankel et al., 2003), was identified among a number of highly up-regulated genes in cultured zygotes (Abiko et al., 2013). Inhibition of OsMET1 expression slightly enhances asymmetric zygote division; however, normal globular embryos were observed during subsequent embryo culture. This finding suggests that asymmetric cell division could be partly affected by the inhibition of OsMET1. As reported, the redundancy of DNA methylation pathways (Xiao et al., 2006; Mathieu et al., 2007; Jullien et al., 2012) may explain the developmental recovery during early embryogenesis after the interruption of asymmetric zygote division in rice. However, it could also be due to the in vitro system limitations. In wheat, for instance, disturbance of or lack of initial polarity was observed in cultured zygotes compared with zygotes generated in planta (Bakos et al., 2009). Interestingly, a similar phenotype was observed in zygotes of met1 mutants in Arabidopsis, where about 13% of two-celled embryos had abnormal cell division patterns, suggesting a similar role for MET1 in eudicot zygote division (Xiao et al., 2006). Thus, epigenetic control likely also represents a key mechanism for early embryogenesis, but more detailed studies need to be conducted involving the studies of many more genes.
INFLUENCES OF THE PARENTAL GENOMES ON EARLY EMBRYOGENESIS
During fertilization, one sperm cell fuses with the egg cell to produce a zygote, and thus the parental genetic information integrates to form the diploid zygotic genome. In animals, early genetic and molecular studies have shown that maternal inherited information deposited in egg cells is usually sufficient to support early embryogenesis toward the midblastula transition stage (Tadros and Lipshitz, 2009). However, recent evidences indicated that different epigenetic mechanisms in sperm cells could also regulate offspring metabolism and health. One of the proposed mechanisms involves the influence of transfer RNAs of sperm cells as paternal epigenetic factors controlling offspring metabolism (Chen et al., 2016; Sharma et al., 2016). Another proposed mechanism suggests the involvement of the histone H3 Lys 4 demethylase KDM1A. Expression of KDM1A in developing sperm cells was shown to be essential for the survivability and development of offspring (Siklenka et al., 2015). These new findings imply that both sperm- and egg cell-delivered information are critical for the development of offspring in animals.
In plants, the influences of parent-inherited genetic information on early embryogenesis are still largely unknown, especially regarding the specific developmental events of early embryogenesis. Although the extent of parental contribution to embryogenesis has been differently elucidated, a common perspective in plants is that both paternal and maternal inherited information are involved in early embryogenesis (Luo et al., 2014). Based on high-throughput sequencing, it was concluded that early embryogenesis is maternally controlled in Arabidopsis (Autran et al., 2011). Down-regulation of the paternal alleles by the maternal chromatin small interfering RNA pathway has been suggested as a controlling mechanism, whereas the activation of the paternal alleles during the course of embryogenesis is thought to be mediated by the maternal histone chaperone complex CAF1 (Autran et al., 2011). Nonetheless, a similar approach concluded that the majority of genes expressed during early embryogenesis were equally derived from both parental genomes (Nodine and Bartel, 2012). In monocots, parental influence during embryogenesis have been less studied; in maize, an equivalent parental contribution in the zygote during early embryo development was observed by studying a limited number of genes (Meyer and Scholten, 2007). More global studies are necessary to elucidate to which extent both parental genomes contribute to early embryogenesis.
The contribution of parental information toward the zygote and thus early embryogenesis is also reflected by the delivery of parental organelles and cytoplasmic factors. Generally, it is thought that mitochondria and plastids are delivered maternally by the egg cell, while sperm organelles degenerate. However, early ultrastructure observations implied that mitochondria could also be transmitted from sperm to the egg cell in tobacco (Yu and Russell, 1994; Fig. 1A). Studies focusing on cytoplasmic factors are limited due to the difficulties associated with distinguishing gamete-delivered from zygote-de novo-synthesized factors. Nonetheless, the first example of a sperm-delivered transcript exerting a known function during early embryogenesis is SSP. Although SSP transcripts could be detected in mature pollen, yellow fluorescent protein (YFP)-tagged SSP protein was not observed in mature sperm cells. Only transient YFP-tagged SSP fluorescence was visible after fertilization in the zygote (Bayer et al., 2009). After transmission to the zygote, SSP transcripts appear to be translated and transiently accumulate to regulate asymmetric zygotic division by activating the downstream YDA signaling pathway (Bayer et al., 2009). In rice, transposon silencing mediated by microRNA and small interfering RNA pathways was found to be more active in egg cells compared with sperm cells, suggesting that the regulation by small RNAs in the zygote is inherited from the egg cells (Anderson et al., 2013). Another example of maternal control was observed in the GAMETE CELL DEFECTIVE1 (GCD1) gene in Arabidopsis. Its mutation leads to the formation of a small and immature egg cell, which is capable of fusing with a sperm cell during fertilization, but subsequent zygotic division was arrested. Expression pattern analysis revealed that both paternal and maternal GCD1 could be detected in the zygote (Fig. 1A), but paternal GCD1 could not rescue the phenotype attributed by the female gcd1 mutation, suggesting that a gcd1 mutant egg is not able to sustain zygote development and initiate embryogenesis after the egg cell fused with a normal sperm cell (Wu et al., 2012). Another way parents exert influences on their offspring is through genomic imprinting, which results in monoallelic gene expression in early embryos dependent on their parent of origin. In plants, genomic imprinting has been primarily characterized in the endosperm. In Arabidopsis, the endosperm is a transient tissue consumed by the embryo during seed development, while in monocots, the endosperm is persistent and nourishes the germinating seedling. The first maternally expressed imprinted gene in the embryo, MEE1, was identified in maize. Allele-specific expression of MEE1 was correlated with differences in allelic methylation levels (Jahnke and Scholten, 2009). Genome-wide approaches using embryos of monocots and eudicots have later identified several additional candidate imprinted genes (Gehring et al., 2011; Hsieh et al., 2011; Luo et al., 2011; Waters et al., 2011; Zhang et al., 2011). In Arabidopsis, analysis of reciprocal F1 embryos identified more than 100 potentially imprinted genes (Autran et al., 2011; Nodine and Bartel, 2012). Most of the imprinted genes confirmed by allele-specific expression and reporter gene assays were of maternal origin, while only one was paternally derived (Raissig et al., 2013). Despite these new findings, the effects of imprinted genes on embryo development are largely unknown. Thus, the role of imprinted genes and the parental control of early embryogenesis in plants still need to be elucidated in more detail in the future. Furthermore, whether epigenetic regulation is one of the major molecular mechanisms in plant embryogenesis is still a remaining question to be explored.
CELL FATE DETERMINATION OF APICAL AND BASAL CELL LINEAGES
Cell fate determination is one of the most critical developmental events for morphogenesis and pattern formation during early embryogenesis. A number of recent studies contributed to our understanding of the potential role of the asymmetric cell division and the interaction between apical cell and basal cell lineages on cell fate determination (see also above). In Arabidopsis, a MAPK cascade was shown to regulate zygote elongation and developmental fate determination of the basal cell lineage. The sperm-specific receptor-like cytoplasmic kinase SSP, the MAPKK kinase YDA, and the RWP-RK transcriptional regulator GROUNDED were found to regulate the development of basal cell lineages (Lukowitz et al., 2004; Jeong et al., 2011). Zygotes of yda mutants fail to elongate and divide symmetrically into a normal-sized apical cell and a considerably smaller basal cell. The apical cell lineage shows a normal division pattern up to the eight-celled octant stage. In contrast, the small basal cell exhibits a random division pattern and fails to form a functional suspensor. Gain-of-function of YDA has an opposite effect on suspensor development, causing exaggerated growth of the suspensor. It is difficult to determine whether these basal cell lineage defects are attributed to the failure of zygote elongation or its symmetrical division since the two events are always coupled in ssp and yda mutants. However, ZAR1 encoding like SSP a RLK/Pelle kinase in Arabidopsis provides a chance to peep at the contribution of zygote elongation and asymmetrical cell division to daughter cell fate determination. Defective zar1 does not affect the process of zygote elongation but disrupts the first asymmetric division. Accordingly, both apical and basal cell fates were impaired, suggesting that intrinsic factors are required for asymmetrical division and the division itself is likely involved in daughter cell fate determination (Yu et al., 2016). Moreover, central cell-derived peptide ESF1 was recently shown to be required for basal cell lineage development acting through the YDA MAPK pathway in Arabidopsis. ESF1 genes are only detected in the central cell and subsequent endosperm, but not in the embryo itself. Down-regulation of ESF1 expression in the endosperm led to patterning defects in the embryo proper and suspensor formation coupled with abnormal spatial expression of suspensor markers, indicating that extrinsic signals from maternal tissues and the developing endosperm also contribute to development of both apical and basal cell lineages (Costa et al., 2014).
Almost nothing is known about cell fate determination during early embryogenesis in grasses. In maize, two hemoglobin-encoding genes, ZmHb1 and ZmHb2 (Table II), were identified as master regulators in determining the developmental fate of specific cells during maize in vitro embryogenesis (Huang et al., 2014b). A ZmHb1 and ZmHb2 proposed model indicates regulation of cell fate by controlling the accumulation of nitric oxide and Zn2+, which triggers a MAPK cascade leading to the accumulation of reactive oxygen species in cells destined to die. Moreover, immunolocalization of ZmPIN1 proteins was affected in immature somatic embryos down-regulating ZmHb1 or ZmHb2, indicating that ZmHbs control cell fate by regulating auxin flux via ZmPIN1 (Huang et al., 2014a). However, neither cell death nor ZmPIN1 expression occurs during early embryogenesis in maize (Chen et al., 2014), indicating that alternative approaches are required to identify genes regulating cell fate determination at the initial stages of embryogenesis.
Table II. Genes regulated during early embryogenesis in monocots.
Please note that in monocots (grasses) only expression pattern have been described.
| Abbreviation | Full Name | Organism | Gene ID | Reference |
|---|---|---|---|---|
| MEE1 | Maternally expressed in embryo 1 | Zea mays | GRMZM2G104572 | Jahnke and Scholten (2009) |
| CycA1;1 | Cell cycle A1 | Zea mays | GRMZM2G387227 | Sauter et al. (1998) |
| CycB1 | Cell cycle B1 | Zea mays | GRMZM2G073003 | Sauter et al. (1998) |
| CycB2 | Cell cycle B2 | Zea mays | GRMZM2G138886 | Sauter et al. (1998) |
| RPL39 | 60S ribosomal protein L39 | Zea mays | GRMZM2G100467 | Dresselhaus et al. (1999) |
| RPP0 | 60S acidic ribosomal protein P0 | Zea mays | GRMZM2G066460 | Dresselhaus et al. (1999) |
| RPS21A/B | 40S ribosomal protein S21A | Zea mays | GRMZM2G134109 | Dresselhaus et al. (1999) |
| ZmHb1 | Plant hemoglobin 1 | Zea mays | AF236080 | Huang et al. (2014b) |
| ZmHb2 | Plant hemoglobin 2 | Zea mays | DQ171946 | Huang et al. (2014b) |
| WOX2 | Wuschel homeobox like 2 | Zea mays | AM234767 | Nardmann et al. (2007) |
| WOX5 | Wuschel related homeobox like 5 | Zea mays | GRMZM2G116063 | Nardmann et al. (2007) |
| DRN | Dornröschen | Zea mays | GRMZM2G120401 | Zimmermann and Werr (2007) |
| Knotted1 | KN1 | Zea mays | GRMZM2G017087 | Smith et al. (1995) |
| CUC | Cup-shaped cotyledon | Zea mays | AJ833968 | Zimmermann and Werr (2005) |
| DSUL | Di-SUMO like | Zea mays | GRMZM2G006324 | Srilunchang et al. (2010) |
| MAB1 | MATH-BTB domain protein 1 | Zea mays | AC195147 | Juranič et al. (2012) |
| MCM6 | Minichromosome maintenance protein 6 | Zea mays | GRMZM2G021069 | Dresselhaus et al. (2006) |
| MAPK 5 | MAPK 5 | Oryza sativa | Os03g0285800 | Abiko et al. (2013) |
| MET1 | Cytosine-5 DNA methyltransferase 1 | Oryza sativa | Os07g0182900 | Abiko et al. (2013) |
| H2A | Histone H2A | Oryza sativa | Os03g0279200 | Abiko et al. (2013) |
| WRK19 | WRKY transcription factor 19 | Oryza sativa | Os05g0571200 | Abiko et al. (2013) |
| WRK42 | WRKY transcription factor 42 | Oryza sativa | Os02g0462800 | Abiko et al. (2013) |
| HTA712 | Histone H2A variant 1 | Oryza sativa | Os03g06670 | Anderson et al. (2013) |
| HTA705 | Histone H2A variant 9 | Oryza sativa | Os10 g28230 | Anderson et al. (2013) |
| HTA713 | Histone H2A.Z | Oryza sativa | Os03g53190 | Anderson et al. (2013) |
| MPK6 | MAPK 6 | Oryza sativa | Os10g0533600 | Yi et al. (2016) |
| TH254 | Histone H2A.4 | Triticum aestivum | Q43208 | Sprunck et al. (2005) |
| CyP3 | Peptidyl-prolyl cis-trans isomerase | Triticum aestivum | Q93W25 | Sprunck et al. (2005) |
| MAB2 | MATH-BTB domain protein 1 | Triticum aestivum | ACA64045.1 | Leljak-Levanić et al. (2013) |
| TDL1 | TAPETUM DETERMINANT-like 1 | Triticum aestivum | CA722121 | Leljak-Levanić et al. (2013) |
Transcriptome analysis comparing apical and basal cells in tobacco and maize revealed that asymmetric zygotic division likely results in the uneven distribution of several transcripts (Okamoto et al., 2005; Ma et al., 2011). For example, NtCYS transcript encoding a Cys protease inhibitor is exclusively located in the basal cell after zygote division. NtCYS exerts its anti-cell death effect by directly inhibiting cathepsin H-like protease NtCP14 to timely control the onset of programmed cell death in the suspensor (Zhao et al., 2013). Comparable transcriptome analysis between apical and basal cells has not been done in Arabidopsis. However, studies on family members of WOX genes (WUSCHEL [WUS]-related homeobox transcriptional regulators) also implied that cell fates of apical and basal cell lineages have been established along with the asymmetric zygotic division (Haecker et al., 2004). Transcripts of WOX family members linked to apical and basal cell fate determination, WOX2 and WOX8, were expressed in apical and basal daughter cells of the zygote, respectively. WOX2 appears to be the main regulator of embryonic shoot patterning. Loss of WOX2 causes failures to properly separate the protoderm layer by periclinal divisions. Concomitant loss of other apical WOX factors in a wox2 background results in the formation of shoot-less structures. Mutations in WOX8 or its relative WOX9 have no visible effect on embryo development, but embryo pattern of wox8 wox9 double mutants is severely disrupted (Breuninger et al., 2008). Similarly, orthologs of WOX genes have been identified in maize and their expression pattern characterized (Nardmann et al., 2007; J. Chen and T. Dresselhaus, unpublished data; Fig. 2B). Expression of ZmWOX2 was detected only transiently in the zygote and later up-regulated in the early transition stage. ZmWOX9A and ZmWOX9B likely encoding the WOX8 and WOX9 homologs are weakly expressed in the zygote, but only ZmWOX9B was later detectable at significant levels in the zygotic daughter cells. These expression patterns indicate that apical and basal cell lineages and cell fate determination in grasses may involve different players or occur at a later embryonic stage. ZmWOX5 expression was detected from the early transition stage in a central basal domain of the embryo proper subtended by vacuolized suspensor cells. According to their Arabidopsis counterparts, ZmWOX2 and ZmWOX5 were suggested as indicators of apical-basal polarity (Nardmann et al., 2007) and could therefore be used as markers for studying early embryogenesis in the grasses. Other maize genes that may be involved in cell fate determination and polarity include Dornröschen (DRN), Knotted1 (KN1), and Cup-shaped cotyledon (CUC). Expression of the DRN homolog was found to begin during the shift of late proembryo to early transition stage, and its function has been linked to auxin signaling during early embryogenesis (Zimmermann and Werr, 2007; Chandler et al., 2008). KN1 expression begins at early transition stage embryos and is associated with the differentiation of small, cytoplasm-rich cells at the adaxial face of the embryo proper (Smith et al., 1995). CUC expression begins in the early transition stage embryo and together with WUS1, WOX5, and DRN homologs (Fig. 2B) may contribute to bilateral symmetry during early embryogenesis (Zimmermann and Werr, 2005). In summary, the above-described gene expression pattern indicates that cell fate determination and patterning occur delayed compared with Arabidopsis and obviously first involve a stage of cell proliferation before embryonic patterns are established. However, this remains to be shown in functional studies.
At least in Arabidopsis, it appears that both zygote elongation and its asymmetric zygote division seem to directly or indirectly depend on cell type-specific transcripts related to cell fate determination of apical and basal cell lineages. However, it remains unclear how and when the apical and basal cell fates are specified. After zygotic division, the basal cell undergoes limited divisions to form a suspensor. Once a suspensor is formed, suspensor cells stop further divisions and display the typical morphology shown in Figure 2A. It seems that cell fate is already determined at this stage. However, several mutants were identified in which suspensor cells do not initiate programmed cell death and start dividing to form embryo-like structures (Schwartz et al., 1994; Vernon and Meinke, 1994; Yadegari et al., 1994; Zhang and Somerville, 1997; Sanmartín et al., 2011). From the phenotype of these mutants, a model was proposed that embryo-proper-like pathways in the suspensor are suppressed by the embryo proper during the process of normal embryo development (Schwartz et al., 1994). More recently, a combination of in vivo living laser cell ablation and microculture techniques allowed to demonstrate that the basal cell lineage has the potential to produce secondary embryos after the embryo proper has been removed (Gooh et al., 2015; Liu et al., 2015), indicating that the embryonic potential of the basal cell lineage has to be suppressed. Interaction between apical cell and basal cell lineage thus plays a critical role in basal cell fate determination, suggesting that there is cell-to-cell communication between apical and basal cell lineage during the process of early embryogenesis. Almost nothing is known about apical-basal cell (lineage) communication in monocots and thus urgently needs more attention from the research community.
COMMONALITIES AND DIFFERENCES DURING EARLY EMBRYOGENESIS OF EUDICOTS AND MONOCOTS
The diversification of the monocot and eudicot clades occurred approximately 140 to 150 MYA (Chaw et al., 2004). Within the window of evolutionary history, striking variations in embryo development evolved in both classes of flowering plants. Cell division pattern, differentiation and cell fate establishment of apical and basal cell lineages, as well as the number of cotyledons show striking differences between both clades during the process of embryogenesis. The major characters of embryogenesis that distinguish monocots from eudicots include lack of a typical suspensor, less predictable cell division patterns, unclear cell tiers in the embryo proper, usually a bent apical-basal axis, a single cotyledon, and an extra dorsoventral axis. Additional embryonic organs, such as a scutellum, coleoptile, and coleorhiza, add to the morphological variation of embryos in monocots. Whether other structures such as a hypophysis, founder cell of the embryonic root meristem in eudicots (ten Hove et al., 2015), are also formed during embryogenesis in monocots is still unclear. Although these variations already occur at early stages of embryo development, many essential processes are comparable between monocots and eudicots. The first zygote division is usually asymmetric and results in a small apical cell and a larger basal cell as in eudicots, which was also found in Platanus racemosa, a basal eudicot (Floyd et al., 1999). This observation indicates a common character in all angiosperms. In addition, both radial and apical-basal patterns are established, and thus the basic embryo architecture is similarly composed of a shoot apical meristem and a root apical meristem connected by a hypocotyl. Additionally, epidermal, ground, and provascular tissues are differentiated during embryogenesis in all angiosperms. These major characteristics of an embryo likely have been inherited from a common embryophyte ancestor.
Accordingly, key molecular players regulating early embryo development appear conserved in both monocots and eudicots as illustrated in Figure 2, although only a few embryonic genes have been studied at the functional level in monocots so far. WOX family genes, for example, marking cell fates along the apical-basal pattern and PIN-mediated polar auxin transport in eudicots indicate two major conserved molecular mechanisms. In Arabidopsis, WOX2 and WOX8 comprise earliest cell fate markers of apical cell and basal cell lineages, respectively (Haecker et al., 2004). WUS is critical for shoot meristem activity (Mayer et al., 1998), and WOX5 is involved in root stem cell organization (Sarkar et al., 2007). As described above, a partly similar expression pattern of several WOX family genes was found in monocots, but the expression of candidate orthologous genes appears significantly delayed. While Arabidopsis AtWOX2 and AtWOX8 mark apical and basal cell fates after asymmetric zygote division, strong expression of their predicted maize homologs ZmWOX2A and ZmWOX9A/B could not be detected in the zygote and its daughter cells, but significant expression occurs at the early transition stage when shoot apical meristem and root apical meristem are established (Fig. 2B; J. Chen and T. Dresselhaus, unpublished data; Nardmann et al., 2007). Similarly, the maize predicted orthologs of WUS and WOX5 (ZmWUS1 and ZmWOX5) marking the shoot quiescent center and the root quiescent center in Arabidopsis show expression when the embryo already consists of more than 100 cells. Thus, despite the delayed expression of candidate orthologous genes, this overall similarity of expression patterns of WOX family genes implies that a relatively conserved mechanism is likely required for shoot apical meristem and root apical meristem establishment in both monocots and eudicots, which may additionally contribute to the similar apical-basal patterning of the embryo.
However, there also exists some variation in WOX family genes between monocots and eudicots: The most striking difference is that no clear homolog of WOX8 (a marker for suspensor cell fate) has been identified in the genomes of monocots to date. ZmWOX9A/B are the closest maize homologs, but their function(s) is not known. This may contribute to the morphological variations of monocot embryos, e.g. that a typical suspensor cell file is not established in early monocot embryos. Auxin-dependent apical-basal axis establishment is another critical event in early embryogenesis. Polar auxin flow mediated by PIN7 and PIN1 was already found very early during embryogenesis (Fig. 2A) in eudicots and contributes to basic embryo architecture establishment at this stage (Friml et al., 2003; Carraro et al., 2006). Again, a significant expression of ZmPINs was not found during early embryogenesis in maize until the transition stage, when polarized localization of ZmPIN1 in the apical region of the embryo likely mediated apical-basal auxin flux toward the future root apical meristem region (Fig. 2B; J. Chen and T. Dresselhaus, unpublished data; Chen et al., 2014; Locascio et al., 2014). Based on animal embryogenesis, a molecular embryonic “hourglass” model was also proposed for plants through phylotranscriptomic analysis, suggesting that embryos from various taxa appear different during early stages of development, converge to a similar form during midembryogenesis, and again diverge at later stages (Quint et al., 2012). This model is very interesting but requires extensive functional studies during monocot embryogenesis to be sustained.
CONCLUSIONS AND PERSPECTIVES
Remarkable progress has been made in understanding the developmental events that regulate early embryogenesis and the molecular mechanisms underlying these processes in the model plant Arabidopsis (see Advances). It is now widely accepted that both paternal and maternal inherited information are involved in early embryo development, although their contributions may differ. The zygotic genome is activated before zygote cell division occurs, and both gamete-deposited and after-fertilization de novo-transcribed genetic factors are required for zygotic cell division. It is also becoming clear that the accurate asymmetrical division of the zygote is critical for daughter cell fate determination. Furthermore, it has been shown that the suspensor cells still have the potential to develop into an embryo, which is suppressed by the embryo proper during normal embryogenesis. This reveals that cell-to-cell communication plays a crucial role in cell fate determination during early embryogenesis.
However, despite the great advances that have been achieved during the last decade many more interesting questions have arisen in this exciting field (see Outstanding Questions). In Arabidopsis, the inaccessibility to egg cells and zygotes was a major barrier to address some of those questions. New sophisticated techniques, including single-cell transcriptome analyses, are now expected to solve some major problems and lead to significant advances in the near future. Among the many open questions, the mechanism(s) of embryogenesis repression in the mature egg cell and the basal cell lineage after fertilization as well as egg cell activation are the most attractive issues to be solved in the future. Understanding the establishment of egg cell and zygote polarity also is expected to contribute to our knowledge of polarity, asymmetric cell division, and cell fate determination in other developmental processes in plants. Information regarding polar distribution of genetic information such as transcripts or proteins has not yet been proven in the zygote, but its pronounced polarity may help to identify such factors.
Cell fate determination strongly depends on communication between apical and basal cell lineages requiring intensive cross talk with the surrounding maternal seed tissues and the developing endosperm. Further studies are needed, especially studies on other model systems such as the grasses, to answer these important questions and to elucidate to which extent early embryogenesis is similar or differentially regulated across different plant species. Novel techniques, including single-cell RNAseq transcriptomics and CRISPR/cas, are now available and could essentially contribute to essentially increase our understanding about early embryogenesis in the near future.
Glossary
- ZGA
zygotic genome activation
Footnotes
This work was supported by National Natural Science Foundation of China Key Project 31430007, the “973” Project 2013CB126900, National Natural Science Foundation of China Project 31400171, and the Collaborative Research Center SFB960 of the German Research Foundation (DFG).
Articles can be viewed without a subscription.
References
- Abiko M, Maeda H, Tamura K, Hara-Nishimura I, Okamoto T (2013) Gene expression profiles in rice gametes and zygotes: identification of gamete-enriched genes and up- or down-regulated genes in zygotes after fertilization. J Exp Bot 64: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SN, Johnson CS, Jones DS, Conrad LJ, Gou X, Russell SD, Sundaresan V (2013) Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J 76: 729–741 [DOI] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. (2011) Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Bakos F, Szabó L, Olmedilla A, Barnabás B (2009) Histological comparison between wheat embryos developing in vitro from isolated zygotes and those developing in vivo. Sex Plant Reprod 22: 15–25 [DOI] [PubMed] [Google Scholar]
- Baroux C, Grossniklaus U (2015) The maternal-to-zygotic transition in flowering plants: Evidence, mechanisms, and plasticity. Curr Top Dev Biol 113: 351–371 [DOI] [PubMed] [Google Scholar]
- Bayer M, Nawy T, Giglione C, Galli M, Meinnel T, Lukowitz W (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Nardmann J, Werr W (2008) Plant development revolves around axes. Trends Plant Sci 13: 78–84 [DOI] [PubMed] [Google Scholar]
- Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441 [DOI] [PubMed] [Google Scholar]
- Chen J, Lausser A, Dresselhaus T (2014) Hormonal responses during early embryogenesis in maize. Biochem Soc Trans 42: 325–331 [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, et al. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351: 397–400 [DOI] [PubMed] [Google Scholar]
- Costa LM, Marshall E, Tesfaye M, Silverstein KA, Mori M, Umetsu Y, Otterbach SL, Papareddy R, Dickinson HG, Boutiller K, et al. (2014) Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172 [DOI] [PubMed] [Google Scholar]
- Del Toro-De Leon G, Garcia-Aguilar M, Gillmor CS (2014) Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514: 624–627 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Cordts S, Heuer S, Sauter M, Lörz H, Kranz E (1999) Novel ribosomal genes from maize are differentially expressed in the zygotic and somatic cell cycles. Mol Gen Genet 261: 416–427 [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Srilunchang KO, Leljak-Levanic D, Schreiber DN, Garg P (2006) The fertilization-induced DNA replication factor MCM6 of maize shuttles between cytoplasm and nucleus, and is essential for plant growth and development. Plant Physiol 140: 512–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Lerner VT, Friedman WE (1999) A developmental and evolutionary analysis of embryology in Platanus (platanaceae), abasal eudicot. Am J Bot 86: 1523–1537 [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S (2011) Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS One 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooh K, Ueda M, Aruga K, Park J, Arata H, Higashiyama T, Kurihara D (2015) Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev Cell 34: 242–251 [DOI] [PubMed] [Google Scholar]
- Guo L, Jiang L, Zhang Y, Lu XL, Xie Q, Weijers D, Liu CM (2016) The anaphase-promoting complex initiates zygote division in Arabidopsis through degradation of cyclin B1. Plant J 86: 161–174 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Hill RD, Stasolla C (2014a) Plant hemoglobin participation in cell fate determination. Plant Signal Behav 9: e29485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Hill RD, Wally OSD, Dionisio G, Ayele BT, Jami SK, Stasolla C (2014b) Hemoglobin control of cell survival/death decision regulates in vitro plant embryogenesis. Plant Physiol 165: 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke S, Scholten S (2009) Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol 19: 1677–1681 [DOI] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Jeong S, Palmer TM, Lukowitz W (2011) The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling. Curr Biol 21: 1268–1276 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830 [DOI] [PubMed] [Google Scholar]
- Juranič M, Srilunchang KO, Krohn NG, Leljak-Levanic D, Sprunck S, Dresselhaus T (2012) Germline-specific MATH-BTB substrate adaptor MAB1 regulates spindle length and nuclei identity in maize. Plant Cell 24: 4974–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ (2003) Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Slane D, Herud O, Kong J, Jürgens G (2012) Early embryogenesis in flowering plants: setting up the basic body pattern. Annu Rev Plant Biol 63: 483–506 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ (2014) Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leljak-Levanić D, Juranić M, Sprunck S (2013) De novo zygotic transcription in wheat (Triticum aestivum L.) includes genes encoding small putative secreted peptides and a protein involved in proteasomal degradation. Plant Reprod 26: 267–285 [DOI] [PubMed] [Google Scholar]
- Liu Y, Li X, Zhao J, Tang X, Tian S, Chen J, Shi C, Wang W, Zhang L, Feng X, Sun MX (2015) Direct evidence that suspensor cells have embryogenic potential that is suppressed by the embryo proper during normal embryogenesis. Proc Natl Acad Sci USA 112: 12432–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Roig-Villanova I, Bernardi J, Varotto S (2014) Current perspectives on the hormonal control of seed development in Arabidopsis and maize: a focus on auxin. Front Plant Sci 5: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Roeder A, Parmenter D, Somerville C (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116: 109–119 [DOI] [PubMed] [Google Scholar]
- Luo A, Shi C, Zhang L, Sun MX (2014) The expression and roles of parent-of-origin genes in early embryogenesis of angiosperms. Front Plant Sci 5: 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Xin H, Qu L, Zhao J, Yang L, Zhao P, Sun M (2011) Transcription profile analysis reveals that zygotic division results in uneven distribution of specific transcripts in apical/basal cells of tobacco. PLoS One 6: e15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mayer U, Buttner G, Jürgens G (1993) Apical-basal pattern-formation in the Arabidopsis embryo - studies on the role of the gnom gene. Development 117: 149–162 [Google Scholar]
- Meyer S, Scholten S (2007) Equivalent parental contribution to early plant zygotic development. Curr Biol 17: 1686–1691 [DOI] [PubMed] [Google Scholar]
- Mol R, Matthysrochon E, Dumas C (1994) The kinetics of cytological events during double fertilization in Zea mays L. Plant J 5: 197–206 [Google Scholar]
- Nardmann J, Zimmermann R, Durantini D, Kranz E, Werr W (2007) WOX gene phylogeny in Poaceae: a comparative approach addressing leaf and embryo development. Mol Biol Evol 24: 2474–2484 [DOI] [PubMed] [Google Scholar]
- Ning J, Peng XB, Qu LH, Xin HP, Yan TT, Sun M (2006) Differential gene expression in egg cells and zygotes suggests that the transcriptome is restructed before the first zygotic division in tobacco. FEBS Lett 580: 1747–1752 [DOI] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Scholten S, Lörz H, Kranz E (2005) Identification of genes that are up- or down-regulated in the apical or basal cell of maize two-celled embryos and monitoring their expression during zygote development by a cell manipulation- and PCR-based approach. Plant Cell Physiol 46: 332–338 [DOI] [PubMed] [Google Scholar]
- Peters JM. (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Quint M, Drost HG, Gabel A, Ullrich KK, Bönn M, Grosse I (2012) A transcriptomic hourglass in plant embryogenesis. Nature 490: 98–101 [DOI] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jurgens G, Weijers D (2012) Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 22: 211–222 [DOI] [PubMed] [Google Scholar]
- Raissig MT, Bemer M, Baroux C, Grossniklaus U (2013) Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet 9: e1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A, Gadea-Vacas J, Guilleminot J, Lincker F, Delorme V, Lahmy S, Pelletier G, Chabouté ME, Devic M (2008) The first zygotic division in Arabidopsis requires de novo transcription of thymidylate kinase. Plant J 53: 776–789 [DOI] [PubMed] [Google Scholar]
- Sanmartín M, Sauer M, Muñoz A, Zouhar J, Ordóñez A, van de Ven WTG, Caro E, de la Paz Sánchez M, Raikhel NV, Gutiérrez C, et al. (2011) A molecular switch for initiating cell differentiation in Arabidopsis. Curr Biol 21: 999–1008 [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Sato A, Toyooka K, Okamoto T (2010) Asymmetric cell division of rice zygotes located in embryo sac and produced by in vitro fertilization. Sex Plant Reprod 23: 211–217 [DOI] [PubMed] [Google Scholar]
- Sauter M, von Wiegen P, Lörz H, Kranz E (1998) Cell cycle regulatory genes from maize are differentially controlled during fertilization and first embryonic cell division. Sex Plant Reprod 11: 41–48 [Google Scholar]
- Scholten S, Lörz H, Kranz E (2002) Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J 32: 221–231 [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW (1994) Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120: 3235–3245 [DOI] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, et al. (2015) Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 350: aab2006. [DOI] [PubMed] [Google Scholar]
- Smith LG, Jackson D, Hake S (1995) Expression of Knotted1 marks shoot meristem formation during maize embryogenesis. Dev Genet 16: 344–348 [Google Scholar]
- Souter M, Lindsey K (2000) Polarity and signalling in plant embryogenesis. J Exp Bot 51: 971–983 [DOI] [PubMed] [Google Scholar]
- Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T (2005) The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). Plant J 41: 660–672 [DOI] [PubMed] [Google Scholar]
- Srilunchang KO, Krohn NG, Dresselhaus T (2010) DiSUMO-like DSUL is required for nuclei positioning, cell specification and viability during female gametophyte maturation in maize. Development 137: 333–345 [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD (2009) The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, Weijers D (2015) Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142: 420–430 [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Jürgens G (1994) Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120: 2967–2978 [DOI] [PubMed] [Google Scholar]
- Ueda M, Zhang Z, Laux T (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell 20: 264–270 [DOI] [PubMed] [Google Scholar]
- Vernon DM, Meinke DW (1994) Embryogenic transformation of the suspensor in twin, a polyembryonic mutant of Arabidopsis. Dev Biol 165: 566–573 [DOI] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM (2011) Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Peng XB, Li WW, He R, Xin HP, Sun MX (2012) Mitochondrial GCD1 dysfunction reveals reciprocal cell-to-cell signaling during the maturation of Arabidopsis female gametes. Dev Cell 23: 1043–1058 [DOI] [PubMed] [Google Scholar]
- Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL (2006) DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang HY, Xie CH, Xue HW, Dijkhuis P, Liu CM (2005) EMBRYONIC FACTOR 1 encodes an AMP deaminase and is essential for the zygote to embryo transition in Arabidopsis. Plant J 42: 743–756 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Paiva G, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB (1994) Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6: 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Lee YS, Lee DY, Cho MH, Jeon JS, An G (2016) OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. J Exp Bot 67: 2425–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Jiang L, Gong H, Liu CM (2012) EMBRYONIC FACTOR 19 encodes a pentatricopeptide repeat protein that is essential for the initiation of zygotic embryogenesis in Arabidopsis. J Integr Plant Biol 54: 55–64 [DOI] [PubMed] [Google Scholar]
- Yu HS, Russell SD (1994) Occurrence of mitochondria in the nuclei of tobacco sperm cells. Plant Cell 6: 1477–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TY, Shi DQ, Jia PF, Tang J, Li HJ, Liu J, Yang WC (2016) The Arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PLoS Genet 12: e1005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Somerville CR (1997) Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Proc Natl Acad Sci USA 94: 7349–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Zhao H, Guan H, Hu X, Jiao Y, et al. (2011) Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci USA 108: 20042–20047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Xin H, Cao L, Huang X, Shi C, Zhao P, Fu Y, Sun MX (2016) NtDRP is necessary for accurate zygotic division orientation and differentiation of basal cell lineage toward suspensor formation. New Phytol 212: 598–612 [DOI] [PubMed] [Google Scholar]
- Zhao J, Xin H, Qu L, Ning J, Peng X, Yan T, Ma L, Li S, Sun MX (2011) Dynamic changes of transcript profiles after fertilization are associated with de novo transcription and maternal elimination in tobacco zygote, and mark the onset of the maternal-to-zygotic transition. Plant J 65: 131–145 [DOI] [PubMed] [Google Scholar]
- Zhao P, Sun MX (2015) The maternal-to-zygotic transition in higher plants: available approaches, critical limitations, and technical requirements. Curr Top Dev Biol 113: 373–398 [DOI] [PubMed] [Google Scholar]
- Zhao P, Zhou XM, Zhang LY, Wang W, Ma LG, Yang LB, Peng XB, Bozhkov PV, Sun MX (2013) A bipartite molecular module controls cell death activation in the basal cell lineage of plant embryos. PLoS Biol 11: e1001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Werr W (2005) Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol Biol 58: 669–685 [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Werr W (2007) Transcription of the putative maize orthologue of the Arabidopsis DORNROSCHEN gene marks early asymmetry in the proembryo and during leaf initiation in the shoot apical meristem. Gene Expr Patterns 7: 158–164 [DOI] [PubMed] [Google Scholar]